Monitoring Individual Tree Phenology in a Multi-Species Forest Using High Resolution UAV Images
Abstract
1. Introduction
2. Materials and Methods
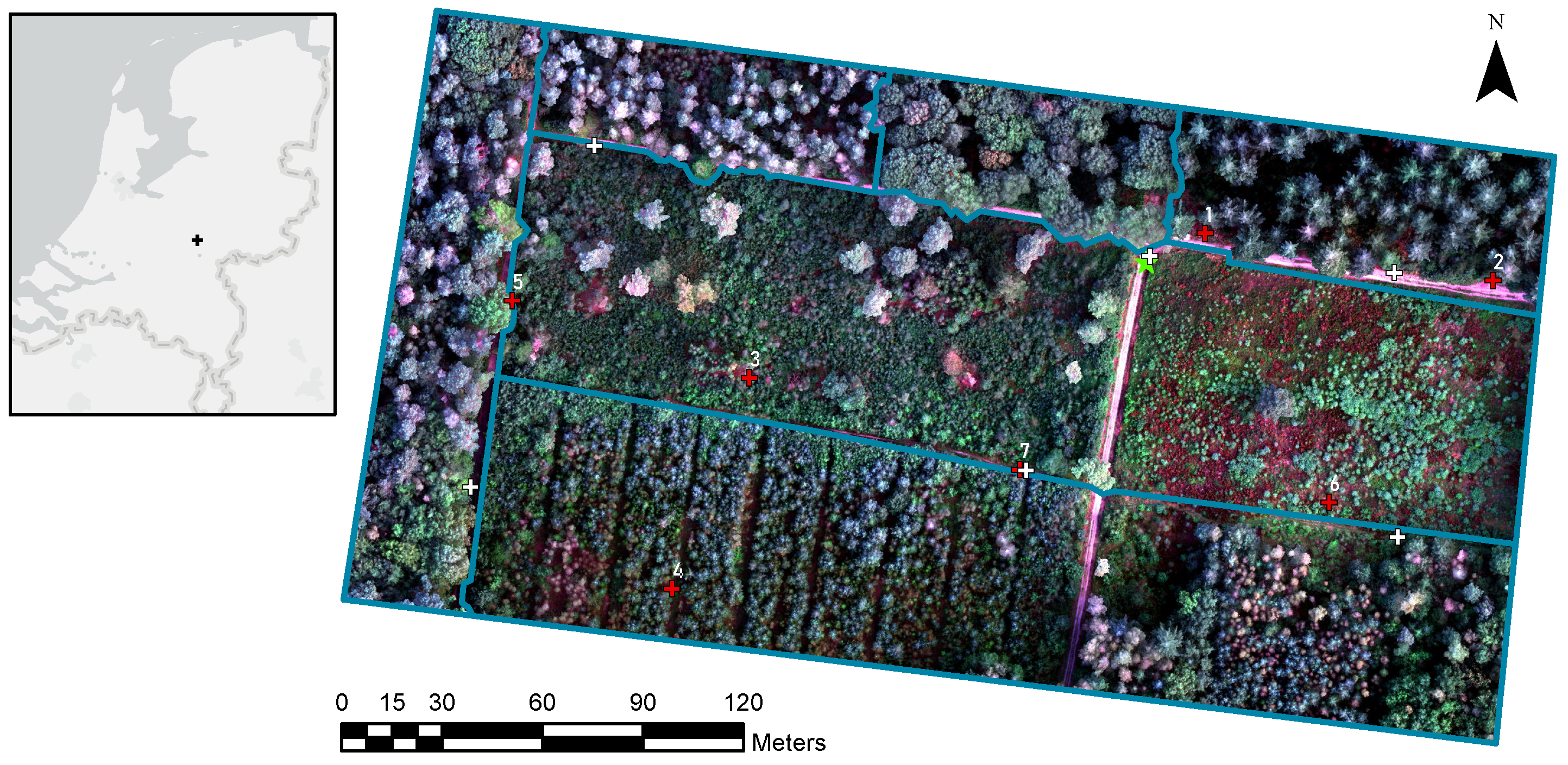
2.1. Study Area
2.2. UAV Data Acquisition and Processing
2.2.1. Preprocessing
2.2.2. Automatic Tree Detection and Crown Delineation
2.3. Vegetation Indices
2.4. Field Reference Data and Manual Tree Classification
2.5. Fit Statistical Model
Evaluate Model Performance
2.6. Validation of the Crown Delineation
- Tree identification
- Tree crown segmentation
3. Results
3.1. Automatic Crown Delineation
3.2. Phenology Monitoring
4. Discussion
4.1. Explaining the Vegetation Index Trends
4.2. Tree Phenology Estimates in Context
4.3. Evaluating the Automatic Tree Crown Delineation
4.4. Considerations for Future Research
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AHN | Actueel Hoogtebestand Nederland (english: Current Height inventory the Netherlands) |
| CCI | Chlorophyll/Carotenoid Index |
| CHM | Canopy Height Model |
| CIre | Chlorophyll Index Red Edge |
| CNN | Convolutional Neural Network |
| DSM | Digital Surface Model |
| DTM | Digital Terrain Model |
| DOY | Day Of the Year |
| EOS | End Of Season |
| EVI2 | Enhanced Vegetation Index 2 |
| FP | False Positive |
| FN | False Negative |
| GCP | Ground Control Point |
| GGC | Green Chromatic Coordinate |
| GSD | Ground Sampling Distance |
| IoU | Intersection over Union |
| IPCC | International Panel for Climate Change |
| ITCD | Individual Tree Crown Delineation |
| LiDAR | Light Detection And Ranging |
| LMF | Local Maximum Filter |
| MCWS | Marker-Controlled WaterShed |
| MTH | Minimum Tree Height |
| MVS | Multi-View Stereo |
| NDVI | Normalised Difference Vegetation Index |
| NIR | Near-Infrared |
| nRMSE | normalised Root Mean Squared Error |
| OBIA | Object-Based Image Analysis |
| OS | Over Segmentation index |
| OSAVI | Optimised Soil-Adjusted Vegetation Index |
| PRI | Photochemical Reflectance Index |
| RMSE | Root Mean Squared Error |
| RTK | Real-Time Kinematic |
| SE | Standard Error |
| SOS | Start Of Season |
| TP | True Positive |
| UAV | Unmanned Aerial Vehicle |
| VI | Vegetation Index |
| US | Under Segmentation index |
Appendix A. UAV Data Acquisition
Appendix A.1. Frequency of Data Collection

Appendix A.2. Geometric Accuracy (Error) of Each Flight
| GCPs | 20/Apr | 03/May | 10/May | 18/May | 25/May | 31/May | 07/Jun | 21/Jun | 28/Jun | 26/Jul | 16/Aug | 04/Oct | 05/Nov | 13/Dec |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3.2 | 2.7 | 1.8 | 2.5 | 2.9 | 3.0 | 7.1 | 2.4 | 2.1 | 46.3 | 2.7 | 3.9 | 0.6 | 6.4 |
| 2 | 2.2 | 2.5 | 2.1 | 1.7 | 1.6 | 0.6 | 3.0 | 2.9 | 3.4 | 12.6 | 2.0 | 5.7 | 2.8 | 3.0 |
| 3 | 3.3 | 2.5 | 0.8 | 2.3 | 0.7 | 2.6 | 5.9 | 2.8 | 2.4 | 28.5 | 3.0 | 4.3 | 5.6 | 3.3 |
| 4 | 1.4 | 0.3 | 1.3 | 0.9 | 2.2 | 0.6 | 4.7 | 2.8 | 1.4 | 39.1 | 1.5 | 2.5 | 9.8 | 0.1 |
| 5 | 1.7 | 2.5 | 2.1 | 0.6 | 1.6 | 1.6 | 3.3 | 0.4 | 1.8 | 30.2 | 1.8 | 0.0 | 6.5 | 6.1 |
| 6 | 61.7 | |||||||||||||
| Average | 2.4 | 2.1 | 1.6 | 1.6 | 1.8 | 1.7 | 4.8 | 2.2 | 2.2 | 36.4 | 2.2 | 4.1 | 5.1 | 3.8 |
Appendix B. UAV Data Processing
Appendix B.1. Metashape Structure from Motion Steps and Settings

Appendix B.2. Workflow of Validation Data Creation and Automatic Crown Segmentation

Appendix B.3. Manually Digitised Validation Data

Appendix C. Classification
Appendix C.1. Decision Tree for Tree Species Classification

Appendix C.2. Classification Map

