New Structural Complexity Metrics for Forests from Single Terrestrial Lidar Scans
Abstract
1. Introduction
1.1. Importance of Structure
1.2. Lidar for Forest Structure
1.3. Objectives
2. Materials and Methods
2.1. Site Selection
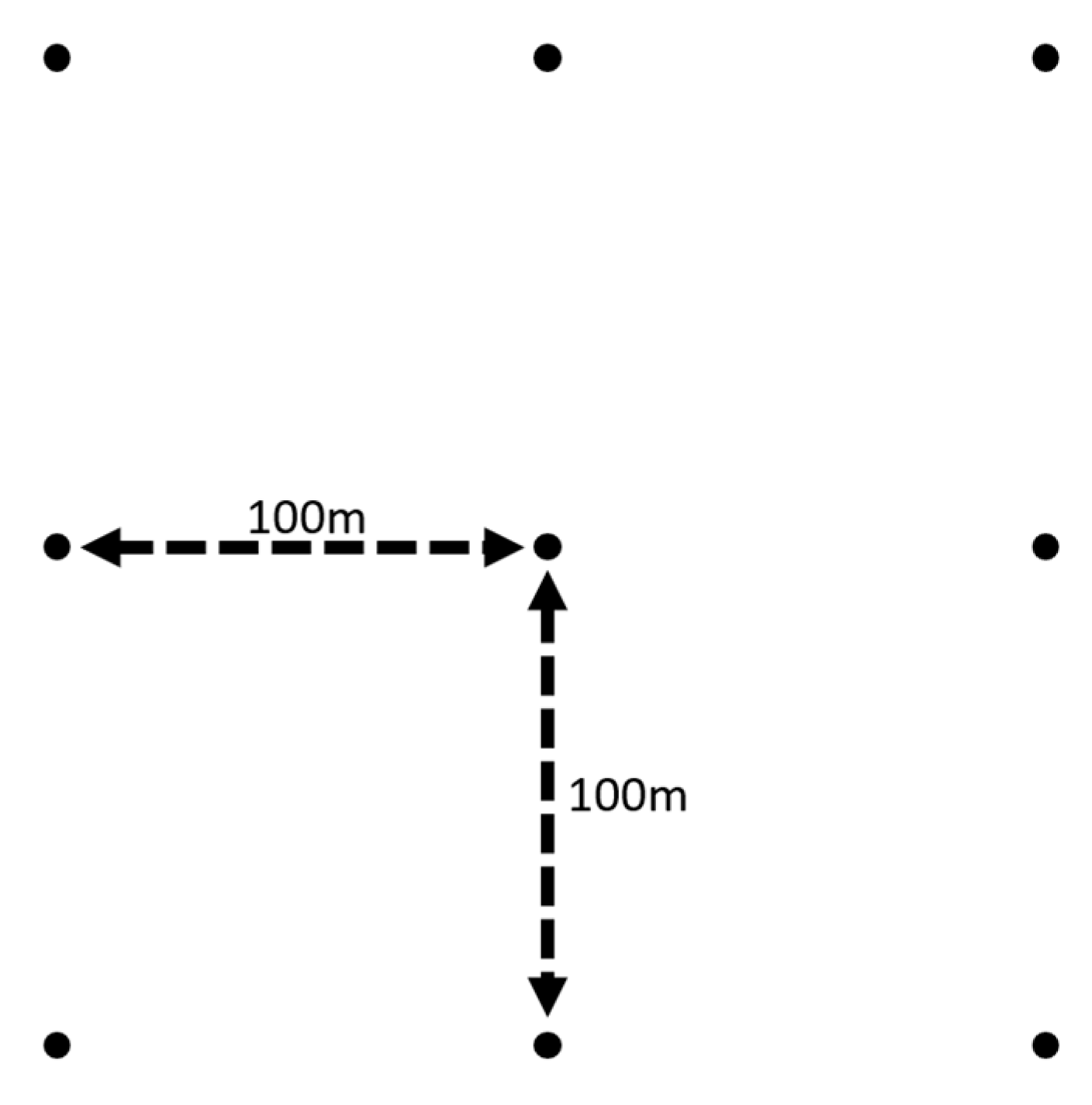
2.2. Scan Acquisition
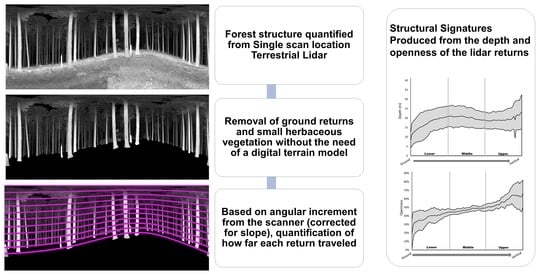
2.3. Scan Processing
