Abstract
The accurate and automated diagnosis of potato late blight disease, one of the most destructive potato diseases, is critical for precision agricultural control and management. Recent advances in remote sensing and deep learning offer the opportunity to address this challenge. This study proposes a novel end-to-end deep learning model (CropdocNet) for accurate and automated late blight disease diagnosis from UAV-based hyperspectral imagery. The proposed method considers the potential disease-specific reflectance radiation variance caused by the canopy’s structural diversity and introduces multiple capsule layers to model the part-to-whole relationship between spectral–spatial features and the target classes to represent the rotation invariance of the target classes in the feature space. We evaluate the proposed method with real UAV-based HSI data under controlled and natural field conditions. The effectiveness of the hierarchical features is quantitatively assessed and compared with the existing representative machine learning/deep learning methods on both testing and independent datasets. The experimental results show that the proposed model significantly improves accuracy when considering the hierarchical structure of spectral–spatial features, with average accuracies of 98.09% for the testing dataset and 95.75% for the independent dataset, respectively.
1. Introduction
Potato late blight disease, caused by Phytophthora infestans (Mont.) de Bary, is one of the most destructive potato diseases, resulting in significant potato yield loss across the major potato growing areas worldwide [1,2]. The yield loss due to the infestation of late blight disease is around to [3,4]. The current control measure mainly relies on the application of fungicides [5], which is expensive and has negative impacts on the environment and human health due to excessive use of pesticides. Therefore, the early, accurate detection of potato late blight disease is vital for effective disease control and management with minimal application of fungicides.
Since late blight disease affects potato leaves, stems and tubers with visible symptoms (e.g., black lesions with granular regions and green halo) [6,7], the current detection of late blight disease in practice is mainly based on visual observation [8,9]. However, this manual inspection method is time consuming and costly and often causes a delay in late blight disease management, especially at an early stage, across large fields [10]. In addition, field surveyors diagnose diseases based on their domain knowledge, which may introduce inconsistency and bias due to individual subjectivity [11]. An automated approach for fast and reliable potato late blight disease diagnosis is important to ensure effective disease management and control.
With the advancements in low-cost sensor technology, computer vision and remote sensing, machine vision technology based on images (such as red, green and blue (RGB) images, thermal images, multispectral and hyperspectral images) has been successfully used in agricultural and engineering fields [12,13,14,15,16,17,18,19,20,21]. For example, Wu et al. [20] developed a deep learning-based model to detect the edge images of flower buds and inflorescence axes and successfully applied this algorithm to the banana bud-cutting robot for real-time operation. Cao et al. [21] developed a multi-objective particle swarm optimizer for a multi-objective trajectory model of the manipulator, which has improved the stability of the fruit picking manipulator and facilitated nondestructive picking. Particularly, in the area of automated crop disease diagnosis [22,23], Unmanned Aerial Vehicles (UAVs) equipped with RGB cameras and thermal sensors have been used for plant physiological monitoring (e.g., transpiration, leaf water, etc.) [13]. Li et al. [24] acquired the potato biomass-associated spatial and spectral features from the UAV-based RGB and hyperspectral imagery, respectively, and then they fed them into a random forest (RF) model to predict the potato yield. Wan et al. [25] fused the spectral and structural information from multispectral imagery into a multi-temporal vegetation index model to predict the rice grain yield.
In addition, with the advancements in remote sensing technologies, remote sensing-based vision technology has shown great potential for agricultural control and management, especially for automatic crop disease diagnosis [22,23]. The existing remote sensing-based computer vision models were developed based on the characteristics of the images (such as the red, green and blue (RGB) images, thermal images, multispectral and hyperspectral images) [12,13,14,15,16]. For instance, Unmanned Aerial Vehicles (UAVs) equipped with RGB cameras and thermal sensors have been used for plant physiological monitoring (e.g., transpiration, leaf water, etc.) [13]. Li et al. [24] acquired potato biomass-associated spatial and spectral features from the UAV-based RGB and hyperspectral imagery, respectively, and then they fed them into a random forest (RF) model to predict the potato yield. Wan et al. [25] fused the spectral and structural information from multispectral imagery into a multi-temporal vegetation index model to predict the rice grain yield.
Benefiting from many more narrow spectral bands over a contiguous spectral range, hyperspectral imagery (HSI) provides spatial information in two dimensions and rich spectral information in the third dimension, capturing detailed spectral–spatial information of the disease infestation and offering the potential to provide better diagnostic accuracy [26,27]. However, extracting effective infestation features from the abundant spectral and spatial information from hyperspectral images is a key challenge for disease diagnosis. Currently, based on the features used in HSI-based disease detection, the existing models can be divided into three categories: spectral feature-based approaches focusing on spectral signatures composed of the associated radiation signal of each pixel of ab image scene in various spectral ranges [28,29,30]; spatial feature-based approaches focusing on features such as shape, texture and geometrical structures [31,32,33,34]; and the joint spectral–spatial feature-based approaches focusing on a combination of spectral and spatial features [35,36,37,38,39,40,41,42]. A detailed discussion of these methods can be found in Section 2.
Despite the fact that existing works are encouraging, the existing models do not consider the hierarchical structure of the spectral and spatial information of the crop diseases (for instance, canopy structural information and reflectance radiation variance of the ground objects hidden in HSI data), which comprises important indicators for crop disease diagnosis. In fact, changes in reflectance due to plant pathogens and plant diseases are highly disease-specific since the optical properties of plant diseases are related to a number of factors such as foliar pathogens, canopy structural information, pigment content, etc.