Appendix C.3. Segments Included and Excluded after Manual Classification

Appendix D. Fit Model
RMSE under Various Iterations

Appendix E. Results
Appendix E.1. Phenology SOS and EOS Estimates

Appendix E.2. EVI2–Derived Spatial Variation SOS and EOS Estimates of the Deciduous Trees

Appendix E.3. OSAVI–Derived Spatial Variation SOS and EOS Estimates of the Deciduous Trees

Appendix E.4. CIre–Derived Spatial Variation SOS and EOS Estimates of the Deciduous Trees

References
- Badeck, F.W.; Bondeau, A.; Böttcher, K.; Doktor, D.; Lucht, W.; Schaber, J.; Sitch, S. Responses of spring phenology to climate change. New Phytol. Found. 2004, 162, 295–309. [Google Scholar] [CrossRef]
- Saxe, H.; Cannell, M.G.R.; Johnsen, Ø.R.; Ryan, M.G.; Vourlitis, G. Tree and forest functioning in response to global warming. New Phytol. 2001, 149, 359–388. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.D.; Keenan, T.F.; Migliavacca, M.; Ryu, Y.; Sonnentag, O.; Toomey, M. Climate change, phenology, and phenological control of vegetation feedbacks to the climate system. Agric. For. Meteorol. 2013, 169, 156–173. [Google Scholar] [CrossRef]
- Peñuelas, J.; Rutishauser, T.; Filella, I. A conceptual framework explains how individual species’ responses to climate warming affect the length of the growing season. Phenol. Feed. Clim. Chang. 2009, 324, 5929. [Google Scholar] [CrossRef]
- Estrella, N.; Menzel, A. Responses of leaf colouring in four deciduous tree species to climate and weather in Germany. Clim. Res. 2006, 32, 253–267. [Google Scholar] [CrossRef]
- Rosenzweig, C.; Casassa, G.; Karoly, D.J.; Imeson, A.; Liu, C.; Menzel, A.; Rawlins, S.; Root, T.L.; Seguin, B.; Tryjanowski, P. Assessment of observed changes and responses in natural and managed systems. In Climate Change 2007: Impacts, Adaptation and Vulnerability. Contribution of Working Group II to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change; Parry, M.L., Ed.; Cambridge Univerisity Press: Cambridge, UK, 2007; pp. 79–113. [Google Scholar]
- Richardson, A.D.; Baily, A.S.; Denny, E.G.; Martin, C.W.; O’Keefe, J. Phenology of a northern hardwood forest canopy. Glob. Chang. Biol. 2006, 12, 1174–1188. [Google Scholar] [CrossRef]
- Melaas, E.K.; Friedl, M.A.; Zhu, Z. Detecting interannual variation in deciduous broadleaf forest phenology using Landsat TM/ETM+ data. Remote Sens. Environ. 2013, 132, 176–185. [Google Scholar] [CrossRef]
- Pastor-Guzman, J.; Dash, J.; Atkinson, P.M. Remote sensing of mangrove forest phenology and its environmental drivers. Remote Sens. Environ. 2018, 205, 71–84. [Google Scholar] [CrossRef]
- Pennec, A.; Gond, V.; Sabatier, D. Tropical forest phenology in French Guiana from MODIS time series. Remote Sens. Lett. 2011, 2, 337–345. [Google Scholar] [CrossRef]
- Walker, J.; de Beurs, K.; Wynne, R.; Gao, F. Evaluation of Landsat and MODIS data fusion products for analysis of dryland forest phenology. Remote Sens. Environ. 2012, 117, 381–393. [Google Scholar] [CrossRef]
- Han, Q. Remote sensing-based quantification of spatial variation in canopy phenology of four dominant tree species in Europe. J. Appl. Remote Sens. 2013, 7, 073485. [Google Scholar] [CrossRef]
- Delbart, N.; Kergoat, L.; Le Toan, T.; Lhermitte, J.; Picard, G. Determination of phenological dates in boreal regions using normalized difference water index. Remote Sens. Environ. 2005, 97, 26–38. [Google Scholar] [CrossRef]