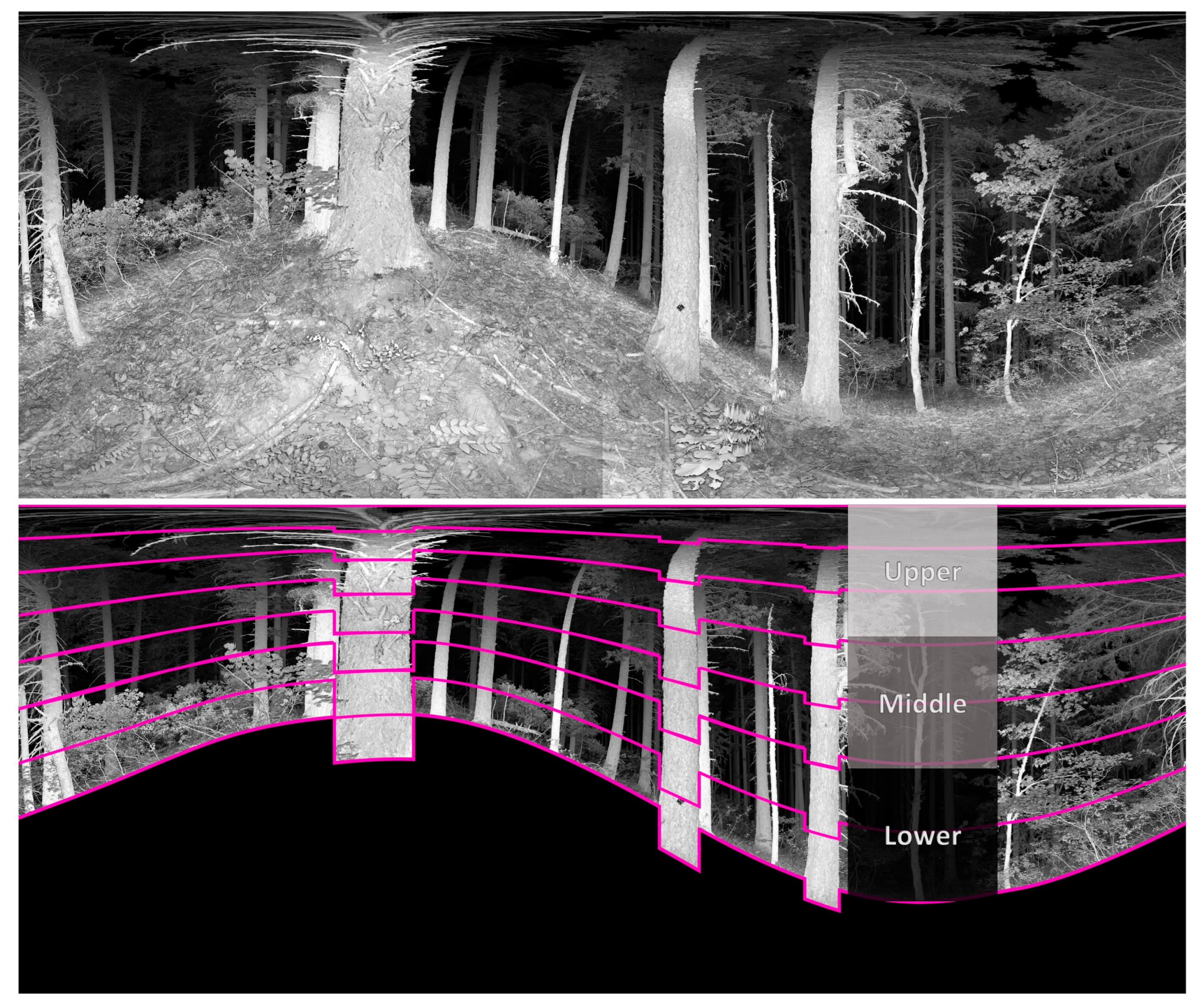
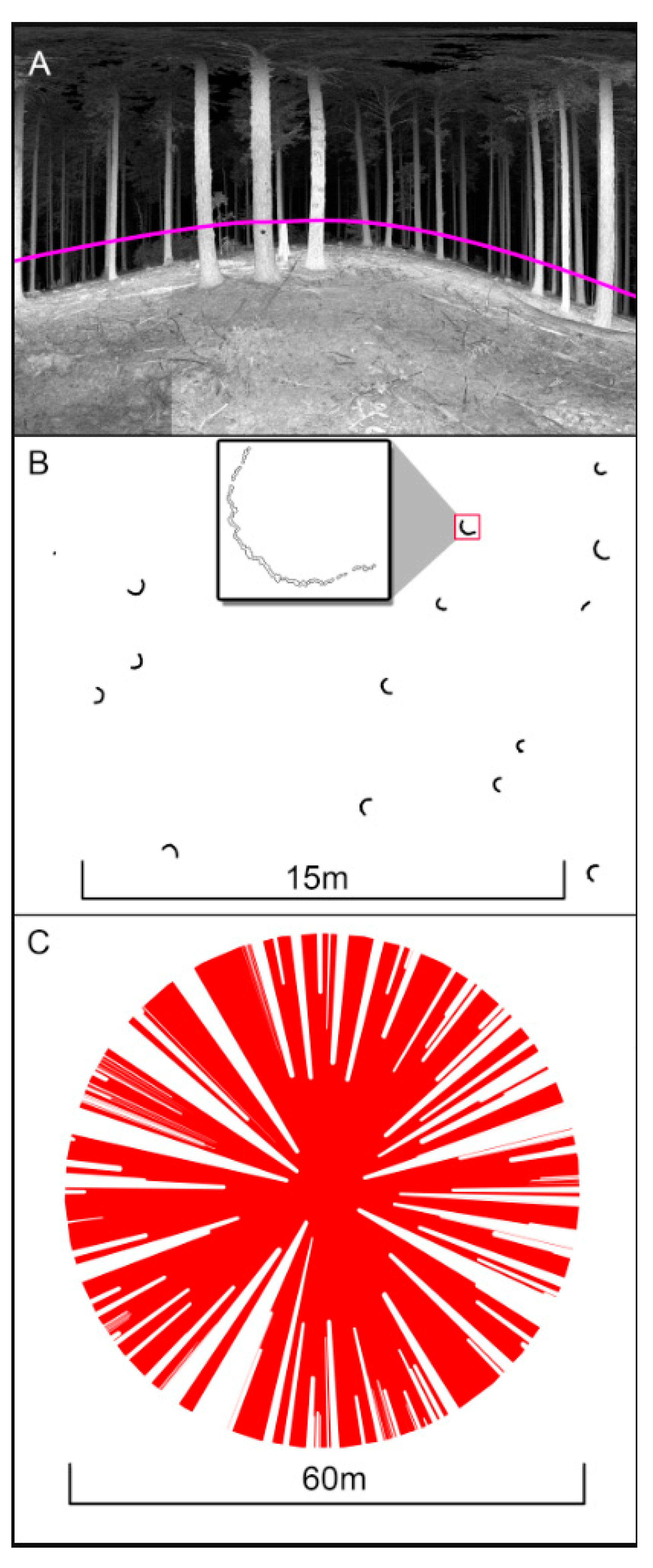
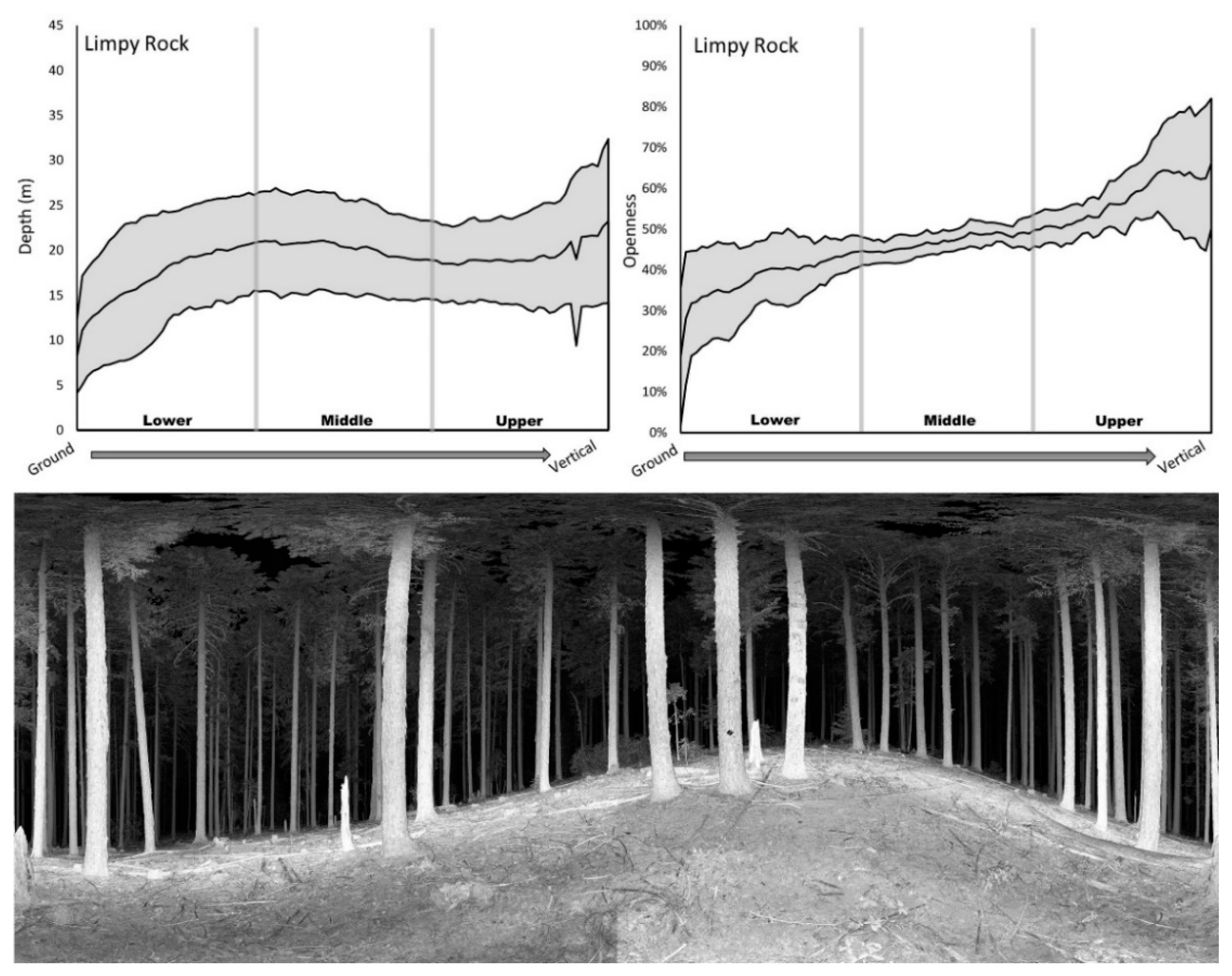
2.4. Depth and Openness Metric Calculation
2.5. Isovists
2.6. Comparisons
2.6.1. Depth and Openness Statistical Tests
2.6.2. Isovist Statistical Tests
3. Results
3.1. Structural Signatures
3.2. Depth and Openness Statistical Tests