Therefore, to address the issue presented above, the hierarchical structure of the spectral–spatial features should be considered in the learning process. In this paper, we propose a novel CropdocNet for the automated detection and discrimination of potato late blight disease. The contributions of the proposed work include the following:
- The development of an end-to-end deep learning framework (CropdocNet) for potato disease detection.
- The proposed introduction of multiple capsule layers to handle the hierarchical structure of the spectral–spatial features extracted from HSIs.
- Combination of the spectral–spatial features to represent the part-to-whole relationship between the deep features and the target classes (i.e., healthy potato and the potato infested with late blight disease).
The remainder of this paper is organized as follows: Section 2 describes the related work; Section 3 describes the study area, data collection, and the proposed model; Section 4 presents the experimental results; Section 5 provides discussions; and Section 6 summarizes this work and highlights future works.
2. Related Work in Crop Disease Detection Based on Hyperspectral Imagery
In this section, we mainly discuss related work in crop disease detection based on hyperspectral imagery (HSI). Based on features used for HSI-based crop disease detection, there are broadly three main categories: spectral feature-based approaches, spatial feature-based approaches and joint spectral–spatial feature-based approaches. Table 1 summarizes the existing models on potato late blight disease detection based on different features used in the machine learning process, which provides a baseline for hyperspectral imagery-based late blight disease detection. The detailed reviews of each class are described below.

Table 1.
Comparison of the existing models for potato late blight disease detection.
The category of spectral feature-based approaches exploits the spectral features associated with plant diseases, which represent the biophysical and biochemical status of the plant leaves from the spectral domain of HSI [28,29,30]. For example, Nagasubramanian et al. [43] found that the spectral bands associated with the depth of chlorophyll absorption are very sensitive to the occurrence of plant diseases, and they extracted the optimal spectral bands as the input of the Genetic Algorithm (GA)-based SVM for the early identification of charcoal rot disease in soybean, with a classification accuracy. Huang et al. [44] extracted 12 sensitive spectral features for Fusarium head blight, which were then fed into a SVM model to diagnose the severity of Fusarium head blight with good performance.
The category of spatial feature-based approaches exploits the spatial texture of the hyperspectral image, which represents the foliar contextual variances, such as the color, density and leaf angle, and is one of the important factors for crop disease diagnosis [31,32,33,34]. For example, Mahlein et al. [45] summarized the spatial features of the RGB, multi-spectral, and hyperspectral images used in the automatic detection of disease detection. Their study showed that the spatial properties of the crop leaves were affected by leaf chemical parameters (e.g., pigments, water, sugars, etc.) and light reflected from internal leaf structures. For instance, the spatial texture of the hyperspectral bands from 400 to 700 nm is mainly influenced by foliar content, and the spatial texture of the bands from 700 to 1100 nm reflects the leaf structure and internal scattering processes. Yuan et al. [46] introduced the spatial texture of the satellite data into the spatial angle mapper (SAM) to monitor wheat powdery mildew at the regional level.
In the category of joint spectral–spatial feature-based approaches, there are two main strategies for extracting joint spectral–spatial features to represent the characteristics of crop diseases in HSI data. The first strategy is to extract spatial and spectral features separately and then combine them together based on 1D or 2D approaches (e.g., feature stacking, convolutional filters, etc.) [40,41,42]. For example, Xie et al. [47] investigated the spectral and spatial features extracted from hyperspectral imagery to detect early blight disease on eggplant leaves, and they then stacked these features as the input of an AdaBoost model to detect healthy and infected samples. The second strategy is to jointly extract the correlated spectral–spatial information of the HSI cube through 3D kernel-based approaches [48,49,50]. For instance, Nguyen et al. [51] tested the performance of the 2D convolutional neural network (2D-CNN) and 3D convolutional neural network (3D-CNN) for the early detection of grapevine viral diseases. Their findings demonstrated that the 3D convolutional filter was able to produce promising results compared with the 2D convolutional filter from hyperspectral cubes. Benefiting from the advanced self-learning performance of the 3D convolutional kernel, the depth of the 3D convolutional kernel has also been investigated for crop disease diagnosis [35,36,37,38,39]. For instance, Suryawati et al. [52] compared the CNN baselines with the depths of 2, 5 and 13 3D convolutional layers, and their findings suggested that the deeper architecture achieved higher accuracy for plant disease detection tasks. Nagasubramanian et al. [53] developed a 3D deep convolutional neural network (DCNN) with eight 3D convolutional layers to extract the deep spectral–spatial features to represent the inoculated stem images from the soybean crops. Kumar et al. [54] proposed a 3D convolutional neural network (CNN) with six 3D convolutional layers to extract the spectral–spatial features for various crop diseases.
However, these existing methods fail to model the various kinds of reflectance radiation of the crop disease and the hierarchical structure of the disease-specific features, which are affected by the particular combination of multiple factors, such as the foliar biophysical variations, the appearance of typical fungal structures and canopy structural information, from region to region [27]. A reason behind this is that the convolutional kernels in the existing CNN methods are independent of each other, making it hard to model the part-to-whole relationship of the spatial–spatial features and to characterize the complexity and diversity of potato late blight disease on HSI data [36]. Therefore, this study proposes a novel end-to-end deep learning model to address the limitations under consideration of the hierarchical structure of the spectral–spatial features associated with plant diseases.