- Jones, M.O.; Jones, L.A.; Kimball, J.S.; McDonald, K.C. Satellite passive microwave remote sensing for monitoring global land surface phenology. Remote Sens. Environ. 2011, 115, 1081–1089. [Google Scholar] [CrossRef]
- Zhang, X.; Friedl, M.A.; Schaaf, C.B.; Strahler, A.H.; Hodges, J.C.; Gao, F.; Reed, B.C.; Huete, A. Monitoring vegetation phenology using MODIS. Remote Sens. Environ. 2003, 84, 471–475. [Google Scholar] [CrossRef]
- Pinzon, J.; Tucker, C. A Non-Stationary 1981–2012 AVHRR NDVI3g Time Series. Remote Sens. 2014, 6, 6929. [Google Scholar] [CrossRef]
- Tucker, C.J.; Pinzon, J.E.; Brown, M.E.; Slayback, D.A.; Pak, E.W.; Mahoney, R.; Vermote, E.F.; El Saleous, N. An extended AVHRR 8-km NDVI dataset compatible with MODIS and SPOT vegetation NDVI data. Int. J. Remote Sens. 2005, 26, 4485–4498. [Google Scholar] [CrossRef]
- White, M.A.; de Beurs, K.M.; Didan, K.; Inouye, D.W.; Richarson, A.D.; Jensen, O.P.; O’Keefe, J.; Zhang, G.; Nemani, R.R.; van Leeuweb, W.J.D.; et al. Intercomparison, interpretation, and assessment of spring phenology in North America estimated from remote sensing for 1982–2006. Glob. Chang. Biol. 2009, 15, 2335–2359. [Google Scholar] [CrossRef]
- Polgar, C.A.; Primack, R.B. Leaf-out phenology of temperate woody plants: From trees to ecosystems. New Phytol. 2011, 191, 926–941. [Google Scholar] [CrossRef]
- Liu, Y.; Hill, M.J.; Zhang, X.; Wang, Z.; Richardson, A.D.; Hufkens, K.; Filippa, G.; Baldocchi, D.D.; Ma, S.; Verfaillie, J.; et al. Using data from Landsat, MODIS, VIIRS and PhenoCams to monitor the phenology of California oak/grass savanna and open grassland across spatial scales. Agric. For. Meteorol. 2017, 237–238, 311–325. [Google Scholar] [CrossRef]
- Berra, E.F.; Gaulton, R.; Barr, S. Assessing spring phenology of a temperate woodland: A multiscale comparison of ground, unmanned aerial vehicle and Landsat satellite observations. Remote Sens. Environ. 2019, 223, 229–242. [Google Scholar] [CrossRef]
- Sparks, T.H.; Menzel, A. Observed changes in seasons: An overview. Int. J. Climatol. 2002, 22, 1715–1725. [Google Scholar] [CrossRef]
- Fitter, A.H.; Fitter, R.S.R. Rapid Changes in Flowering Time in British Plants. Science 2002, 296, 1689–1691. [Google Scholar] [CrossRef] [PubMed]
- Denéchère, R.; Delpierre, N.; Apostol, E.N.; Berveiller, D.; Bonne, F.; Cole, E.; Delzon, S.; Dufrêne, E.; Gressler, E.; Jean, F.; et al. The within-population variability of leaf spring and autumn phenology is influenced by temperature in temperate deciduous trees. Int. J. Biometeorol. 2021, 65, 369–379. [Google Scholar] [CrossRef]
- Delpierre, N.; Guillemot, J.; Dufrêne, E.; Cecchini, S.; Nicolas, M. Tree phenological ranks repeat from year to year and correlate with growth in temperate deciduous forests. Agric. For. Meteorol. 2017, 234–235, 1–10. [Google Scholar] [CrossRef]
- D’Odorico, P.; Besik, A.; Wong, C.Y.; Isabel, N.; Ensminger, I. High-throughput drone-based remote sensing reliably tracks phenology in thousands of conifer seedlings. New Phytol. 2020, 226, 1667–1681. [Google Scholar] [CrossRef]
- Borra-Serrano, I.; Swaef, T.D.; Quataert, P.; Aper, J.; Saleem, A.; Saeys, W.; Somers, B.; Roldán-Ruiz, I.; Lootens, P. Closing the phenotyping gap: High resolution UAV time series for soybean growth analysis provides objective data from field trials. Remote Sens. 2020, 12, 1644. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; Partel, V. UAV-based high throughput phenotyping in citrus utilizing multispectral imaging and artificial intelligence. Remote Sens. 2019, 11, 410. [Google Scholar] [CrossRef]
