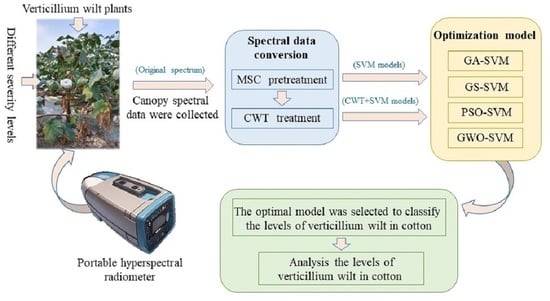
Detection of Cotton Verticillium Wilt Disease Severity Based on Hyperspectrum and GWO-SVM
Abstract
1. Introduction
2. Materials and Methods
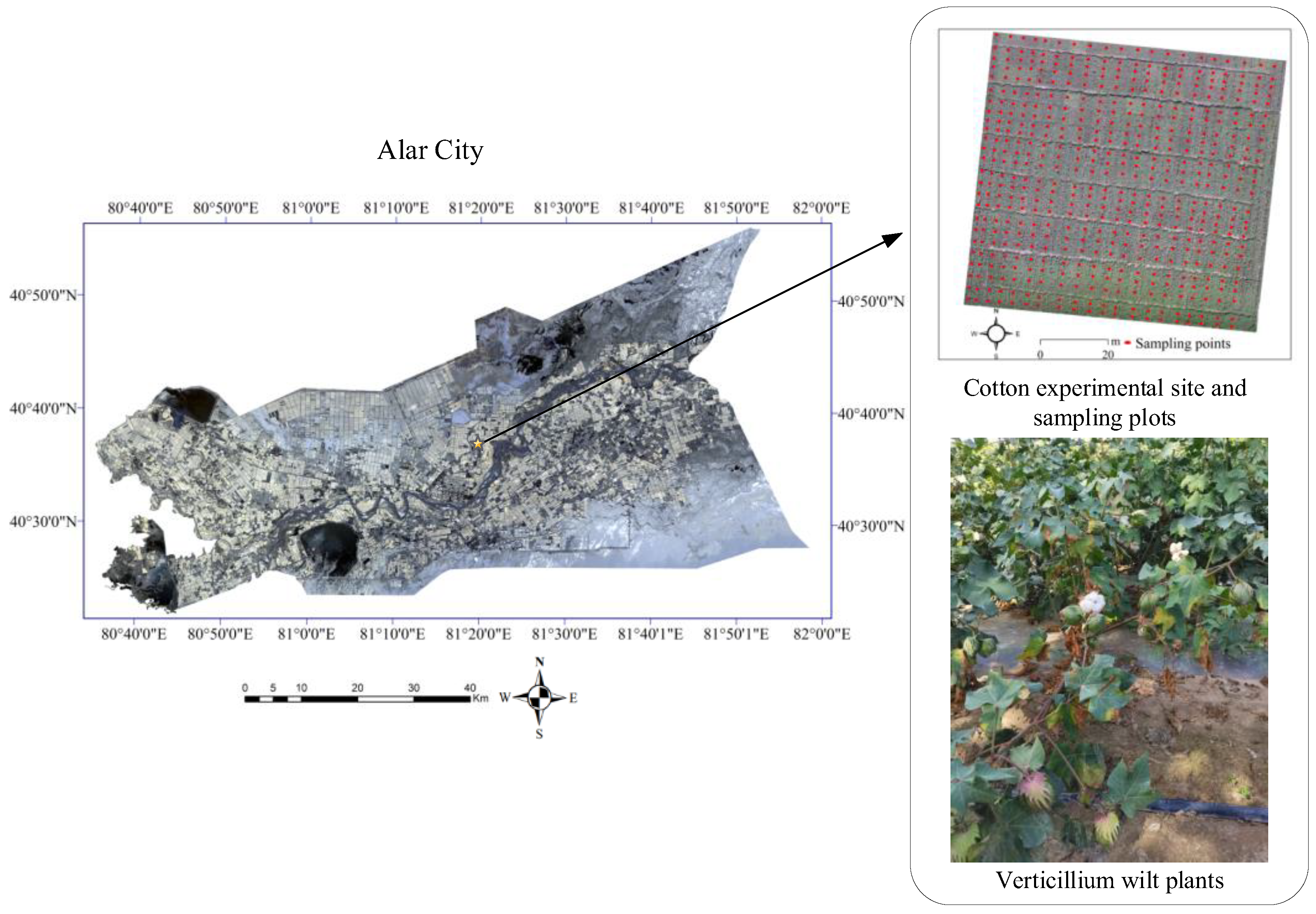
2.1. Sample
2.2. Data Acquisition
2.3. Data Processing
2.4. Continuous Wavelet Analysis
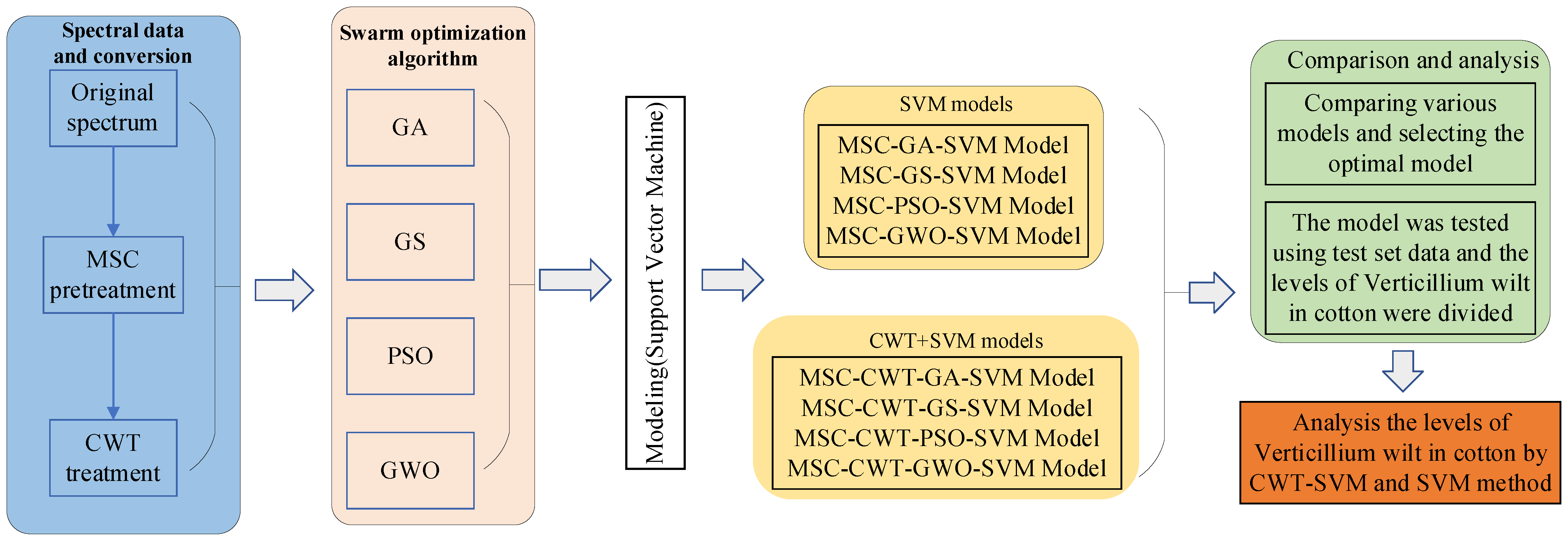
2.5. SVM Algorithm
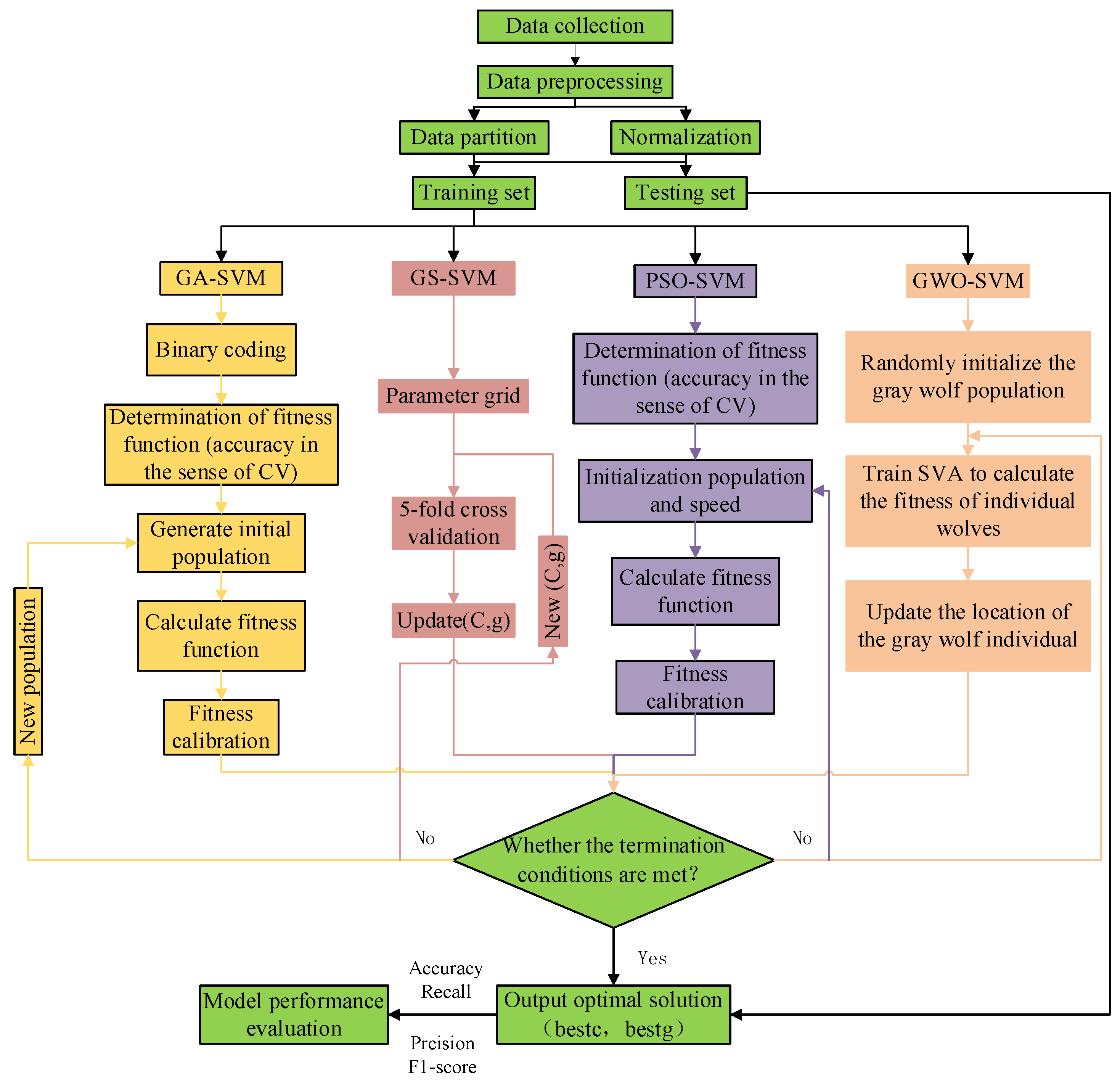
2.5.1. GA
2.5.2. PSO
2.5.3. GS
2.5.4. GWO
2.6. Model Evaluation Methods
3. Results
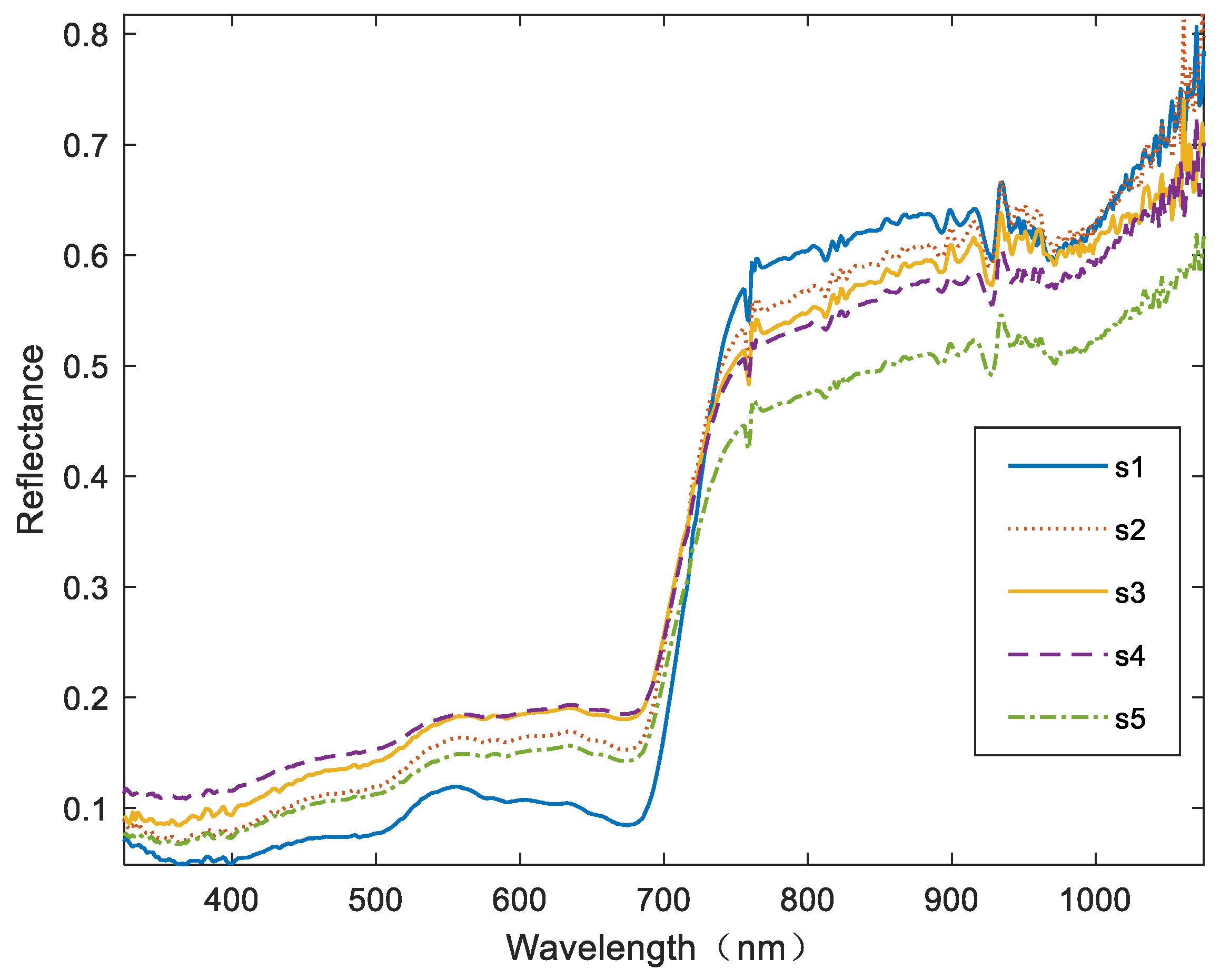
3.1. Spectrum Processing and Analysis