3. Materials and Methods
3.1. Data Acquisition
3.1.1. Study Site
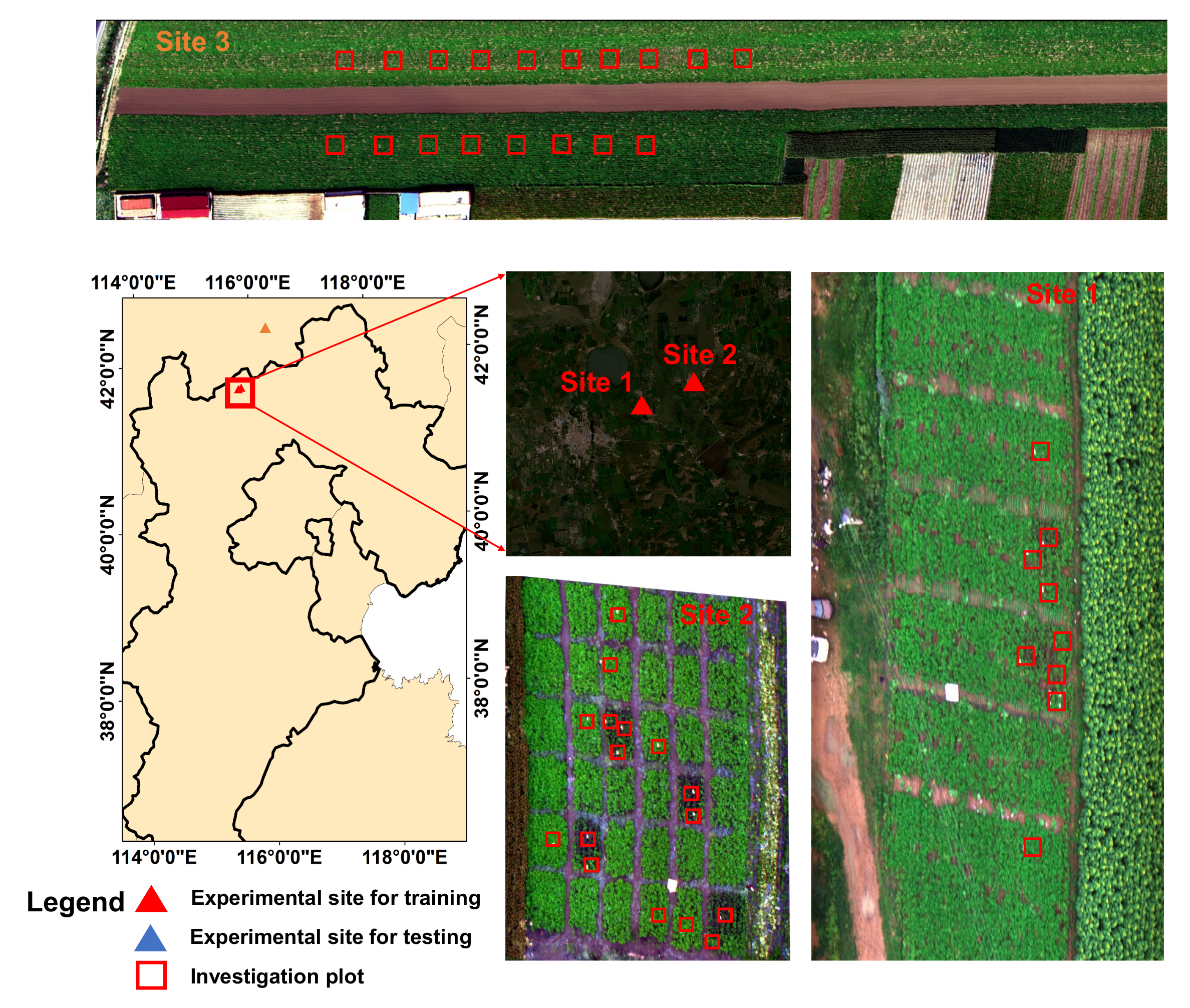
The field experiments were conducted at three experimental sites (see Figure 1), with experiments in the first two sites conducted under controlled conditions to collect high-quality labelled data for model training and the experiment in the third site conducted under natural conditions to obtain an independent dataset for model evaluation. All of the experiments were performed in Guyuan county, Hebei province, China. The detailed information for each experimental site is described below.
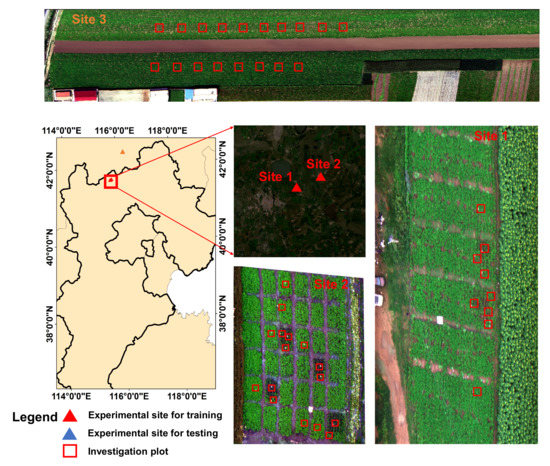
Figure 1.
The experimental sites in Guyuan, Hebei province, China.
Site 1 was located at (N, E). The potato cultivars ‘Yizhangshu No.12’ and ‘Shishu No.1’ were selected due to their different susceptibility to late blight infestation. Two control groups and four infected groups of late blight were applied. Each field group occupied 410 m of field campaigns. Seedlings of these cultivars were inoculated with late blight on 13 May 2020. A spore concentration of 9 mg mL was used. A total of nine 1 m × 1 m observation plots were set for the ground truth data investigation (see Figure 1). There were two reasons for using 1 m × 1 m observation plots: (1) they allowed for the collection of the canopy spectral–spatial variations of the potato leaves; (2) they enabled easy identification of the same patches on hyperspectral images to ensure the right match between the ground truth investigation patches and the pixel-level labels. The field observations were conducted on 16 August 2020.