- Mahlein, A.K. Plant Disease Detection by Imaging Sensors – Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef]
- Shakoor, N.; Lee, S.; Mockler, T.C. High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 2017, 38, 184–192. [Google Scholar] [CrossRef]
- Fawcett, D.; Bennie, J.; Anderson, K. Monitoring spring phenology of individual tree crowns using drone-acquired NDVI data. Remote Sens. Ecol. Conserv. 2021, 7, 227–244. [Google Scholar] [CrossRef]
- Budianti, N.; Mizunaga, H.; Iio, A. Crown structure explains the discrepancy in leaf phenology metrics derived from ground-and uav-based observations in a japanese cool temperate deciduous forest. Forests 2021, 12, 425. [Google Scholar] [CrossRef]
- Dingle Robertson, L.; King, D.J. Comparison of pixel- and object-based classification in land cover change mapping. Int. J. Remote Sens. 2011, 32, 1505–1529. [Google Scholar] [CrossRef]
- Cai, S.; Liu, D. A comparison of object-based and contextual pixel-based classifications using high and medium spatial resolution images. Remote Sens. Lett. 2013, 4, 998–1007. [Google Scholar] [CrossRef]
- Pande-Chhetri, R.; Abd-Elrahman, A.; Liu, T.; Morton, J.; Wilhelm, V.L. Object-based classification of wetland vegetation using very high-resolution unmanned air system imagery. Eur. J. Remote Sens. 2017, 50, 564–576. [Google Scholar] [CrossRef]
- Hamylton, S.; Morris, R.; Carvalho, R.; Roder, N.; Barlow, P.; Mills, K.; Wang, L. Evaluating techniques for mapping island vegetation from unmanned aerial vehicle (UAV) images: Pixel classification, visual interpretation and machine learning approaches. Int. J. Appl. Earth Obs. Geoinf. 2020, 89, 102085. [Google Scholar] [CrossRef]
- Hossain, M.D.; Chen, D. Segmentation for Object-Based Image Analysis (OBIA): A review of algorithms and challenges from remote sensing perspective. ISPRS J. Photogramm. Remote Sens. 2019, 150, 115–134. [Google Scholar] [CrossRef]
- Ke, Y.; Quackenbush, L.J. A review of methods for automatic individual tree-crown detection and delineation from passive remote sensing. Int. J. Remote Sens. 2011, 32, 4725–4747. [Google Scholar] [CrossRef]
- Wagner, F.H.; Ferreira, M.P.; Sanchez, A.; Hirye, M.C.; Zortea, M.; Gloor, E.; Phillips, O.L.; de Souza Filho, C.R.; Shimabukuro, Y.E.; Aragão, L.E. Individual tree crown delineation in a highly diverse tropical forest using very high resolution satellite images. ISPRS J. Photogramm. Remote Sens. 2018, 145, 362–377. [Google Scholar] [CrossRef]
- Gu, J.; Grybas, H.; Congalton, R.G. A comparison of forest tree crown delineation from unmanned aerial imagery using canopy height models vs. spectral lightness. Forests 2020, 11, 605. [Google Scholar] [CrossRef]
- Li, D.; Zhang, G.; Wu, Z.; Yi, L. An Edge Embedded Marker-Based Watershed Algorithm for High Spatial Resolution Remote Sensing Image Segmentation. IEEE Trans. Image Process. 2010, 19, 2781–2787. [Google Scholar] [CrossRef]
- Cai, Y.; Tong, X.; Shu, R. Multi-scale segmentation of remote sensing image based on watershed transformation. In Proceedings of the 2009 Joint Urban Remote Sensing Event, Shanghai, China, 20–22 May 2009; pp. 1–6. [Google Scholar] [CrossRef]
- Wang, M.; Li, R. Segmentation of High Spatial Resolution Remote Sensing Imagery Based on Hard-Boundary Constraint and Two-Stage Merging. IEEE Trans. Geosci. Remote Sens. 2014, 52, 5712–5725. [Google Scholar] [CrossRef]