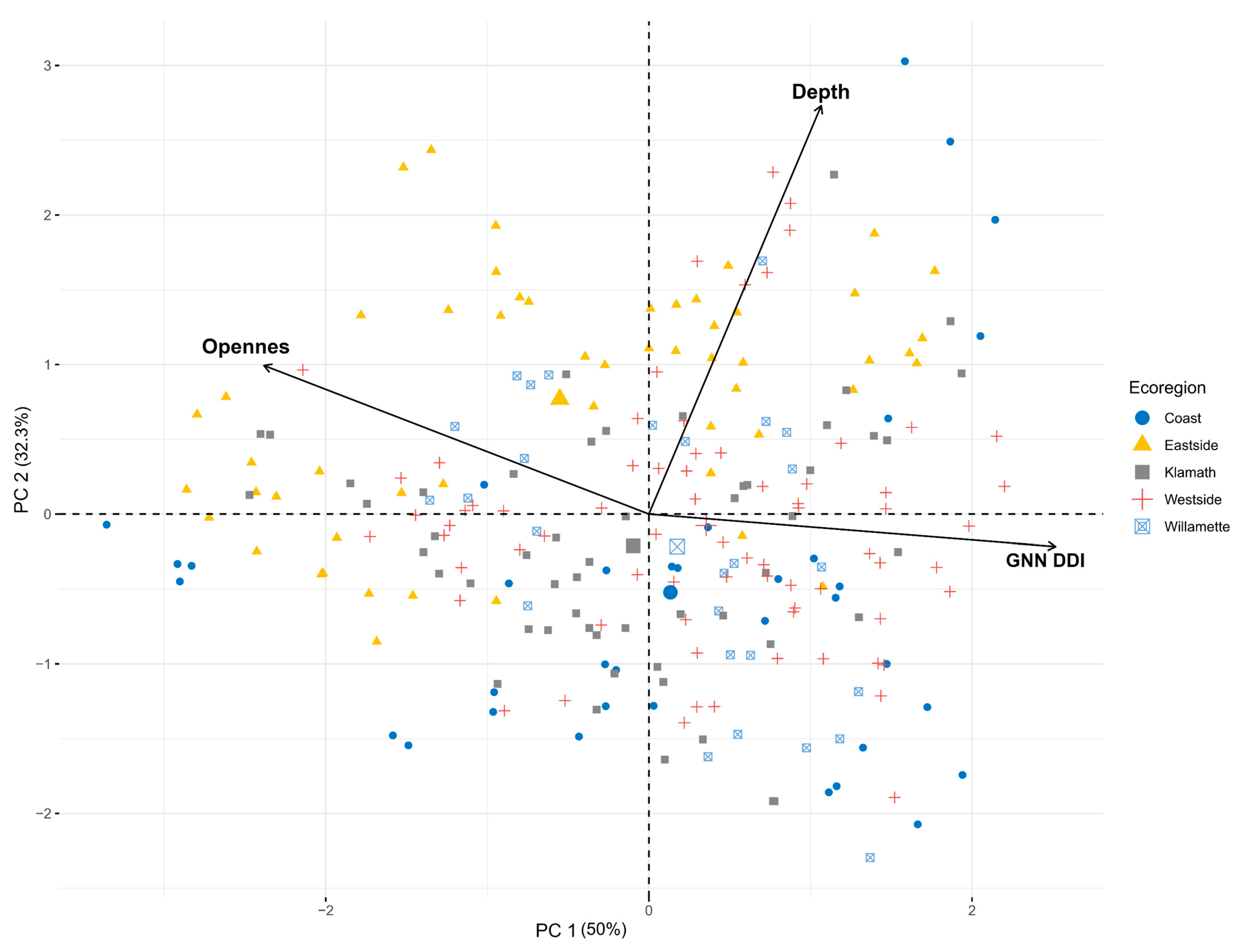
3.2.1. Ordination
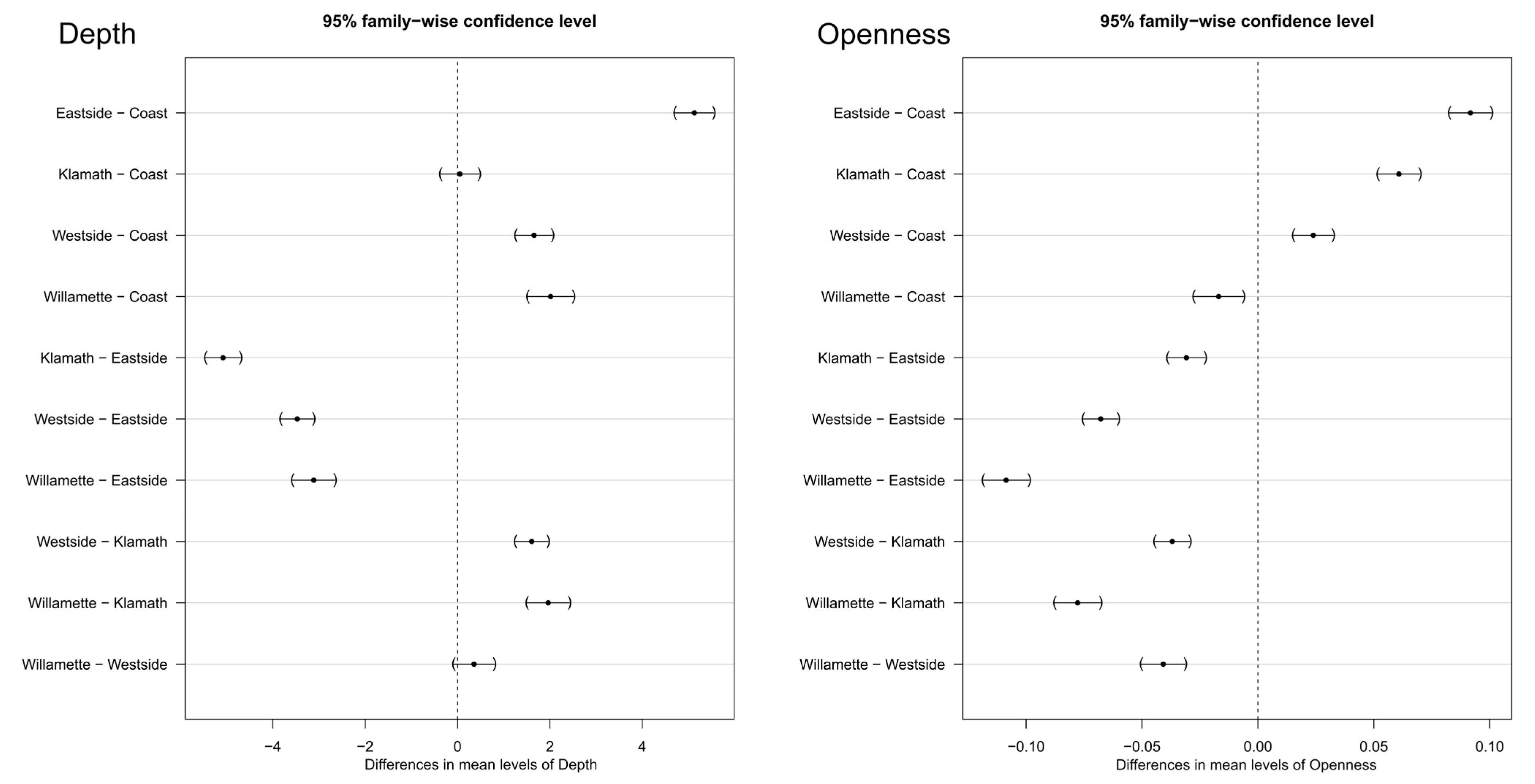
3.2.2. Depth and Openness ANOVA
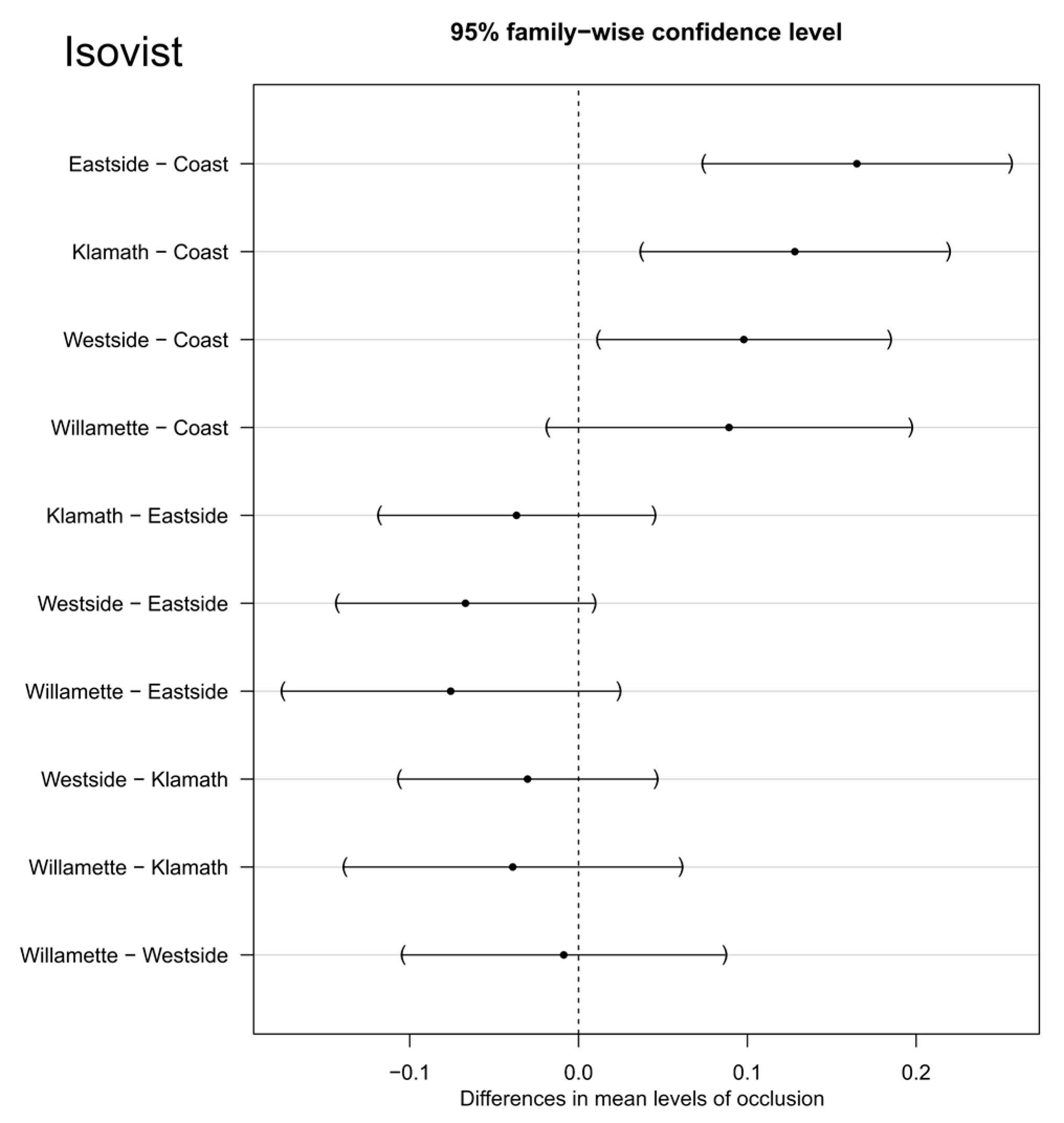
3.3. Isovist Statistical Tests
4. Discussion
4.1. Depth and Openness
4.2. Isovists
4.3. Potential Applications
4.4. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindenmayer, D.B.; Franklin, J.F. Conserving Forest Biodiversity: A Comprehensive Multiscaled Approach; Island Press: Washington, DC, USA, 2002. [Google Scholar]
- Carey, A.B. AIMing for Healthy Forests: Active, Intentional Management for Multiple Values; General Technical Report PNW-GTR-721; U.S. Department of Agriculture, Forest Service, Pacific Northwest Research Station: Portland, OR, USA, 2007.