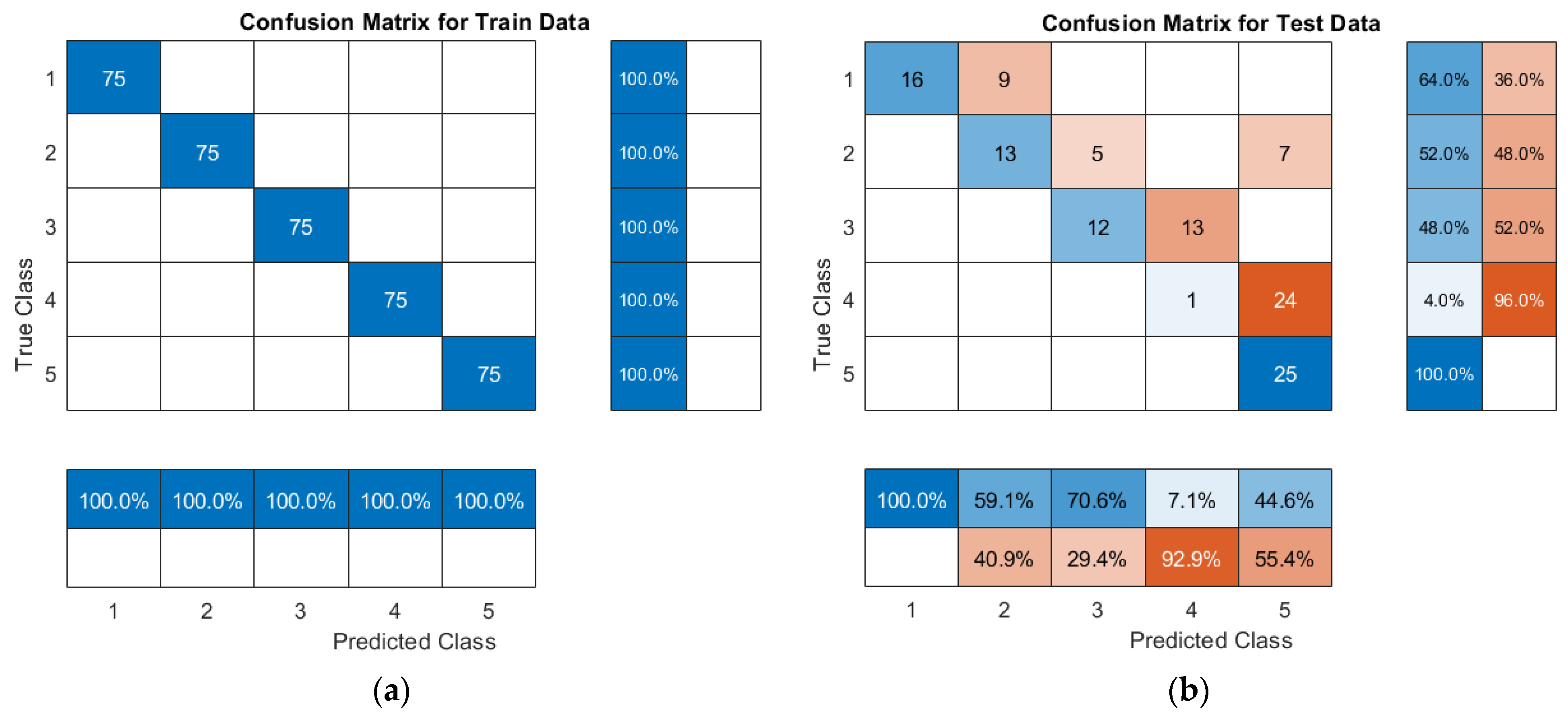
3.2. Grading of Cotton Crown Wilt Disease Based on the SVM Model
3.3. Grading of Cotton Wilt Disease with a Combination of Continuous Wavelet Analysis and SVM Models
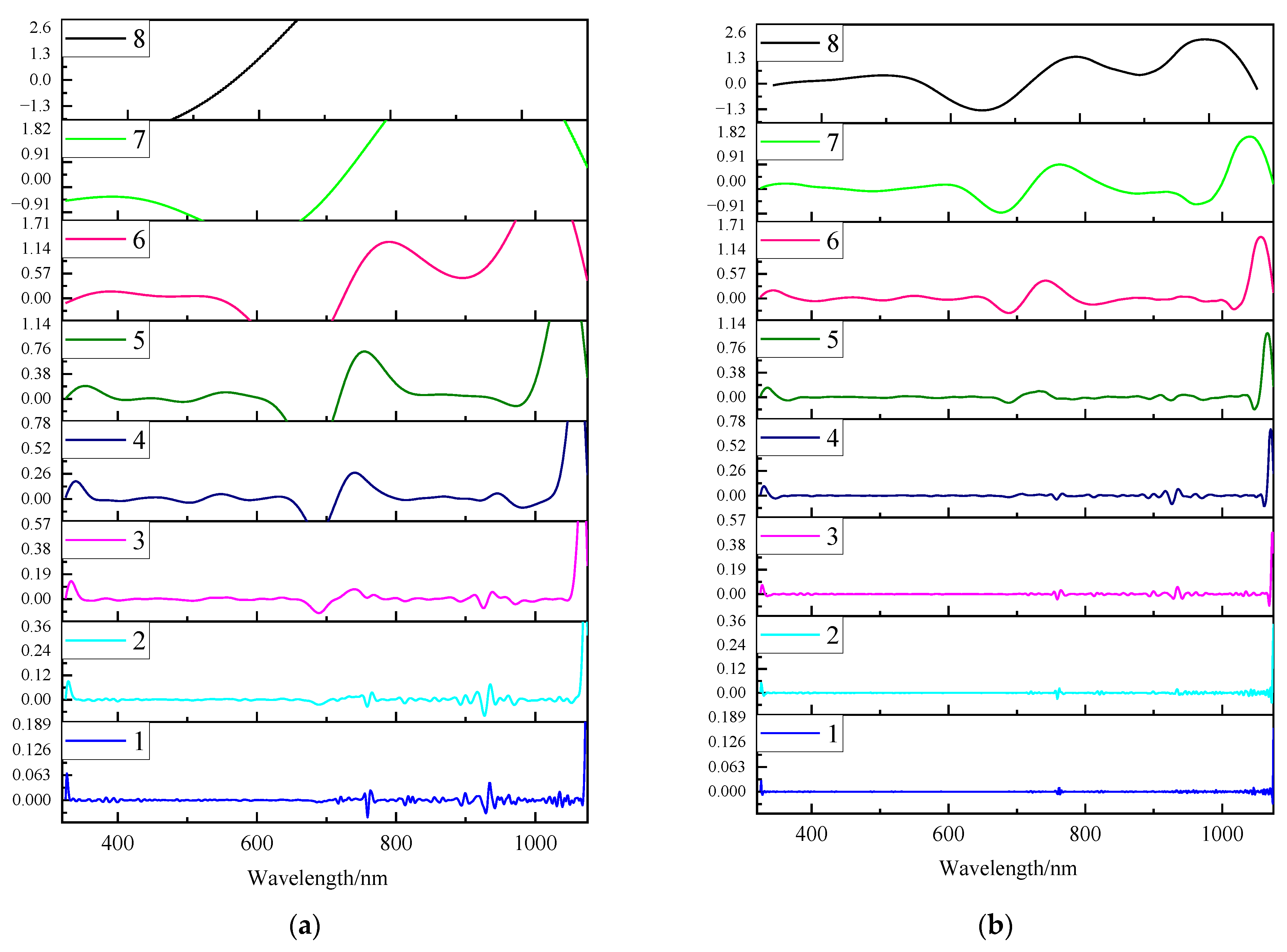
3.3.1. Analysis of Wavelet Coefficient Curves at Different Decomposition Levels
3.3.2. Establishment and Comparison of Cotton Wilt Disease Grading Models Based on the Continuous Wavelet Analysis and the SVM Model
4. Discussion
4.1. Analysis of Spectrum Features of Cotton Verticillium Wilt Disease
4.2. Performance Comparison of Different Optimization Algorithms
4.3. Improving Model Performance through CWT De-Noising at Different Decomposition Levels
4.4. Limitations of This Study and Future Work
5. Conclusions
- (1)
- Based on the cotton crown spectral data, the SVM models combined with the GA, GS, PSO, GWO optimization algorithms can be used to classify the cotton wilt disease severity. The MSC-PSO-SVM model can achieve good classification results with a relatively long running time. The GWO-SVM model has the shortest running time with relatively low parameters, but the results generated through this model are not satisfactory.
- (2)
- After different CWT processing, the accuracy, macro precision, macro recall, and macro F1-score indicators under all eight models have obtained better values. Among these eight models, those four indicators under the MSC-db3(23)-PSO-SVM and MSC-db3(23)-GWO-SVM models have obtained the same and highest values. After the wavelet (db3) processing, the accuracy, macro precision, macro recall, and macro F1-score indicators under the GWO-SVM model have achieved the biggest increase in their values. The algorithm running time of this model is relatively short.