- Mylonas, S.; Stavrakoudis, D.; Theocharis, J.; Mastorocostas, P. A Region-Based GeneSIS Segmentation Algorithm for the Classification of Remotely Sensed Images. Remote Sens. 2015, 7, 2474–2508. [Google Scholar] [CrossRef]
- Gaetano, R.; Masi, G.; Poggi, G.; Verdoliva, L.; Scarpa, G. Marker-controlled watershed-based segmentation of multiresolution remote sensing images. IEEE Trans. Geosci. Remote Sens. 2015, 53, 2987–3004. [Google Scholar] [CrossRef]
- Klosterman, S.; Melaas, E.; Wang, J.; Martinez, A.; Frederick, S.; O’Keefe, J.; Orwig, D.A.; Wang, Z.; Sun, Q.; Schaaf, C.; et al. Fine-scale perspectives on landscape phenology from unmanned aerial vehicle (UAV) photography. Agric. For. Meteorol. 2018, 248, 397–407. [Google Scholar] [CrossRef]
- Park, J.Y.; Muller-Landau, H.C.; Lichstein, J.W.; Rifai, S.W.; Dandois, J.P.; Bohlman, S.A. Quantifying leaf phenology of individual trees and species in a tropical forest using unmanned aerial vehicle (UAV) images. Remote Sens. 2019, 11, 1534. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Marcial-Pablo, M.d.J.; Gonzalez-Sanchez, A.; Jimenez-Jimenez, S.I.; Ontiveros-Capurata, R.E.; Ojeda-Bustamante, W. Estimation of vegetation fraction using RGB and multispectral images from UAV. Int. J. Remote Sens. 2019, 40, 420–438. [Google Scholar] [CrossRef]
- Yin, G.; Verger, A.; Filella, I.; Descals, A.; Peñuelas, J. Divergent Estimates of Forest Photosynthetic Phenology Using Structural and Physiological Vegetation Indices. Geophys. Res. Lett. 2020, 47, e2020GL089167. [Google Scholar] [CrossRef]
- Wong, C.Y.S.; D’Odorico, P.; Bhathena, Y.; Arain, M.A.; Ensminger, I. Carotenoid based vegetation indices for accurate monitoring of the phenology of photosynthesis at the leaf-scale in deciduous and evergreen trees. Remote Sens. Environ. 2019, 233, 111407. [Google Scholar] [CrossRef]
- Thapa, S.; Garcia Millan, V.E.; Eklundh, L. Assessing forest phenology: A multi-scale comparison of near-surface (UAV, spectral reflectance sensor, phenocam) and satellite (MODIS, sentinel-2) remote sensing. Remote Sens. 2021, 13, 1597. [Google Scholar] [CrossRef]
- Huang, S.; Tang, L.; Hupy, J.P.; Wang, Y.; Shao, G. A commentary review on the use of normalized difference vegetation index (NDVI) in the era of popular remote sensing. J. For. Res. 2021, 32, 1–6. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- KNMI. Weerstations: Dagwaarnemingen. Temperature and Precipitation Were Derived from Historic Daily Temperature (TG) and Rainfall (RH) Measurements Conducted by the KNMI at Location of Deelen Which Is 15 km from the Study Area. Historic Temperature Measurements Date Back to 1960 and Rainfall to 1983. 2022. Available online: https://www.daggegevens.knmi.nl/klimatologie/daggegevens (accessed on 2 April 2022).
- Tu, Y.H.; Phinn, S.; Johansen, K.; Robson, A. Assessing radiometric correction approaches for multi-spectral UAS imagery for horticultural applications. Remote Sens. 2018, 10, 1684. [Google Scholar] [CrossRef]
- Assmann, J.J.; Kerby, J.T.; Cunliffe, A.M.; Myers-Smith, I.H. Vegetation monitoring using multispectral sensors—Best practices and lessons learned from high latitudes. J. Unmanned Veh. Syst. 2019, 7, 54–75. [Google Scholar] [CrossRef]
- Agisoft LLC. Metashape, 2019. Available online: https://www.agisoft.com/downloads/installer/ (accessed on 9 March 2022).
- ESRI. ArcGIS Pro, 2021. Available online: https://www.esri.com/en-us/arcgis/products/arcgis-pro/overview (accessed on 9 March 2022).