Site 2 was located at (N, E). The same potato cultivars as in site 1 were selected. There were 6 control groups, and 30 infected groups of late blight were applied. Each field group occupied 81 m of field campaigns. Seedlings of these cultivars were inoculated with late blight on 14 May 2020. In the infected groups, a spore concentration of 9 mg mL was used. A total of 18 1 m × 1 m observation plots were set for the ground truth data investigation. The field observations were conducted on 18 August 2020.
Site 3 was located at (N, E). The potato cultivar ‘Shishu No.1’ was selected. The late blight disease naturally occurred in this experimental site under natural conditions. A total of 18 1 m × 1 m in-situ observation plots were set for the ground truth data investigation. The field observations were conducted on 20 August 2020.
3.1.2. Ground Truth Disease Investigation
Four types (classes) of ground truth data were investigated: healthy potato, late blight disease, soil and background (i.e., the roof, road and other facilities). Of these, the classes of soil and background could be easily labelled based on visual investigation from the UAV HSI. For the classes of healthy potato and late blight disease, we firstly investigated the disease ratio (i.e., the diseased area/the total leaf area) of the experiment sites based on National Rules for Investigation and Forecast Technology of the Potato Late Blight (NY/T1854-2010). Then, we labeled the diseased ratio in a sampling plot lower than as a healthy potato class; otherwise, it was labeled as a diseased class. The reason for choosing the threshold of was mainly because the hyperspectral signal and the spatial texture of the potato leaves with a disease ratio lower than were indistinguishable from the healthy leaves in our HSI data (with the spatial resolution of 2.5 cm).
3.1.3. UAV-Based HSI Collection
The UAV-based HSIs were collected by Dajiang (DJI) S1000 (ShenZhen (SZ) DJI Technology Co., Ltd., Gungdong, China) equipped with a UHD-185 Imaging spectrometer (Cubert GmbH, Ulm, Baden-Warttemberg, Germany). The collected HSI imagery covered the wavelength range from 450 nm to 950 nm with 125 bands. In the measurements, a total of 23 HSIs (the overlap rate was set as to avoid mosaicking errors [55]) were mosaicked to cover experiment site 1, and the full size for experimental site 1 was 16,382 × 8762 pixels. A total of 16 HSIs were mosaicked to cover experiment site 2, and the full size for experimental site 2 was pixels. A total of 14 HSIs were mosaicked to cover experiment site 3, and the full size for experimental site 2 was 15,822 × 6256 pixels. All of the UAV-based HSI data were collected between 11:30 a.m. and 13:30 p.m. under a cloud-free condition. The spatial resolution of the HSI was 2.5 cm, with a height of 30 m. HSI data were manually labeled based on the ground truth investigations. The HSIs for experimental site 1 and site 2 were used as a training dataset for model training and cross-validation, while the HSI for experimental site 3 was used as an independent dataset for model evaluation.
3.2. The Proposed CropdocNet Model
Since the traditional convolutional neural networks extract spectral–spatial features without considering the hierarchical structure representations among the features, this may lead to suboptimal performance in terms of characterizing the part-to-whole relationship between the features and the target classes. In this study, inspired by the dynamic routing mechanism of capsules [56], the proposed CropdocNet model introduces multiple capsule layers (see below) with the aim of modeling the effective hierarchical structure of spectral–spatial details and generating encapsulated features to represent the various classes and the rotation invariance of the disease attributes in the feature space for accurate disease detection.
Essentially, the design rationale behind our proposed approach is that, unlike the traditional CNN methods, which extract the abstract scalar features to predict the classes, the spectral–spatial information extracted by the convolutional filters in the form of scalars is encapsulated into a series of hierarchical class-capsules to generate the deep vector features, representing the specific combination of the spectral–spatial features for the target classes. Based on this rationale, the length of the encapsulated vector features represents the membership degree of an input belonging to a class, and the direction of the encapsulated vector features represents the consistency of the spectral–spatial feature combination between the labeled classes and the predicted classes.