- (3)
- Under the MSC-db3(23)-GWO-SVM model, the best results have been obtained on the classification of cotton crown wilt disease severity, and the prediction accuracy rates of the prediction set in this model on disease levels 1, 2, 3, 4, and 5 are 100%, 88%, 84%, 84%, and 100%, respectively.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kang, X.; Huang, C.; Zhang, L.; Yang, M.; Zhang, Z.; Lyu, X. Assessing the Severity of Cotton Verticillium Wilt Disease from in Situ Canopy Images and Spectra Using Convolutional Neural Networks. Crop J. 2022, 12, 933–940. [Google Scholar] [CrossRef]
- Kaur, K.; Vyas, A. Suppression of Verticillium Wilt of Cotton through Liquid Material and Antagonistic Fungal Strains under Natural Field Conditions. Mater. Today Proc. 2022, 60, 1186–1198. [Google Scholar] [CrossRef]
- Chen, B.; Li, S.; Wang, K.; Zhou, G.; Bai, J. Evaluating the Severity Level of Cotton Verticillium Using Spectral Signature Analysis. J. Remote Sens. 2012, 33, 2706–2724. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhao, M.; Li, T.; Wang, L.; Liao, C.; Liu, D.; Zhang, H.; Zhao, Y.; Liu, L.; Ge, X.; et al. Interactions between Verticillium Dahliae and Cotton: Pathogenic Mechanism and Cotton Resistance Mechanism to Verticillium Wilt. Front. Plant Sci. 2023, 14, 1174281. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Li, S.; Wang, K.; Wang, J.; Wang, F.; Xiao, C.; Lai, J.; Wang, N. Spectrum Characteristics of Cotton Canopy Infected with Verticillium Wilt and Applications. Agric. Sci. China 2008, 7, 561–569. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, X.; Ran, W.; Xu, Y.; Shen, Q. Screening of bacteria antagonistic against soil-borne cotton Verticillium wilt and their biological effects on the soil-cotton system. Acta Pedol. Sin. 2008, 45, 1095–1101. [Google Scholar] [CrossRef]
- Bock, C.H.; Barbedo, J.G.A.; Del Ponte, E.M.; Bohnenkamp, D.; Mahlein, A.-K. From Visual Estimates to Fully Automated Sensor-Based Measurements of Plant Disease Severity: Status and Challenges for Improving Accuracy. Phytopathol. Res. 2020, 2, 9. [Google Scholar] [CrossRef]
- Chen, B. Study on Monitoring Cotton Infected with Verticillium Wilt Based on Multi-platform Remote Sensing. Ph.D. Thesis, Shihezi University, Shihezi, China, 2010. [Google Scholar]
- Feng, L.; Wu, B.; Zhu, S.; Wang, J.; Su, Z.; Liu, F.; He, Y.; Zhang, C. Investigation on Data Fusion of Multisource Spectral Data for Rice Leaf Diseases Identification Using Machine Learning Methods. Front. Plant Sci. 2020, 11, 577063. [Google Scholar] [CrossRef]
- Feng, Z.-H.; Wang, L.-Y.; Yang, Z.-Q.; Zhang, Y.-Y.; Li, X.; Song, L.; He, L.; Duan, J.-Z.; Feng, W. Hyperspectral Monitoring of Powdery Mildew Disease Severity in Wheat Based on Machine Learning. Front. Plant Sci. 2022, 13, 828454. [Google Scholar] [CrossRef]
- Galieni, A.; Nicastro, N.; Pentangelo, A.; Platani, C.; Cardi, T.; Pane, C. Surveying Soil-Borne Disease Development on Wild Rocket Salad Crop by Proximal Sensing Based on High-Resolution Hyperspectral Features. Sci. Rep. 2022, 12, 5098. [Google Scholar] [CrossRef]
- Jing, X.; Du, K.; Duan, W.; Zou, Q.; Zhao, T.; Li, B.; Ye, Q.; Yan, L. Quantifying the Effects of Stripe Rust Disease on Wheat Canopy Spectrum Based on Eliminating Non-Physiological Stresses. Crop J. 2022, 10, 1284–1291. [Google Scholar] [CrossRef]
- Liu, Z.; Huang, J.; Shi, J.; Tao, R.; Zhou, W.; Zhang, L. Characterizing and Estimating Rice Brown Spot Disease Severity Using Stepwise Regression, Principal Component Regression and Partial Least-Square Regression. J. Zhejiang Univ. Sci. B 2007, 8, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Feng, S.; Cao, Y.; Yu, F.; Guan, Q.; Li, J.; Zhang, G.; Xu, T. Study on the Classification Method of Rice Leaf Blast Levels Based on Fusion Features and Adaptive-Weight Immune Particle Swarm Optimization Extreme Learning Machine Algorithm. Front. Plant Sci. 2022, 13, 879668. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yuan, L.; Wang, J.; Luo, J.; Du, S.; Huang, W. Research progress of crop diseases and pests monitoring based on remote sensing. Trans. CSAE 2012, 28, 1–11. [Google Scholar]
- Chen, Q.; Fan, Y.; Wu, H.; Yang, Q.; Wang, D.; Deng, Y.; Wang, C. Spectral Characteristics Analysis of Cotton Verticillium Wilt Canopy and Establishment of Its Severity Estimation Model. J. Xinjiang Agric. Univ. 2020, 43, 261–269. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, J.; Chen, Y.; Wan, S.; Zhang, L. Detection of Peanut Leaf Spots Disease Using Canopy Hyperspectral Reflectance. Comput. Electron. Agric. 2019, 156, 677–683. [Google Scholar] [CrossRef]