- Berra, E.F.; Gaulton, R.; Barr, S. Commercial Off-the-Shelf Digital Cameras on Unmanned Aerial Vehicles for Multitemporal Monitoring of Vegetation Reflectance and NDVI. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4878–4886. [Google Scholar] [CrossRef]
- Panagiotidis, D.; Abdollahnejad, A.; Surový, P.; Chiteculo, V. Determining tree height and crown diameter from high-resolution UAV imagery. Int. J. Remote Sens. 2017, 38, 2392–2410. [Google Scholar] [CrossRef]
- AHN. AHN 3 DTM, 2019. Available online: https://app.pdok.nl/ahn3-downloadpage/ (accessed on 10 March 2022).
- Mohan, M.; Leite, R.V.; Broadbent, E.N.; Wan Mohd Jaafar, W.S.; Srinivasan, S.; Bajaj, S.; Dalla Corte, A.P.; do Amaral, C.H.; Gopan, G.; Saad, S.N.M.; et al. Individual tree detection using UAV-lidar and UAV-SfM data: A tutorial for beginners. Open Geosci. 2021, 13, 1028–1039. [Google Scholar] [CrossRef]
- Roussel, J.R.; Auty, D. lidR, 2021. Available online: https://cran.r-project.org/web/packages/lidR/index.html (accessed on 11 March 2022).
- Doruska, P.F.; Burkhart, H.E. Modeling the diameter and locational distribution of branches within the crowns of loblolly pine trees in unthinned plantations. Can. J. For. Res. 1994, 24, 2362–2376. [Google Scholar] [CrossRef]
- Popescu, S.C.; Wynne, R.H. Seeing the Trees in the Forest. Photogramm. Eng. Remote Sens. 2004, 70, 589–604. [Google Scholar] [CrossRef]
- Plowright, A.; Roussel, J.R. ForestTools, 2021. Available online: https://cran.r-project.org/web/packages/ForestTools/index.html (accessed on 13 March 2022).
- Meyer, F.; Beucher, S. Morphological segmentation. J. Vis. Commun. Image Represent. 1990, 1, 21–46. [Google Scholar] [CrossRef]
- Strimas-Mackey, M. Smoothr, 2021. Available online: https://cran.r-project.org/web/packages/smoother/index.html (accessed on 14 March 2022).
- Zeng, L.; Wardlow, B.D.; Xiang, D.; Hu, S.; Li, D. A review of vegetation phenological metrics extraction using time-series, multispectral satellite data. Remote Sens. Environ. 2020, 237, 111511. [Google Scholar] [CrossRef]
- Beck, P.S.; Atzberger, C.; Høgda, K.A.; Johansen, B.; Skidmore, A.K. Improved monitoring of vegetation dynamics at very high latitudes: A new method using MODIS NDVI. Remote Sens. Environ. 2006, 100, 321–334. [Google Scholar] [CrossRef]
- Forkel, W.M. Greenbrown, 2015. Available online: http://greenbrown.r-forge.r-project.org/ (accessed on 16 March 2022).
- Cao, R.; Chen, J.; Shen, M.; Tang, Y. An improved logistic method for detecting spring vegetation phenology in grasslands from MODIS EVI time-series data. Agric. For. Meteorol. 2015, 200, 9–20. [Google Scholar] [CrossRef]
- Clinton, N.; Holt, A.; Scarborough, J.; Yan, L.; Gong, P. Accuracy Assessment Measures for Object-based Image Segmentation Goodness. Photogramm. Eng. Remote Sens. 2010, 76, 289–299. [Google Scholar] [CrossRef]
- Winter, S. Location similarity of regions. ISPRS J. Photogramm. Remote Sens. 2000, 55, 189–200. [Google Scholar] [CrossRef]
- Sellers, P.J. Canopy reflectance, photosynthesis and transpiration. Int. J. Remote Sens. 1985, 6, 1335–1372. [Google Scholar] [CrossRef]
- Vitasse, Y.; Lenz, A.; Körner, C. The interaction between freezing tolerance and phenology in temperate deciduous trees. Front. Plant Sci. 2014, 5, 541. [Google Scholar] [CrossRef]
- Dong, T.; Meng, J.; Shang, J.; Liu, J.; Wu, B. Evaluation of Chlorophyll-Related Vegetation Indices Using Simulated Sentinel-2 Data for Estimation of Crop Fraction of Absorbed Photosynthetically Active Radiation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 4049–4059. [Google Scholar] [CrossRef]
- Nguy-Robertson, A.L.; Peng, Y.; Gitelson, A.A.; Arkebauer, T.J.; Pimstein, A.; Herrmann, I.; Karnieli, A.; Rundquist, D.C.; Bonfil, D.J. Estimating green LAI in four crops: Potential of determining optimal spectral bands for a universal algorithm. Agric. For. Meteorol. 2014, 192-193, 140–148. [Google Scholar] [CrossRef]
- Schlemmer, M.; Gitelson, A.; Schepers, J.; Ferguson, R.; Peng, Y.; Shanahan, J.; Rundquist, D. Remote estimation of nitrogen and chlorophyll contents in maize at leaf and canopy levels. Int. J. Appl. Earth Obs. Geoinf. 2013, 25, 47–54. [Google Scholar] [CrossRef]