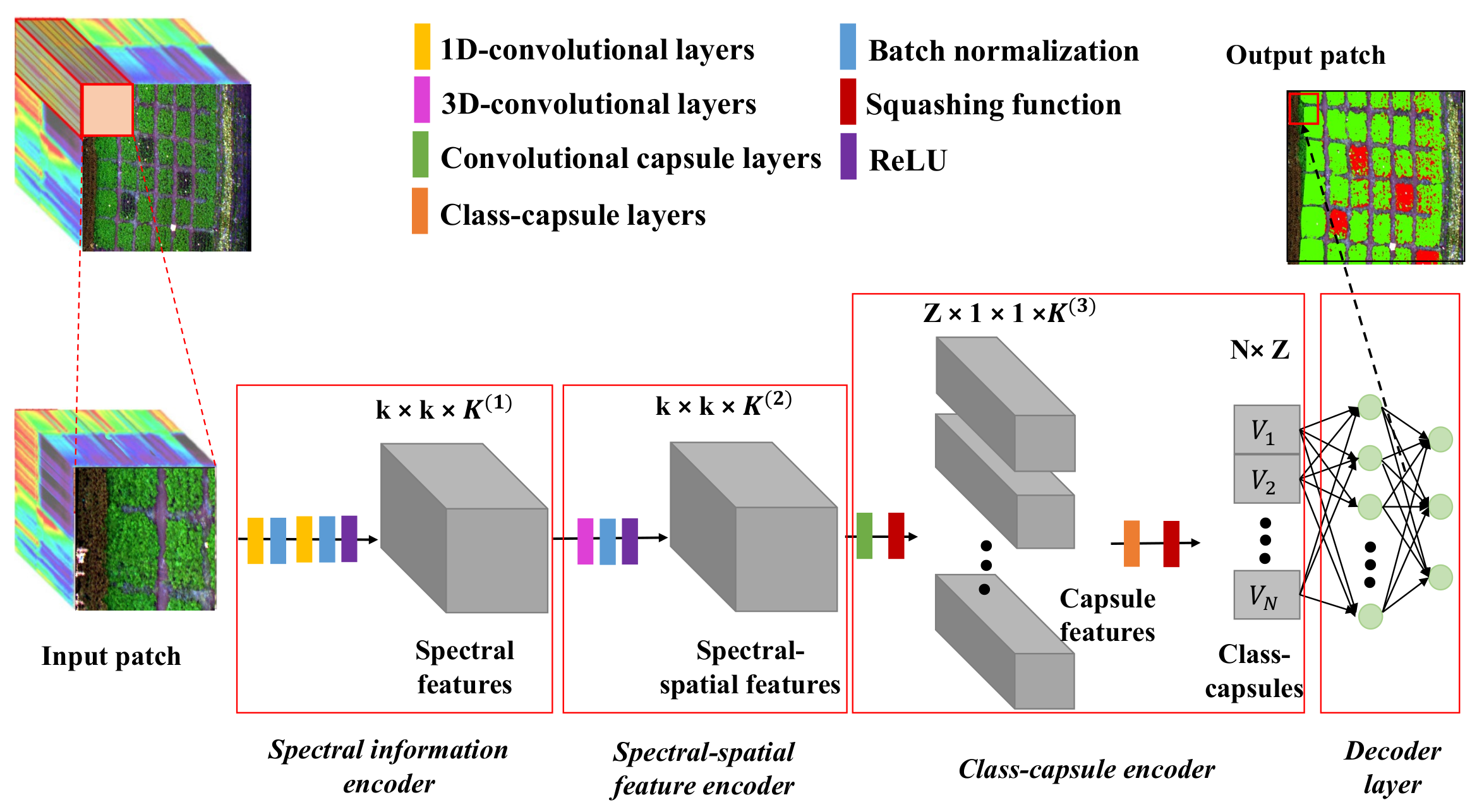
Figure 2 shows the proposed framework, which consists of a spectral information encoder, a spectral–spatial feature encoder, a class-capsule encoder and a decoder.
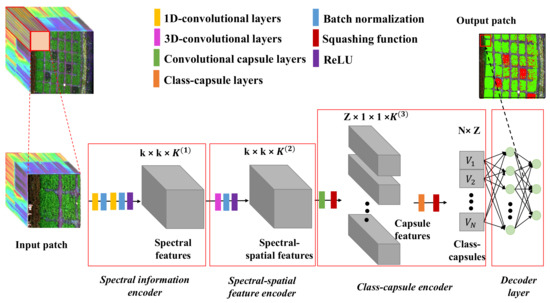
Figure 2.
The workflow of the CropdepcNet framework for potato late blight disease diagnosis (k is the spatial size of the convolutional kernel, K is the number of the channel of the convolutional kernel, Z is the dimensionality of the class-capsule, N is the number of the class-capsule, and V represents a vector of the high-level features).
Specifically, the proposed CropdocNet firstly extracts the effective information from the spectral domain based on the 1D convolutional blocks and then encodes the spectral–spatial details around the central pixels by using the 3D convolutional blocks. Subsequently, these spectral–spatial features are sent to the hierarchical structure of the class-capsule blocks in order to build the part-to-whole relationship and to generate the hierarchical vector features for representing the specific classes. Finally, a decoder is employed to predict the classes based on the length and direction of the hierarchical vector features in the feature space. The detailed information for the model blocks is described below.
3.2.1. Spectral Information Encoder
The spectral information encoder, located at the beginning of the model, is set to extract the effective spectral information from the input HSI data patches. It is composed of a serial connection of two 1D convolutional layers, two batch normalization layers and a ReLu layer.
Specifically, as shown in Figure 2, the HSI data with H rows, W columns and B bands, denoted as , can be viewed as a sample set with pixel vectors. Each of the pixels represents a class. Then, the 3D patches with a size of around each pixel are extracted as the model input, where d is the patch size. In this study, d is set as 13 so that the input patch is able to capture at least one intact potato leaf. These patches are labeled with the same classes as their central pixels.
Subsequently, the joint 1D convolution and batch normalization series, which receive the data patch from the input HSI cube, are introduced to extract the radiation magnitude of the central band and their neighboring bands. A total of convolutional kernels with a size of are employed by the 1D convolutional layer, where is the length of the receptive field for the spectral domain. The 1D convolutional layer is calculated as follows:
where is the intermediate output of the pth neuron with the jth kernel, is the weight for the lth unit of the jth kernel, and is the feature value of the lth unit corresponding to the pth neuron.
The second 1D convolution and batch normalization series are used to extract the abstract spectral details from the low-level spectral features. Finally, a ReLu activation function is used to obtain a spectral feature output denoted as .
3.2.2. Spectral–Spatial Feature Encoder
The spectral–spatial feature encoder is located after the spectral information encoder and aims to arrange the extracted spectral features in into the joint spectral–spatial features that are fed to the subsequent capsule encoder. Firstly, a total of global convolutional operations are used on the with a kernel size of , where c is the kernel size, which is set as 13 in order to match the size of the input patch. Then, the batch normalization step and a ReLu activation function are used to generate the output volume .