- Shugart, H.; Saatchi, S.; Hall, F. Importance of Structure and Its Measurement in Quantifying Function of Forest Ecosystems. J. Geophys. Res. Biogeosci. 2010, 115. [Google Scholar] [CrossRef]
- Chazdon, R.L. Sunflecks and Their Importance to Forest Understorey Plants. Adv. Ecol. Res. 1988, 18, 1–63. [Google Scholar]
- Beier, P.; Drennan, J.E. Forest Structure and Prey Abundance in Foraging Areas of Northern Goshawks. Ecol. Appl. 1997, 7, 564–571. [Google Scholar] [CrossRef]
- Pardini, R.; de Souza, S.M.; Braga-Neto, R.; Metzger, J.P. The Role of Forest Structure, Fragment Size and Corridors in Maintaining Small Mammal Abundance and Diversity in an Atlantic Forest Landscape. Biol. Conserv. 2005, 124, 253–266. [Google Scholar] [CrossRef]
- Musselman, K.N.; Molotch, N.P.; Margulis, S.A.; Kirchner, P.B.; Bales, R.C. Influence of Canopy Structure and Direct Beam Solar Irradiance on Snowmelt Rates in a Mixed Conifer Forest. Agric. For. Meteorol. 2012, 161, 46–56. [Google Scholar] [CrossRef]
- Davis, C.R.; Belote, T.; Williamson, M.; Larson, A.; Esch, B. A Rapid Forest Assessment Method for Multiparty Monitoring across Landscapes. J. For. 2016, 114, 125–133. [Google Scholar] [CrossRef]
- Everett, R.L.; Leader, A.T. Eastside Forest Ecosystem Health Assessment; General Technical Report PNW-GTR-330; U.S. Department of Agriculture, Forest Service, Pacific Northwest Research Station: Portland, OR, USA, 1994.
- Massie, M. Assessment of the Vulnerability of Oregon and Washington’s Natural Areas to Climate Change. Master’s Thesis, Oregon State University, Portland, OR, USA, 2014. [Google Scholar]
- Rapp, V. Northwest Forest Plan—The First 10 Years (1994–2003): First-Decade Results of the Northwest Forest Plan; General Technical Report PNW-GTR-720; U.S. Department of Agriculture, Forest Service, Pacific Northwest Research Station: Portland, OR, USA, 2008.
- Acker, S.A.; Sabin, T.E.; Ganio, L.M.; McKee, W.A. Development of Old-Growth Structure and Timber Volume Growth Trends in Maturing Douglas-Fir Stands. For. Ecol. Manag. 1998, 104, 265–280. [Google Scholar] [CrossRef]
- McComb, W.C.; McGrath, M.T.; Spies, T.A.; Vesely, D. Models for Mapping Potential Habitat at Landscape Scales: An Example Using Northern Spotted Owls. For. Sci. 2002, 48, 203–216. [Google Scholar]
- McElhinny, C.; Gibbons, P.; Brack, C.; Bauhus, J. Forest and Woodland Stand Structural Complexity: Its Definition and Measurement. For. Ecol. Manag. 2005, 218, 1–24. [Google Scholar] [CrossRef]
- Staudhammer, C.L.; LeMay, V.M. Introduction and Evaluation of Possible Indices of Stand Structural Diversity. Can. J. For. Res. 2001, 31, 1105–1115. [Google Scholar] [CrossRef]
- Tuchmann, E.T.; Connaughton, K.P. The Northwest Forest Plan: A Report to the President and Congress; DIANE Publishing: Collingdale, PA, USA, 1998. [Google Scholar]
- Pommerening, A. Approaches to Quantifying Forest Structures. Forestry 2002, 75, 305–324. [Google Scholar] [CrossRef]
- Bruggisser, M.; Hollaus, M.; Kükenbrink, D.; Pfeifer, N. Comparison of Forest Structure Metrics Derived from UAV Lidar and ALS Data. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 4, 325–332. [Google Scholar] [CrossRef]
- Moeser, D.; Morsdorf, F.; Jonas, T. Novel Forest Structure Metrics from Airborne LiDAR Data for Improved Snow Interception Estimation. Agric. For. Meteorol. 2015, 208, 40–49. [Google Scholar] [CrossRef]
- Uuemaa, E.; Antrop, M.; Roosaare, J.; Marja, R.; Mander, Ü. Landscape Metrics and Indices: An Overview of Their Use in Landscape Research. Living Rev. Landsc. Res. 2009, 3, 1–28. [Google Scholar] [CrossRef]
- Frey, J.; Joa, B.; Schraml, U.; Koch, B. Same Viewpoint Different Perspectives—A Comparison of Expert Ratings with a TLS Derived Forest Stand Structural Complexity Index. Remote Sens. 2019, 11, 1137. [Google Scholar] [CrossRef]
- Vales, D.J.; Bunnell, F.L. Comparison of Methods for Estimating Forest Overstory Cover. I. Observer Effects. Can. J. For. Res. 1988, 18, 606–609. [Google Scholar]
- Beland, M.; Parker, G.; Sparrow, B.; Harding, D.; Chasmer, L.; Phinn, S.; Antonarakis, A.; Strahler, A. On Promoting the Use of Lidar Systems in Forest Ecosystem Research. For. Ecol. Manag. 2019, 450, 117484. [Google Scholar] [CrossRef]
- Camarretta, N.; Harrison, P.A.; Bailey, T.; Potts, B.; Lucieer, A.; Davidson, N.; Hunt, M. Monitoring Forest Structure to Guide Adaptive Management of Forest Restoration: A Review of Remote Sensing Approaches. New For. 2020, 51, 573–596. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Næsset, E.; Ørka, H.O.; Coops, N.C.; Hilker, T.; Bater, C.W.; Gobakken, T. Lidar Sampling for Large-Area Forest Characterization: A Review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef]
- Calders, K.; Lewis, P.; Disney, M.; Verbesselt, J.; Herold, M. Investigating Assumptions of Crown Archetypes for Modelling LiDAR Returns. Remote Sens. Environ. 2013, 134, 39–49. [Google Scholar] [CrossRef]
- Zimble, D.A.; Evans, D.L.; Carlson, G.C.; Parker, R.C.; Grado, S.C.; Gerard, P.D. Characterizing Vertical Forest Structure Using Small-Footprint Airborne LiDAR. Remote Sens. Environ. 2003, 87, 171–182. [Google Scholar] [CrossRef]
- Ackers, S.H.; Davis, R.J.; Olsen, K.A.; Dugger, K.M. The Evolution of Mapping Habitat for Northern Spotted Owls (Strix Occidentalis Caurina): A Comparison of Photo-Interpreted, Landsat-Based, and Lidar-Based Habitat Maps. Remote Sens. Environ. 2015, 156, 361–373. [Google Scholar] [CrossRef]
- Coops, N.C.; Tompalski, P.; Goodbody, T.R.; Queinnec, M.; Luther, J.E.; Bolton, D.K.; White, J.C.; Wulder, M.A.; van Lier, O.R.; Hermosilla, T. Modelling Lidar-Derived Estimates of Forest Attributes over Space and Time: A Review of Approaches and Future Trends. Remote Sens. Environ. 2021, 260, 112477. [Google Scholar] [CrossRef]
- Lister, A.J.; Andersen, H.; Frescino, T.; Gatziolis, D.; Healey, S.; Heath, L.S.; Liknes, G.C.; McRoberts, R.; Moisen, G.G.; Nelson, M. Use of Remote Sensing Data to Improve the Efficiency of National Forest Inventories: A Case Study from the United States National Forest Inventory. Forests 2020, 11, 1364. [Google Scholar] [CrossRef]
- Tinkham, W.T.; Mahoney, P.R.; Hudak, A.T.; Domke, G.M.; Falkowski, M.J.; Woodall, C.W.; Smith, A.M. Applications of the United States Forest Inventory and Analysis Dataset: A Review and Future Directions. Can. J. For. Res. 2018, 48, 1251–1268. [Google Scholar] [CrossRef]