- Adams, W.; Zarter, R.; Ebbert, V.; Demmig-Adams, B. Photoprotective Strategies of Overwintering Evergreens. BioScience 2004, 54, 41–49. [Google Scholar] [CrossRef]
- Fu, Y.H.; Piao, S.; Op de Beeck, M.; Cong, N.; Zhao, H.; Zhang, Y.; Menzel, A.; Janssens, I.A. Recent spring phenology shifts in western Central Europe based on multiscale observations. Glob. Ecol. Biogeogr. 2014, 23, 1255–1263. [Google Scholar] [CrossRef]
- Jeong, S.J.; Schimel, D.; Frankenberg, C.; Drewry, D.T.; Fisher, J.B.; Verma, M.; Berry, J.A.; Lee, J.E.; Joiner, J. Application of satellite solar-induced chlorophyll fluorescence to understanding large-scale variations in vegetation phenology and function over northern high latitude forests. Remote Sens. Environ. 2017, 190, 178–187. [Google Scholar] [CrossRef]
- Badgley, G.; Field, C.B.; Berry, J.A. Canopy near-infrared reflectance and terrestrial photosynthesis. Sci. Adv. 2017, 3, 1602244. [Google Scholar] [CrossRef]
- Tanaka, S.; Kawamura, K.; Maki, M.; Muramoto, Y.; Yoshida, K.; Akiyama, T. Spectral Index for Quantifying Leaf Area Index of Winter Wheat by Field Hyperspectral Measurements: A Case Study in Gifu Prefecture, Central Japan. Remote Sens. 2015, 7, 5329–5346. [Google Scholar] [CrossRef]
- Hashimoto, N.; Saito, Y.; Maki, M.; Homma, K. Simulation of reflectance and vegetation indices for unmanned aerial vehicle (UAV) monitoring of paddy fields. Remote Sens. 2019, 11, 2119. [Google Scholar] [CrossRef]
- Kikuzawa, K. Phenological and morphological adaptations to the light environment in two woody and two herbaceous plant species. Funct. Ecol. 2003, 17, 29–38. [Google Scholar] [CrossRef]
- Mariën, B.; Balzarolo, M.; Dox, I.; Leys, S.; Lorène, M.J.; Geron, C.; Portillo-Estrada, M.; AbdElgawad, H.; Asard, H.; Campioli, M. Detecting the onset of autumn leaf senescence in deciduous forest trees of the temperate zone. New Phytol. 2019, 224, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Medvigy, D.; Jeong, S.J.; Clark, K.L.; Skowronski, N.S.; Schäfer, K.V.R. Effects of seasonal variation of photosynthetic capacity on the carbon fluxes of a temperate deciduous forest. J. Geophys. Res. Biogeosci. 2013, 118, 1703–1714. [Google Scholar] [CrossRef]
- de Natuurkalender. Natuurkalender. 2021. Available online: https://www.naturetoday.com/intl/nl/observations/natuurkalender/sightings/view-sightings (accessed on 30 March 2022).
- Wu, S.; Wang, J.; Yan, Z.; Song, G.; Chen, Y.; Ma, Q.; Deng, M.; Wu, Y.; Zhao, Y.; Guo, Z.; et al. Monitoring tree-crown scale autumn leaf phenology in a temperate forest with an integration of PlanetScope and drone remote sensing observations. ISPRS J. Photogramm. Remote Sens. 2021, 171, 36–48. [Google Scholar] [CrossRef]
- Kuster, T.M.; Dobbertin, M.; Günthardt-Goerg, M.S.; Schaub, M.; Arend, M. A Phenological Timetable of Oak Growth under Experimental Drought and Air Warming. PLoS ONE 2014, 9, e89724. [Google Scholar] [CrossRef] [PubMed]
- Leslie, A.; Mencuccini, M.; Perks, M. A resource capture efficiency index to compare differences in early growth of four tree species in northern England. iForest-Biogeosci. For. 2017, 10, 397–405. [Google Scholar] [CrossRef]
- Vander Mijnsbrugge, K.; Janssens, A. Differentiation and Non-Linear Responses in Temporal Phenotypic Plasticity of Seasonal Phenophases in a Common Garden of Crataegus monogyna Jacq. Forests 2019, 10, 293. [Google Scholar] [CrossRef]
- Lim, Y.S.; La, P.H.; Park, J.S.; Lee, M.H.; Pyeon, M.W.; Kim, J.I. Calculation of Tree Height and Canopy Crown from Drone Images Using Segmentation. J. Korean Soc. Surv. Geod. Photogramm. Cartogr. 2015, 33, 605–614. [Google Scholar] [CrossRef]
- Xu, Z.; Shen, X.; Cao, L.; Coops, N.C.; Goodbody, T.R.; Zhong, T.; Zhao, W.; Sun, Q.; Ba, S.; Zhang, Z.; et al. Tree species classification using UAS-based digital aerial photogrammetry point clouds and multispectral imageries in subtropical natural forests. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102173. [Google Scholar] [CrossRef]