3.2.3. Class-Capsule Encoder
The class-capsule encoder, the most important module of the proposed network, is introduced to generate the hierarchical features to represent the translational and rotational correlations between the low-level spectral–spatial information and the target classes of healthy and diseased potato. It comprises two layers: a feature encapsulation layer and a class-capsule layer.
Specifically, the feature encapsulation layer consists of Z convolutional-based capsule units, where each of the capsule units is composed of K convolutional filters, and the size of each filter is . In the training process, the from the spectral–spatial feature encoder os input into a series of capsules units to learn the potential translational and rotational structure of the features in . An output vector is generated by the K convolutional kernel of the mth capsule. The orientation of the output vector represents the class-specific hierarchical structure characteristics, while its length represents the degree to which a capsule corresponds to a class (e.g., healthy or diseased). To measure the length of the output vector as a probability value, a nonlinear squash function is used as follows:
where is the scaled vector of . This function compresses the short vector features to zero and enlarges the long vector features to a value close to 1. The final output is denoted as .
Subsequently, the class-capsule layer is introduced to encode the encapsulated vector features in to the class-capsule vectors corresponding to the target classes. The length of the class-capsule vectors indicates the probability of belonging to corresponding classes. Here, a dynamic routing algorithm is introduced to iteratively update the parameters between the class-capsule vectors with the previous capsule vectors. The dynamic routing algorithm provides a well-designed learning mechanism between the feature vectors, which reinforces the connection coefficients between the layers and highlights the part-to-whole correlation relationship between the generated capsule features. Mathematically, the class-capsule is calculated as
where is a transformation matrix connecting layer with layer l, where is the mth feature of layer and is the biases. This function allows the vector features at a low level to make predictions for the rotation invariance of high-level features corresponding to the target classes. After that, the prediction agreement can be computed by a dynamic routing coefficient :
where is a dynamic routing coefficient measuring the weight of the mth capsule feature of layer activating the nth class-capsule of layer l; the sum of all the coefficients would be 1, and the dynamic routing coefficient can be calculated as
where is the prior representing the correlation between layer and layer l, which is initialized as 0 and is iteratively updated as follows:
where is the activated capsule of layer l, which can be calculated based on the function as follows:
Updated by the dynamic routing algorithm, the capsule features with similar predictions are clustered, and a robust prediction based on these capsule clusters is performed. Finally, the the loss function (L) is defined as follows:
where is set as 1 when class i is currently classified in the data; otherwise, it is 0. The , set as 0.9, and , set as 0.1, are defined to force the into a series of small interval values to update the loss function. , defined as 0.5, is a regularization parameter used to avoid over-fitting and to reduce the effect of the negative activity vectors.
3.2.4. The Decoder Layer
The decoder layer, composed of two fully connected layers, is designed to reconstruct the classification map from the output vector features. The final output of this model is regarded as . To update the model, the model loss aims to minimize the difference between the labeled map, , and the output map, . The final loss function is defined as follows:
where is the mean square error (MSE) loss between the labelled map and the output map, and is the learning rate, in this study, is set to 0.0005 in order to trade-off the contribution of and , and an Adam optimizer is used to optimize the learning process.
3.3. Model Evaluation for the Detection of Potato Late Blight Disease
3.3.1. Experimental Design
In order to evaluate the performance of the proposed CropdocNet on the detection of potato late blight disease, three experiments were conducted: (1) determining the model’s sensitivity to the network depth, (2) an accuracy comparison study between CropdocNet and the existing machine/deep learning models for potato late blight disease detection and (3) accuracy evaluation at both pixel and patch scales. The detailed experimental settings are described as follow.
- (1)
- Experiment 1: Determining the model’s sensitivity to the depth of the network
The depth of the network is an important parameter that determines the model’s performance in spectral–spatial feature extraction. To investigate the effect of the depth of the network, we change the number of the 1D convolutional layers and the 3D convolutional layers in the proposed model to control the model depth. For each of the configurations, we compare the model’s performance in potato late blight disease detection and show the best accuracy.
- (2)
- Experiment 2: An accuracy comparison study between CropdocNet and the existing machine/deep learning models
In order to evaluate the effectiveness of the hierarchical structure of the spectral–spatial information in our model for the detection of potato late blight disease, we compare the proposed CropdocNet considering the hierarchical structure of the spectral–spatial information with the existing representative machine/deep learning approaches using (a) spectral features only, (b) the spatial features only and (c) joint spectral–spatial features only. Based on the literature review, SVM, random forest (RF) and 3D-CNN are selected as existing representative machine learning/deep learning models for comparison study. For the spectral feature-based models, the works in [43,44,57] have reported the support vectors machine (SVM) to be an effective classifier for plant disease diagnosis based on spectral features. For the spatial feature-based models, the works in [27,33,34] have demonstrated that random forest (RF) is an effective classifier for the analysis of plant stress-associated spatial information in disease diagnosis. For joint spectral–spatial feature based models, a number of deep learning models have been proposed to extract the spectral–spatial features from the HSI data, among which 3D convolutional neural network (3D-CNN)-based models [39,50,53] are the most commonly used in plant disease detection. All these existing methods do not consider the hierarchical structure of the spectral–spatial information.