- Cao, X.; Luo, Y.; Zhou, Y.; Duan, X.; Cheng, D. Detection of Powdery Mildew in Two Winter Wheat Cultivars Using Canopy Hyperspectral Reflectance. Crop Prot. 2013, 45, 124–131. [Google Scholar] [CrossRef]
- Li, L.; Geng, S.; Lin, D.; Su, G.; Zhang, Y.; Chang, L.; Ji, Y.; Wang, Y.; Wang, L. Accurate Modeling of Vertical Leaf Nitrogen Distribution in Summer Maize Using in Situ Leaf Spectroscopy via CWT and PLS-Based Approaches. Eur. J. Agron. 2022, 140, 126607. [Google Scholar] [CrossRef]
- Luo, J.; Huang, W.; Yuan, L.; Zhao, C.; Du, S.; Zhang, J.; Zhao, J. Evaluation of Spectral Indices and Continuous Wavelet Analysis to Quantify Aphid Infestation in Wheat. Precis. Agric. 2013, 14, 151–161. [Google Scholar] [CrossRef]
- Mustafa, G.; Zheng, H.; Khan, I.H.; Tian, L.; Jia, H.; Li, G.; Cheng, T.; Tian, Y.; Cao, W.; Zhu, Y.; et al. Hyperspectral Reflectance Proxies to Diagnose In-Field Fusarium Head Blight in Wheat with Machine Learning. Remote Sens. 2022, 14, 2784. [Google Scholar] [CrossRef]
- Huang, W.; Lu, J.; Ye, H.; Kong, W.; Hugh Mortimer, A.; Shi, Y. Quantitative Identification of Crop Disease and Nitrogen-Water Stress in Winter Wheat Using Continuous Wavelet Analysis. Int. J. Agric. Biol. Eng. 2018, 11, 145–152. [Google Scholar] [CrossRef]
- Tang, J.; Liu, G.; Pan, Q. A Review on Representative Swarm Intelligence Algorithms for Solving Optimization Problems: Applications and Trends. IEEE/CAA J. Autom. Sinica 2021, 8, 1627–1643. [Google Scholar] [CrossRef]
- Li, L.; Ustin, S.L.; Riano, D. Retrieval of Fresh Leaf Fuel Moisture Content Using Genetic Algorithm Partial Least Squares (GA-PLS) Modeling. IEEE Geosci. Remote Sens. Lett. 2007, 4, 216–220. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, F.; Kung, H.; Johnson, V.C.; Latif, A. Extracting Soil Salinization Information with a Fractional-Order Filtering Algorithm and Grid-Search Support Vector Machine (GS-SVM) Model. J. Remote Sens. 2020, 41, 953–973. [Google Scholar] [CrossRef]
- Bonah, E.; Huang, X.; Yi, R.; Aheto, J.H.; Yu, S. Vis-NIR Hyperspectral Imaging for the Classification of Bacterial Foodborne Pathogens Based on Pixel-Wise Analysis and a Novel CARS-PSO-SVM Model. Infrared Phys. Technol. 2020, 105, 103220. [Google Scholar] [CrossRef]
- Saremi, S.; Mirjalili, S.; Lewis, A. Grasshopper Optimisation Algorithm: Theory and Application. Adv. Eng. Softw. 2017, 105, 30–47. [Google Scholar] [CrossRef]
- Zhong, L.; Zhou, Y.; Zhou, G.; Luo, Q. Enhanced Discrete Dragonfly Algorithm for Solving Four-Color Map Problems. Appl. Intell. 2023, 53, 6372–6400. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey Wolf Optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Zhang, Y.; Kang, C.; Liu, Y.; Fu, X.; Zhang, J.; Wang, M.; Yang, L. Rapidly Detection of Total Nitrogen and Phosphorus Content in Water by Surface Enhanced Raman Spectroscopy and GWO-SVR Algorithm. Spectrosc. Spectr. Anal. 2021, 41, 3147–3152. [Google Scholar]
- Gao, Q.; Wang, P.; Niu, T.; He, D.; Wang, M.; Yang, H.; Zhao, X. Soluble Solid Content and Firmness Index Assessment and Maturity Discrimination of Malus Micromalus Makino Based on Near-Infrared Hyperspectral Imaging. Food Chem. 2022, 370, 131013. [Google Scholar] [CrossRef]
- Wu, S.; Wang, L.; Zhou, G.; Liu, C.; Ji, Z.; Li, Z.; Li, W. Strategies for the Content Determination of Capsaicin and the Identification of Adulterated Pepper Powder Using a Hand-Held near-Infrared Spectrometer. Food Res. Int. 2023, 163, 112192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, X.; Cui, X.; Cao, W.; Zhang, X.; Zhang, Y. Classification of Qianxi Tomatoes by Visible/Near Infrared Spectroscopy Combined with GMO-SVM. Spectrosc. Spectr. Anal. 2022, 42, 3291–3297. [Google Scholar] [CrossRef]
- Du, L.; Yang, H.; Song, X.; Wei, N.; Yu, C.; Wang, W.; Zhao, Y. Estimating Leaf Area Index of Maize Using UAV-Based Digital Imagery and Machine Learning Methods. Sci. Rep. 2022, 12, 15937. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, S.; Fernández-Novales, J.; Diago, M.P.; Tardaguila, J. On-The-Go Hyperspectral Imaging Under Field Conditions and Machine Learning for the Classification of Grapevine Varieties. Front. Plant Sci. 2018, 9, 1102. [Google Scholar] [CrossRef]
- He, Y.; Zhang, W.; Ma, Y.; Li, J.; Ma, B. The Classification of Rice Blast Resistant Seed Based on Ranman Spectroscopy and SVM. Molecules 2022, 27, 4091. [Google Scholar] [CrossRef]
- Wang, Z.; Fan, S.; Wu, J.; Zhang, C.; Xu, F.; Yang, X.; Li, J. Application of Long-Wave near Infrared Hyperspectral Imaging for Determination of Moisture Content of Single Maize Seed. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 254, 119666. [Google Scholar] [CrossRef]