- Campbell, M.J.; Dennison, P.E.; Hudak, A.T.; Parham, L.M.; Butler, B.W. Quantifying Understory Vegetation Density Using Small-Footprint Airborne Lidar. Remote Sens. Environ. 2018, 215, 330–342. [Google Scholar] [CrossRef]
- Hilker, T.; van Leeuwen, M.; Coops, N.C.; Wulder, M.A.; Newnham, G.J.; Jupp, D.L.B.; Culvenor, D.S. Comparing Canopy Metrics Derived from Terrestrial and Airborne Laser Scanning in a Douglas-Fir Dominated Forest Stand. Trees 2010, 24, 819–832. [Google Scholar] [CrossRef]
- Ruiz, L.Á.; Crespo-Peremarch, P.; Torralba, J. Modelling Canopy Fuel Properties and Understory Vegetation with Full-Waveform LiDAR. In Proceedings of the International Conference on Smart Geoinformatics Applications (ICSGA 2021), Phuket, Thailand, 24–25 February 2021; pp. 29–32. [Google Scholar]
- Calders, K.; Adams, J.; Armston, J.; Bartholomeus, H.; Bauwens, S.; Bentley, L.P.; Chave, J.; Danson, F.M.; Demol, M.; Disney, M.; et al. Terrestrial Laser Scanning in Forest Ecology: Expanding the Horizon. Remote Sens. Environ. 2020, 251, 112102. [Google Scholar] [CrossRef]
- Henning, J.G.; Radtke, P.J. Detailed Stem Measurements of Standing Trees from Ground-Based Scanning Lidar. For. Sci. 2006, 52, 67–80. [Google Scholar]
- Palace, M.; Sullivan, F.B.; Ducey, M.; Herrick, C. Estimating Tropical Forest Structure Using a Terrestrial Lidar. PLoS ONE 2016, 11, e0154115. [Google Scholar] [CrossRef]
- Loudermilk, E.L.; Hiers, J.K.; O’Brien, J.J.; Mitchell, R.J.; Singhania, A.; Fernandez, J.C.; Cropper, W.P.; Slatton, K.C. Ground-Based LIDAR: A Novel Approach to Quantify Fine-Scale Fuelbed Characteristics. Int. J. Wildland Fire 2009, 18, 676–685. [Google Scholar] [CrossRef]
- Rowell, E.; Loudermilk, E.L.; Hawley, C.; Pokswinski, S.; Seielstad, C.; Queen, L.; O’Brien, J.J.; Hudak, A.T.; Goodrick, S.; Hiers, J.K. Coupling Terrestrial Laser Scanning with 3D Fuel Biomass Sampling for Advancing Wildland Fuels Characterization. For. Ecol. Manag. 2020, 462, 117945. [Google Scholar] [CrossRef]
- Wilson, N.; Bradstock, R.; Bedward, M. Detecting the Effects of Logging and Wildfire on Forest Fuel Structure Using Terrestrial Laser Scanning (TLS). For. Ecol. Manag. 2021, 488, 119037. [Google Scholar] [CrossRef]
- Ashcroft, M.B.; Gollan, J.R.; Ramp, D. Creating Vegetation Density Profiles for a Diverse Range of Ecological Habitats Using Terrestrial Laser Scanning. Methods Ecol. Evol. 2014, 5, 263–272. [Google Scholar] [CrossRef]
- Kazakova, A.N. Quantifying Vertical and Horizontal Stand Structure Using Terrestrial LiDAR in Pacific Northwest Forests. Master’s Thesis, University of Washington, Seattle, WA, USA, 2014. [Google Scholar]
- Olsoy, P.J.; Forbey, J.S.; Rachlow, J.L.; Nobler, J.D.; Glenn, N.F.; Shipley, L.A. Fearscapes: Mapping Functional Properties of Cover for Prey with Terrestrial LiDAR. BioScience 2015, 65, 74–80. [Google Scholar] [CrossRef][Green Version]
- Shokirov, S.; Levick, S.R.; Jucker, T.; Yeoh, P.; Youngentob, K. Comparison of TLS and ULS Data for Wildlife Habitat Assessments in Temperate Woodlands. In Proceedings of the IGARSS 2020—2020 IEEE International Geoscience and Remote Sensing Symposium, Online, 26 September–2 October 2020; pp. 6097–6100. [Google Scholar]
- Soma, M.; Pimont, F.; Allard, D.; Fournier, R.; Dupuy, J.-L. Mitigating Occlusion Effects in Leaf Area Density Estimates from Terrestrial LiDAR through a Specific Kriging Method. Remote Sens. Environ. 2020, 245, 111836. [Google Scholar] [CrossRef]
- Wan, P.; Wang, T.; Zhang, W.; Liang, X.; Skidmore, A.K.; Yan, G. Quantification of Occlusions Influencing the Tree Stem Curve Retrieving from Single-Scan Terrestrial Laser Scanning Data. For. Ecosyst. 2019, 6, 43. [Google Scholar] [CrossRef]
- Litkey, P.; Liang, X.; Kaartinen, H.; Hyyppä, J.; Kukko, A.; Holopainen, M.; Hill, R.; Rosette, J.; Suárez, J. Single-Scan TLS Methods for Forest Parameter Retrieval. In Proceedings of the SilviLaser, Edinburgh, UK, 17–19 September 2008. [Google Scholar]
- Xia, S.; Wang, C.; Pan, F.; Xi, X.; Zeng, H.; Liu, H. Detecting Stems in Dense and Homogeneous Forest Using Single-Scan TLS. Forests 2015, 6, 3923–3945. [Google Scholar] [CrossRef]
- Pokswinski, S.; Gallagher, M.R.; Skowronski, N.S.; Loudermilk, E.L.; Hawley, C.; Wallace, D.; Everland, A.; Wallace, J.; Hiers, J.K. A Simplified and Affordable Approach to Forest Monitoring Using Single Terrestrial Laser Scans and Transect Sampling. MethodsX 2021, 8, 101484. [Google Scholar] [CrossRef]
- Moskal, L.M.; Zheng, G. Retrieving Forest Inventory Variables with Terrestrial Laser Scanning (TLS) in Urban Heterogeneous Forest. Remote Sens. 2011, 4, 1–20. [Google Scholar] [CrossRef]
- Richardson, J.J.; Moskal, L.M.; Bakker, J.D. Terrestrial Laser Scanning for Vegetation Sampling. Sensors 2014, 14, 20304–20319. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Moskal, L.M.; Batchelor, J.L.; Thau, D.; Hudak, A.T. Relationships between Satellite-Based Spectral Burned Ratios and Terrestrial Laser Scanning. Forests 2019, 10, 444. [Google Scholar] [CrossRef]
- Gallagher, M.R.; Maxwell, A.E.; Guillén, L.A.; Everland, A.; Loudermilk, E.L.; Skowronski, N.S. Estimation of Plot-Level Burn Severity Using Terrestrial Laser Scanning. Remote Sens. 2021, 13, 4168. [Google Scholar] [CrossRef]
- Tan, K.; Ke, T.; Tao, P.; Liu, K.; Duan, Y.; Zhang, W.; Wu, S. Discriminating Forest Leaf and Wood Components in TLS Point Clouds at Single-Scan Level Using Derived Geometric Quantities. IEEE Trans. Geosci. Remote Sens. 2021, 60, 1–17. [Google Scholar] [CrossRef]
- Anderson, C.T.; Dietz, S.L.; Pokswinski, S.M.; Jenkins, A.M.; Kaeser, M.J.; Hiers, J.K.; Pelc, B.D. Traditional Field Metrics and Terrestrial LiDAR Predict Plant Richness in Southern Pine Forests. For. Ecol. Manag. 2021, 491, 119118. [Google Scholar] [CrossRef]
- Wallace, L.; Hillman, S.; Hally, B.; Taneja, R.; White, A.; McGlade, J. Terrestrial Laser Scanning: An Operational Tool for Fuel Hazard Mapping? Fire 2022, 5, 85. [Google Scholar] [CrossRef]
- Murgoitio, J.; Shrestha, R.; Glenn, N.; Spaete, L. Airborne LiDAR and Terrestrial Laser Scanning Derived Vegetation Obstruction Factors for Visibility Models. Trans. GIS 2014, 18, 147–160. [Google Scholar] [CrossRef]
- Omernik, J.M. Ecoregions of the Conterminous United States. Ann. Assoc. Am. Geogr. 1987, 77, 118–125. [Google Scholar] [CrossRef]
- Wilson, T.M. Pacific Northwest Interagency Natural Areas Network. Available online: http://www.fsl.orst.edu/rna/index.html (accessed on 28 March 2015).