- (3)
- Experiment 3: Accuracy evaluation at both pixel and patch scales
To evaluate the model’s performance regarding the mapping of potato blight disease occurrence under different observation scales, two evaluation methods were used: (1) pixel-scale evaluation, which focuses on the performance evaluation of the proposed model for the detection of the detailed late blight disease occurrence at the pixel-level based on the pixel-wised ground truth data—in addition, to validate the model’s robustness and generalizability, we also compared the classification maps of all four models based on the independent dataset—and (2) patch-scale evaluation, which focuses on performance evaluation at the patch level by the aggregation of the pixel-wised classification into the patches with a given size. For instance, in our case, the field is divided into 1 m × 1 m patches/grids, and the disease predictions at the pixel level are aggregated into the 1 m × 1 m patches, which are compared against the corresponding real disease occurrence within that given patch area. In this study, the patch size of 1 m × 1 m was used for two reasons: (1) to enable easy pixel-level data labeling and (2) to enable the easy identification of the patches on HSIs to ensure the right match between the ground truth investigation patches and the pixel-level labels. This patch-scale evaluation further indicates the classification robustness of the disease detection at different observation scales.
3.3.2. Evaluation Metrics
A set of widely used evaluation metrics was introduced to evaluate the accuracy of the detection of potato late blight disease: the confusion matrix, sensitivity, specificity, overall accuracy (OA), average accuracy (AA), and Kappa coefficient. These evaluation metrics were computed based on the statistics of the positive condition (P), negative condition (N), true positive (TP), false positive (FP), true negative (TN) and false negative (FN). Specifically, for a given class (e.g., late blight disease), the real P indicates the samples labeled as late blight disease and the real N indicates the samples labeled as non-late blight disease. TP, TN, FP and FN are obtained from the model output. The detailed definition of the metrics are set in Table 2 and their mathematic formats are listed as follows.

Table 2.
The definition of the confusion matrix: P = positive condition; N = negative condition; TP = true positive; FP = false positive; TN = true negative; FN = False Negative; UA = user’s accuracy; PA = producer’s accuracy. The producer’s accuracy refers to the probability that a certain class is classified correctly, and the user’s accuracy refers to the reliability of a certain class.
3.3.3. Model Training
In this study, a slide window approach was used to extract the input samples for model training. Here, the slide window size was set as . A total of 3200 (i.e., 800 for each class) HSI blocks with a size of were randomly extracted from the HSI data collected from the controlled field conditions (i.e., experimental site 1 and 2). In order to prevent over-fitting in the training process, five-fold cross validation was used. For model optimization, an Adam optimizer, with a batch size of 64, was used to train the proposed model. The learning rate was initially set as and iteratively increased with a step of .
The hardware environment for model training consisted of an Intel (R) Xeon (R) CPU E5-2650, NVIDIA TITAN X (Pascal) and 64 GB memory. The software environment was the Tensorflow 2.2.0 framework with Python 3.5.2 as the programming language.
4. Results
4.1. The CropdocNet Model’s Sensitivity to the Depth of the Convolutional Filters
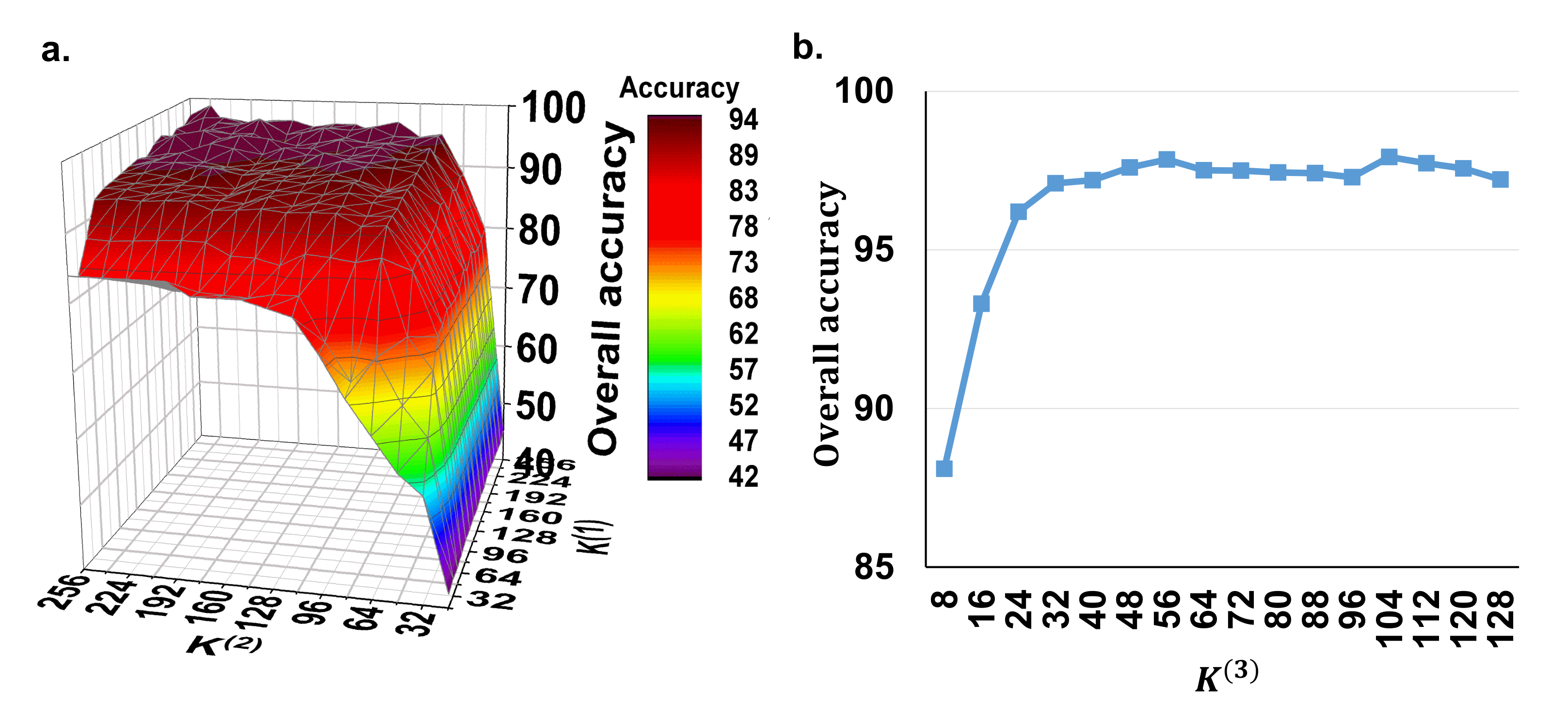
In the proposed method, we need to set the parameters , and , which represent the depth of the 1D convolutional layers for the spectral feature extraction, the depth of the 3D convolutional layers for the spectral–spatial feature extraction and the number of the capsule vector features, respectively. Due to the fact that, in our model, the high-level capsule vector features are derived from the low-level spectral–spatial scalar features, the depth of the convolutional filters is the main factor that influences this process. Therefore, we firstly set the to a fixed value of 16 to evaluate the effect of using different depths of and for spectral–spatial scalar feature extraction. Figure 3a shows the overall accuracy of the potato late blight disease classification using the the various and values from 32 to 256 with a step of 16. It can be seen that both and have positive effects on the classification accuracy. The accuracy convergence is more sensitive to than to . This is because controls the joint spectral–spatial features with more correlation with the plant stress and affects the final disease recognition accuracy. Overall, the classification accuracy reaches convergence (approximately ) when and . Thus, in the following experiments, we set and for optimal model performance and computing efficiency.