- Zhao, G.; Pei, Y.; Yang, R.; Xiang, L.; Fang, Z.; Wang, Y.; Yin, D.; Wu, J.; Gao, D.; Yu, D.; et al. A Non-Destructive Testing Method for Early Detection of Ginseng Root Diseases Using Machine Learning Technologies Based on Leaf Hyperspectral Reflectance. Front. Plant Sci. 2022, 13, 1031030. [Google Scholar] [CrossRef]
- Zhu, H.; Chu, B.; Zhang, C.; Liu, F.; Jiang, L.; He, Y. Hyperspectral Imaging for Presymptomatic Detection of Tobacco Disease with Successive Projections Algorithm and Machine-Learning Classifiers. Sci. Rep. 2017, 7, 4125. [Google Scholar] [CrossRef]
- Zhang, G.; Xu, T.; Tian, Y. Hyperspectral Imaging-Based Classification of Rice Leaf Blast Severity over Multiple Growth Stages. Plant Methods 2022, 18, 123. [Google Scholar] [CrossRef]
- Zhao, J.; Fang, Y.; Chu, G.; Yan, H.; Hu, L.; Huang, L. Identification of Leaf-Scale Wheat Powdery Mildew (Blumeria graminis f. Sp. tritici) Combining Hyperspectral Imaging and an SVM Classifier. Plants 2020, 9, 936. [Google Scholar] [CrossRef]
- Liu, S.; Yu, H.; Sui, Y.; Zhou, H.; Zhang, J.; Kong, L.; Dang, J.; Zhang, L. Classification of Soybean Frogeye Leaf Spot Disease Using Leaf Hyperspectral Reflectance. PLoS ONE 2021, 16, e0257008. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Manohara, K.K.; Mahajan, G.R.; Sahoo, R.N. Spectroscopy Based Novel Spectral Indices, PCA- and PLSR-Coupled Machine Learning Models for Salinity Stress Phenotyping of Rice. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2020, 229, 117983. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhao, X.; Zhang, W.; He, X.; Tong, L. Study on the Identification of Resistance of Rice Blast Based on near Infrared Spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 266, 120439. [Google Scholar] [CrossRef] [PubMed]
- Geetharamani, G.; Pandian, A. Identification of Plant Leaf Diseases Using a Nine-Layer Deep Convolutional Neural Network. Comput. Electr. Eng. 2019, 76, 323–338. [Google Scholar] [CrossRef]
- Lu, J.; Tan, L.; Jiang, H. Review on Convolutional Neural Network (CNN) Applied to Plant Leaf Disease Classification. Agriculture 2021, 11, 707. [Google Scholar] [CrossRef]
- Kang, L.; Yuan, J.; Gao, R.; Kong, Q.; Jia, Y.; Su, Z. Early Detection and Identification of Rice Blast Based on Hyperspectral Image. Spectrosc. Spectr. Anal. 2021, 41, 898–902. [Google Scholar]
- Zhang, Z.; Wang, P.; Yao, Z.; Qin, L.; He, D.; Xu, Y.; Zhang, J.; Hu, J. Early Detection of Downy Mildew on Grape Leaves Using Multicolor Fluorescence Imaging and Model SVM. Spectrosc. Spectr. Anal. 2021, 41, 828–834. [Google Scholar]
- Han, Y.; Liu, H.; Zhang, X.; Yu, Z.; Meng, X.; Kong, F.; Song, S.; Han, J. Prediction Model of Rice Panicles Blast Disease Degree Based on Canopy Hyperspectral Reflectance. Spectrosc. Spectr. Anal. 2021, 41, 1220–1226. [Google Scholar]
- Liu, Y.; Lin, X.; Gao, H.; Wang, S.; Gao, X. Research on Tea Cephaleuros Virescens Kunze Model Based on Chlorophyll Fluorescence Spectroscopy. Spectrosc. Spectr. Anal. 2021, 41, 2129–2134. [Google Scholar]
- Kok, Z.H.; Mohamed Shariff, A.R.; Alfatni, M.S.M.; Khairunniza-Bejo, S. Support Vector Machine in Precision Agriculture: A Review. Comput. Electron. Agric. 2021, 191, 106546. [Google Scholar] [CrossRef]
- Li, F.; Wang, L.; Liu, J.; Wang, Y.; Chang, Q. Evaluation of Leaf N Concentration in Winter Wheat Based on Discrete Wavelet Transform Analysis. Remote Sens. 2019, 11, 1331. [Google Scholar] [CrossRef]
- Le Maire, G.; François, C.; Dufrêne, E. Towards Universal Broad Leaf Chlorophyll Indices Using PROSPECT Simulated Database and Hyperspectral Reflectance Measurements. Remote Sens. Environ. 2004, 89, 1–28. [Google Scholar] [CrossRef]
- Zhao, J.; Xiong, Z.; Ning, J.; Xie, D. Wavelet transform combined with spa to optimize the near-infrared analysis model of caffeine in tea. J. Anal. Sci. 2021, 37, 611–617. [Google Scholar] [CrossRef]
- Ding, Y.; Yan, Y.; Li, J.; Chen, X.; Jiang, H. Classification of Tea Quality Levels Using Near-Infrared Spectroscopy Based on CLPSO-SVM. Foods 2022, 11, 1658. [Google Scholar] [CrossRef]
- Nkongolo, M.; Van Deventer, J.P.; Kasongo, S.M.; Zahra, S.R.; Kipongo, J. A Cloud Based Optimization Method for Zero-Day Threats Detection Using Genetic Algorithm and Ensemble Learning. Electronics 2022, 11, 1749. [Google Scholar] [CrossRef]
- Huang, C.-L.; Dun, J.-F. A Distributed PSO–SVM Hybrid System with Feature Selection and Parameter Optimization. Appl. Soft Comput. 2008, 8, 1381–1391. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.; Fan, Y.; Cheng, Y.; Wu, X.; Zhang, J.; Wang, B.; Wang, X.; Yong, T.; Liu, W.; et al. Evaluating Photosynthetic Pigment Contents of Maize Using UVE-PLS Based on Continuous Wavelet Transform. Comput. Electron. Agric. 2020, 169, 105160. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The Advantages of the Matthews Correlation Coefficient (MCC) over F1 Score and Accuracy in Binary Classification Evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Guo, W.; Zhu, Y.; Wang, H.; Zhang, J.; Dong, P.; Qiao, H. Monitoring Model of Winter Wheat Take-all Based on UAV Hyperspectral Imaging. Trans. Chin. Soc. Agric. Mach. 2019, 50, 162–169. [Google Scholar] [CrossRef]