- LEMMA Landscape Ecology, Modeling, Mapping & Analysis Home Page. Available online: http://lemma.forestry.oregonstate.edu/ (accessed on 30 March 2015).
- ESRI. ArcGIS Desktop; Version 10; ESRI: Redlands, CA, USA, 2014. [Google Scholar]
- FARO Scene [Computer Software]; Version 5.3; FARO: Lake Mary, FL, USA. 2014. Available online: http://www.faro.com (accessed on 31 March 2015).
- Cao, N.; Zhu, C.; Kai, Y.; Yan, P. A Method of Background Noise Reduction in Lidar Data. Appl. Phys. B 2013, 113, 115–123. [Google Scholar] [CrossRef]
- Stovall, A.E.L.; Atkins, J.W. Assessing Low-Cost Terrestrial Laser Scanners for Deriving Forest Structure Parameters. Preprints 2021, 2021070690. [Google Scholar] [CrossRef]
- Olsen, M.J.; Ponto, K.; Kimball, J.; Seracini, M.; Kuester, F. 2D Open-Source Editing Techniques for 3D Laser Scans. In Proceedings of the 38th Annual Conference on Computer Applications and Quantitative Methods in Archaeology, Granada, Spain, 6–9 April 2010; pp. 455–461. [Google Scholar]
- Mathworks MATLAB R2015a [Computer Program]. 2015. Available online: HTTP://www.mathworks.Com/products/matlab/ (accessed on 30 March 2015).
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Tukey’s Honestly Significant Difference (HSD) Test. Encycl. Res. Des. 2010, 3, 1–5. [Google Scholar]
- Aben, J.; Pellikka, P.; Travis, J.M. A Call for Viewshed Ecology: Advancing Our Understanding of the Ecology of Information through Viewshed Analysis. Methods Ecol. Evol. 2018, 9, 624–633. [Google Scholar] [CrossRef]
- Davies, A.B.; Tambling, C.J.; Kerley, G.I.; Asner, G.P. Effects of Vegetation Structure on the Location of Lion Kill Sites in African Thicket. PLoS ONE 2016, 11, e0149098. [Google Scholar] [CrossRef] [PubMed]
- Mowat, G.; Slough, B. Habitat Preference of Canada Lynx through a Cycle in Snowshoe Hare Abundance. Can. J. Zool. 2003, 81, 1736–1745. [Google Scholar] [CrossRef]
- Poole, K.G. A Review of the Canada Lynx, Lynx Canadensis, in Canada. Can. Field-Nat. 2003, 117, 360–376. [Google Scholar] [CrossRef]
- Fekety, P.A.; Sadak, R.B.; Sauder, J.D.; Hudak, A.T.; Falkowski, M.J. Predicting Forest Understory Habitat for Canada Lynx Using LIDAR Data. Wildl. Soc. Bull. 2019, 43, 619–629. [Google Scholar] [CrossRef]
- Galluzzi, M.; Puletti, N.; Armanini, M.; Chirichella, R.; Mustoni, A. Mobile Laser Scanner Understory Characterization: An Exploratory Study on Hazel Grouse in Italian Alps. bioRxiv 2022. [Google Scholar] [CrossRef]
- Burgett, S.; Rachlow, J.; Stein, R. Unexpected Properties of Habitat Altered by Ecosystem Engineers: A Pygmy Rabbit Case Study. 2021. Available online: https://scholarworks.boisestate.edu/icur/2021/poster_session/12/ (accessed on 16 December 2022).