- Belcore, E.; Wawrzaszek, A.; Wozniak, E.; Grasso, N.; Piras, M. Individual tree detection from UAV imagery using Hölder exponent. Remote Sens. 2020, 12, 2407. [Google Scholar] [CrossRef]
- Fawcett, D.; Azlan, B.; Hill, T.C.; Kho, L.K.; Bennie, J.; Anderson, K. Unmanned aerial vehicle (UAV) derived structure-from-motion photogrammetry point clouds for oil palm ( <i>Elaeis guineensis</i> ) canopy segmentation and height estimation. Int. J. Remote Sens. 2019, 40, 7538–7560. [Google Scholar] [CrossRef]
- Duncanson, L.; Cook, B.; Hurtt, G.; Dubayah, R. An efficient, multi-layered crown delineation algorithm for mapping individual tree structure across multiple ecosystems. Remote Sens. Environ. 2014, 154, 378–386. [Google Scholar] [CrossRef]
- van Iersel, W.; Straatsma, M.; Addink, E.; Middelkoop, H. Monitoring height and greenness of non-woody floodplain vegetation with UAV time series. ISPRS J. Photogramm. Remote Sens. 2018, 141, 112–123. [Google Scholar] [CrossRef]
- Atkins, J.W.; Stovall, A.E.; Yang, X. Mapping temperate forest phenology using tower, UAV, and ground-based sensors. Drones 2020, 4, 1–15. [Google Scholar] [CrossRef]
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping forest tree species in high resolution UAV-based RGB-imagery by means of convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Zhang, C.; Atkinson, P.M.; George, C.; Wen, Z.; Diazgranados, M.; Gerard, F. Identifying and mapping individual plants in a highly diverse high-elevation ecosystem using UAV imagery and deep learning. ISPRS J. Photogramm. Remote Sens. 2020, 169, 280–291. [Google Scholar] [CrossRef]







| Tree Detection | Segmentation | |||||
|---|---|---|---|---|---|---|
| Recall | Precision | F-score | OS | US | IoU | |
| Dense mixed (29) | 0.55 | 0.84 | 0.66 | 0.15 | 0.54 | 0.41 |
| Coniferous mixed (21) | 0.95 | 0.95 | 0.95 | 0.08 | 0.24 | 0.71 |
| Deciduous (12) | 0.92 | 0.92 | 0.92 | 0.11 | 0.29 | 0.64 |
| Coniferous (15) | 1 | 1 | 1 | 0.13 | 0.29 | 0.65 |
| Sparse mixed (11) | 1 | 0.48 | 0.65 | 0.34 | 0.16 | 0.58 |
| Small (45) | 0.67 | 0.79 | 0.73 | 0.19 | 0.44 | 0.45 |
| Overall (134) | 0.78 | 0.81 | 0.79 | 0.15 | 0.34 | 0.58 |
| Start of Season | End of Season | |||||||
|---|---|---|---|---|---|---|---|---|
| NDVI | EVI2 | OSAVI | CIre | NDVI | EVI2 | OSAVI | CIre | |
| American oak (56) | 135 (4) | 148 (11) | 135 (9) | 171 (39) | 317 (24) | 305 (9) | 310 (13) | 284 (47) |
| Beech (18) | 127 (5) | 127 (10) | 124 (3) | 150 (2) | 313 (26) | 305 (16) | 310 (14) | 279 (14) |
| Common oak (89) | 132 (16) | 150 (28) | 129 (20) | 184 (45) | 329 (30) | 306 (26) | 330 (25) | 268 (47) |
| Silver birch (151) | 118 (18) | 122 (13) | 120 (4) | 152 (12) | 271 (22) | 288 (16) | 295 (18) | 227 (39) |
| Douglas fir (189) | 161 (28) | 174 (53) | 141 (47) | 186 (32) | 197 (74) | 321 (41) | 279 (103) | 180 (68) |
| Hemlock (2) | 150 (2) | 111 (16) | 106 (9) | 153 (4) | 166 (74) | 353 (6) | 361 (18) | 133 (8) |
| Scots pine (98) | 143 (24) | 223 (48) | 185 (57) | 164 (30) | 193 (78) | 297 (29) | 289 (67) | 177 (60) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleinsmann, J.; Verbesselt, J.; Kooistra, L. Monitoring Individual Tree Phenology in a Multi-Species Forest Using High Resolution UAV Images. Remote Sens. 2023, 15, 3599. https://doi.org/10.3390/rs15143599
Kleinsmann J, Verbesselt J, Kooistra L. Monitoring Individual Tree Phenology in a Multi-Species Forest Using High Resolution UAV Images. Remote Sensing. 2023; 15(14):3599. https://doi.org/10.3390/rs15143599
Chicago/Turabian StyleKleinsmann, Jasper, Jan Verbesselt, and Lammert Kooistra. 2023. "Monitoring Individual Tree Phenology in a Multi-Species Forest Using High Resolution UAV Images" Remote Sensing 15, no. 14: 3599. https://doi.org/10.3390/rs15143599
APA StyleKleinsmann, J., Verbesselt, J., & Kooistra, L. (2023). Monitoring Individual Tree Phenology in a Multi-Species Forest Using High Resolution UAV Images. Remote Sensing, 15(14), 3599. https://doi.org/10.3390/rs15143599