- Song, L.; Liang, Q.; Chen, H.; Hu, H.; Luo, Y.; Luo, Y. A New Approach to Optimize SVM for Insulator State Identification Based on Improved PSO Algorithm. Sensors 2022, 23, 272. [Google Scholar] [CrossRef]
- Li, D.; Wang, X.; Zheng, H.; Zhou, K.; Yao, X.; Tian, Y.; Zhu, Y.; Cao, W.; Cheng, T. Estimation of Area- and Mass-Based Leaf Nitrogen Contents of Wheat and Rice Crops from Water-Removed Spectra Using Continuous Wavelet Analysis. Plant Methods 2018, 14, 76. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Cheng, T.; Jia, M.; Zhou, K.; Lu, N.; Yao, X.; Tian, Y.; Zhu, Y.; Cao, W. PROCWT: Coupling PROSPECT with Continuous Wavelet Transform to Improve the Retrieval of Foliar Chemistry from Leaf Bidirectional Reflectance Spectra. Remote Sens. Environ. 2018, 206, 1–14. [Google Scholar] [CrossRef]
- Ma, H.; Huang, W.; Dong, Y.; Liu, L.; Guo, A. Using UAV-Based Hyperspectral Imagery to Detect Winter Wheat Fusarium Head Blight. Remote Sens. 2021, 13, 3024. [Google Scholar] [CrossRef]
- Chen, X.; Li, F.; Chang, Q. Combination of Continuous Wavelet Transform and Successive Projection Algorithm for the Estimation of Winter Wheat Plant Nitrogen Concentration. Remote Sens. 2023, 15, 997. [Google Scholar] [CrossRef]




| Level | Training Set | Testing Set | Number of Samples |
|---|---|---|---|
| 1 | 75 | 25 | 100 |
| 2 | 75 | 25 | 100 |
| 3 | 75 | 25 | 100 |
| 4 | 75 | 25 | 100 |
| 5 | 75 | 25 | 100 |
| Entire sample set | 375 | 125 | 500 |
| Model | Dataset | Accuracy (%) | Macro Precision (%) | Macro Recall (%) | Macro F1-Score (%) | Time (s) |
|---|---|---|---|---|---|---|
| MSC-GA-SVM | Training set | 100 | 100 | 100 | 100 | 50.74 |
| Testing set | 53.6 | 56.28 | 53.6 | 51.46 | ||
| MSC-GS-SVM | Training set | 100 | 100 | 100 | 100 | 146.53 |
| Testing set | 66.4 | 68.12 | 66.4 | 64.67 | ||
| MSC-PSO-SVM | Training set | 100 | 100 | 100 | 100 | 79.72 |
| Testing set | 80 | 81.26 | 80 | 79.57 | ||
| MSC-GWO-SVM | Training set | 100 | 100 | 100 | 100 | 5.33 |
| Testing set | 64 | 66.2 | 64 | 63.48 |
| Model | Dataset | Accuracy (%) | Macro Precision (%) | Macro Recall (%) | Macro F1-Score (%) | Time (s) |
|---|---|---|---|---|---|---|
| MSC-mexh(21)-GA-SVM | Training set | 100 | 100 | 100 | 100 | 126.48 |
| Testing set | 81.6 | 84.14 | 82.4 | 82.18 | ||
| MSC- mexh(21)-GS-SVM | Training set | 100 | 100 | 100 | 100 | 319.02 |
| Testing set | 88.8 | 90.28 | 88.8 | 88.67 | ||
| MSC-mexh(21)-PSO-SVM | Training set | 100 | 100 | 100 | 100 | 178.6 |
| Testing set | 89.6 | 90.7 | 89.6 | 89.53 | ||
| MSC-mexh(21)-GWO-SVM | Training set | 100 | 100 | 100 | 100 | 30.39 |
| Testing set | 87.2 | 87.94 | 87.2 | 87.16 |
| Model | Dataset | Accuracy (%) | Macro Precision (%) | Macro Recall (%) | Macro F1-Score (%) | Time (s) |
|---|---|---|---|---|---|---|
| MSC-db3(23)-GA-SVM | Training set | 100 | 100 | 100 | 100 | 266 |
| Testing set | 89.6 | 91.26 | 90.4 | 90.42 | ||
| MSC-db3(23)-GS-SVM | Training set | 100 | 100 | 100 | 100 | 389.38 |
| Testing set | 88.8 | 91.5 | 90.4 | 90.33 | ||
| MSC-db3(23)-PSO-SVM | Training set | 99.73 | 99.74 | 99.74 | 99.74 | 135.3 |
| Testing set | 91.2 | 92.02 | 91.2 | 91.16 | ||
| MSC-db3(23)-GWO-SVM | Training set | 97.6 | 97.68 | 97.6 | 97.61 | 41.68 |
| Testing set | 91.2 | 92.02 | 91.2 | 91.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Zhang, X.; Shang, P.; Ma, R.; Yuan, X.; Li, L.; Bai, T. Detection of Cotton Verticillium Wilt Disease Severity Based on Hyperspectrum and GWO-SVM. Remote Sens. 2023, 15, 3373. https://doi.org/10.3390/rs15133373
Zhang N, Zhang X, Shang P, Ma R, Yuan X, Li L, Bai T. Detection of Cotton Verticillium Wilt Disease Severity Based on Hyperspectrum and GWO-SVM. Remote Sensing. 2023; 15(13):3373. https://doi.org/10.3390/rs15133373
Chicago/Turabian StyleZhang, Nannan, Xiao Zhang, Peng Shang, Rui Ma, Xintao Yuan, Li Li, and Tiecheng Bai. 2023. "Detection of Cotton Verticillium Wilt Disease Severity Based on Hyperspectrum and GWO-SVM" Remote Sensing 15, no. 13: 3373. https://doi.org/10.3390/rs15133373
APA StyleZhang, N., Zhang, X., Shang, P., Ma, R., Yuan, X., Li, L., & Bai, T. (2023). Detection of Cotton Verticillium Wilt Disease Severity Based on Hyperspectrum and GWO-SVM. Remote Sensing, 15(13), 3373. https://doi.org/10.3390/rs15133373