- Lecigne, B.; Eitel, J.; Rachlow, J. Viewshed3d: An R Package for Quantifying 3D Visibility Using Terrestrial Lidar Data. Methods Ecol. Evol. 2020, 11, 733–738. [Google Scholar] [CrossRef]
- Fan, G.; Nan, L.; Dong, Y.; Su, X.; Chen, F. AdQSM: A New Method for Estimating Above-Ground Biomass from TLS Point Clouds. Remote Sens. 2020, 12, 3089. [Google Scholar] [CrossRef]
- Hackenberg, J.; Wassenberg, M.; Spiecker, H.; Sun, D. Non Destructive Method for Biomass Prediction Combining TLS Derived Tree Volume and Wood Density. Forests 2015, 6, 1274–1300. [Google Scholar] [CrossRef]
- Holopainen, M.; Vastaranta, M.; Kankare, V.; Räty, M.; Vaaja, M.; Liang, X.; Yu, X.; Hyyppä, J.; Hyyppä, H.; Viitala, R. Biomass Estimation of Individual Trees Using Stem and Crown Diameter TLS Measurements. ISPRS—Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2011, 3812, 91–95. [Google Scholar] [CrossRef]
- Coomes, D.A.; Dalponte, M.; Jucker, T.; Asner, G.P.; Banin, L.F.; Burslem, D.F.R.P.; Lewis, S.L.; Nilus, R.; Phillips, O.L.; Phua, M.-H.; et al. Area-Based vs Tree-Centric Approaches to Mapping Forest Carbon in Southeast Asian Forests from Airborne Laser Scanning Data. Remote Sens. Environ. 2017, 194, 77–88. [Google Scholar] [CrossRef]
- Duncanson, L.; Neuenschwander, A.; Hancock, S.; Thomas, N.; Fatoyinbo, T.; Simard, M.; Silva, C.A.; Armston, J.; Luthcke, S.B.; Hofton, M.; et al. Biomass Estimation from Simulated GEDI, ICESat-2 and NISAR across Environmental Gradients in Sonoma County, California. Remote Sens. Environ. 2020, 242, 111779. [Google Scholar] [CrossRef]
- Ene, L.T.; Gobakken, T.; Andersen, H.-E.; Næsset, E.; Cook, B.D.; Morton, D.C.; Babcock, C.; Nelson, R. Large-Area Hybrid Estimation of Aboveground Biomass in Interior Alaska Using Airborne Laser Scanning Data. Remote Sens. Environ. 2018, 204, 741–755. [Google Scholar] [CrossRef]
- Mahoney, M.J.; Johnson, L.K.; Bevilacqua, E.; Beier, C.M. Filtering Ground Noise from LiDAR Returns Produces Inferior Models of Forest Aboveground Biomass in Heterogenous Landscapes. GISci. Remote Sens. 2022, 59, 1266–1280. [Google Scholar] [CrossRef]
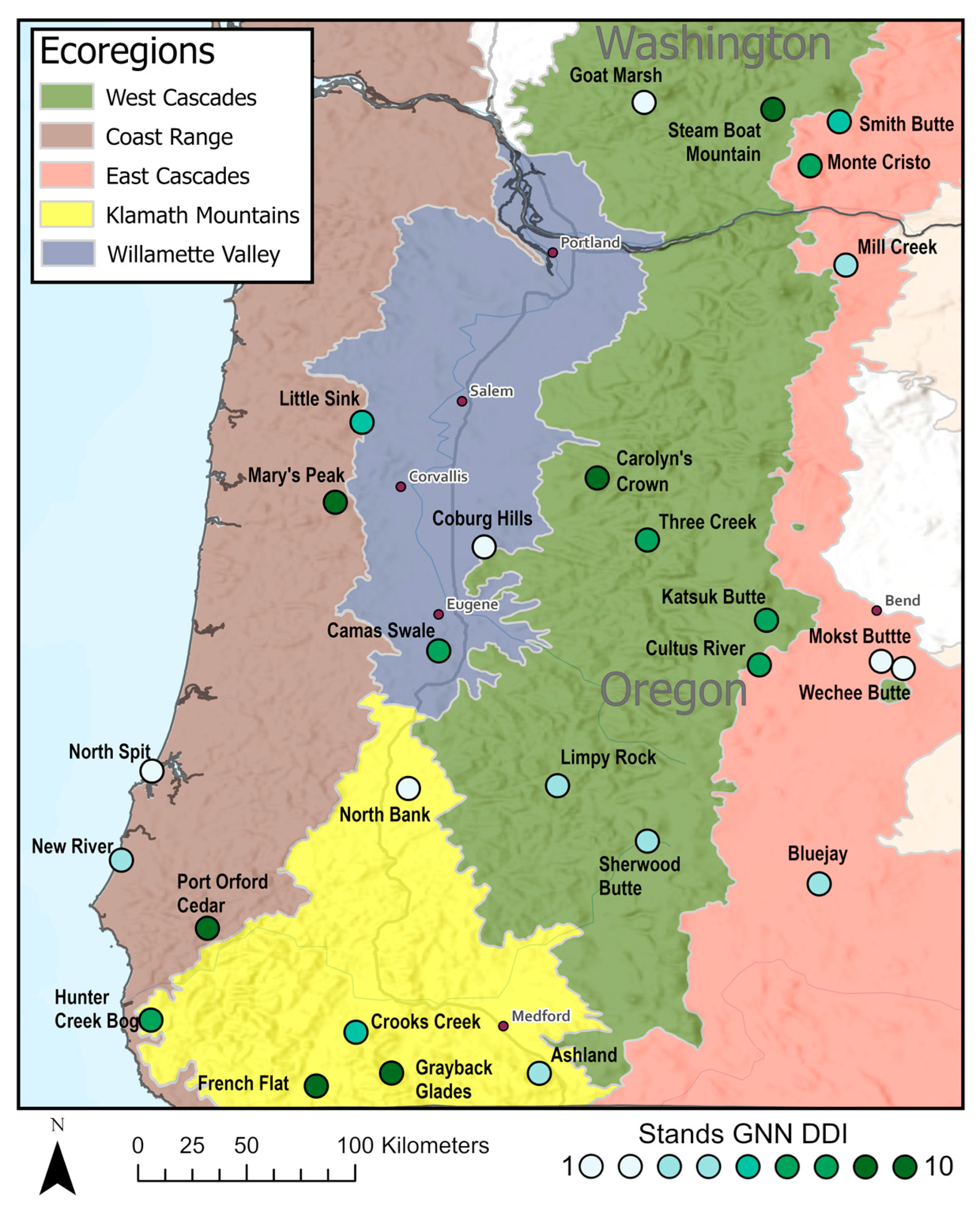




| Ecoregion | Name | GNN DDI |
|---|---|---|
| Coast Range | North Spit | 1–2 |
| New River | 3–4 | |
| Mary’s Peak | 8–9 | |
| Port Orford Cedar | 8–9 | |
| East Cascades | Wechee Butte | 1–2 |
| Mokst Butte | 2–3 | |
| Bluejay | 3–4 | |
| Mill Creek | 4–5 | |
| Smith Butte | 5–6 | |
| Monte Cristo | 7–8 | |
| Klamath Mountains | North Bank | 2–3 |
| Ashland | 4–5 | |
| Crooks Creek | 5–6 | |
| Hunter Creek Bog | 7–8 | |
| Grayback Glades | 8–9 | |
| French Flat | 9–10 | |
| West Cascades | Goat Marsh | 2–3 |
| Sherwood Butte | 3–4 | |
| Limpy Rock | 4–5 | |
| Cultus River | 6–7 | |
| Katsuk Butte | 6–7 | |
| Three Creek | 7–8 | |
| Carolyn’s Crown | 9–10 | |
| Steamboat Mountain | 9–10 | |
| Willamette Valley | Coburg Hills | 1–2 |
| Little Sink | 5–6 | |
| Camas Swale | 6–7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batchelor, J.L.; Wilson, T.M.; Olsen, M.J.; Ripple, W.J. New Structural Complexity Metrics for Forests from Single Terrestrial Lidar Scans. Remote Sens. 2023, 15, 145. https://doi.org/10.3390/rs15010145
Batchelor JL, Wilson TM, Olsen MJ, Ripple WJ. New Structural Complexity Metrics for Forests from Single Terrestrial Lidar Scans. Remote Sensing. 2023; 15(1):145. https://doi.org/10.3390/rs15010145
Chicago/Turabian StyleBatchelor, Jonathan L., Todd M. Wilson, Michael J. Olsen, and William J. Ripple. 2023. "New Structural Complexity Metrics for Forests from Single Terrestrial Lidar Scans" Remote Sensing 15, no. 1: 145. https://doi.org/10.3390/rs15010145
APA StyleBatchelor, J. L., Wilson, T. M., Olsen, M. J., & Ripple, W. J. (2023). New Structural Complexity Metrics for Forests from Single Terrestrial Lidar Scans. Remote Sensing, 15(1), 145. https://doi.org/10.3390/rs15010145