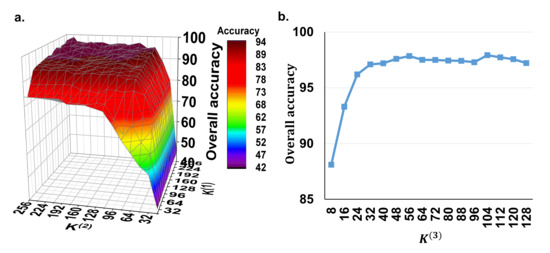
Figure 3.
The model’s sensitivity to the depth of the convolutional filters. (a) Overall accuracy of using different and values with a fixed of 16. (b) Overall accuracy of using different values under the fixed and values of 128 and 64. Here, is the depth of the 1D convolutional layers for the spectral feature extraction, is the depth of the 3D convolutional layers for the spectral–spatial feature extraction, and is the number of the capsule vector features.
Subsequently, we test the effect of the parameter with the fixed and values of 128 and 64. Figure 3b shows that the classification accuracy increases when increases from 8 to 32 and then converges to approximately when is greater than 32. These findings suggest that the number of 32 capsule vector blocks is the minimum configuration for our model for the detection of potato late blight disease. Therefore, in order to achieve a trade-off between the model performance and computing performance, is set as 32 in the subsequent experiments.
4.2. Accuracy Comparison Study between CropdocNet and Existing Machine Learning-Based Approaches for Potato Disease Diagnosis
In this experiment, we quantitatively investigated the performance of the proposed model considering the hierarchical structure of the spectral–spatial information and the representative machine/deep learning approaches without considering it (i.e., SVM with the spectral features only, RF with the spatial features only and 3D-CNN with the joint spectral–spatial features only) for potato late blight disease detection with different feature extraction strategies. In contrast, for SVM, we used the Radial Basis Function (RBF) kernel to learn the non-linear classifier, where the two kernel parameters C and were set to 1000 and 1, respectively [43,44]. For RF, a quantity of 500 decision trees was employed because this value has been proven to be effective in crop disease detection tasks [33,34]. For 3D-CNN, we employed the model architecture and configurations reported in Nagasubramanian et al. [53]’s study. All of the models were trained on the training dataset and validated on both of the testing and independent datasets.
Table 3 shows the accuracy comparison between the proposed model and the competitors using the test dataset and the independent dataset. The results suggest that the proposed model using the hierarchical vector features consistently outperforms the representative machine/deep learning approaches with scalar features in all of the classes. The OA and AA of the proposed model are and , respectively, with a Kappa value of on the test dataset, which is on average higher than the second-best model (i.e., the 3D-CNN model with joint spectral–spatial scalar features). In addition, the classification accuracy of the proposed model is found to be , which is higher than the second-best model. For the independent test dataset, the OA and AA of the proposed model were found to be and , respectively, with a Kappa value of , which is the best classifier. The classification accuracy is found to be , which is higher than the second best model. These findings demonstrate that the proposed model with the hierarchical structure of the spectral–spatial information outperforms scalar spectral–spatial feature-based models in terms of the classification accuracy of late blight disease detection.

Table 3.
The accuracy comparison between the proposed model and existing representative machine/deep learning models in potato late blight disease detection.
To further explore the classification difference significance between the proposed method and the existing machine models, the McNemar’s Chi-squared () test was conducted between two-paired models. The significant statistics are shown in Table 4. Our results show that the overall accuracy improvement of the proposed model is statistically significant with for SVM, for RF and for 3D-CNN.

Table 4.
The McNemar’s Chi-squared test of the proposed model and the existing representative machine/deep learning models for potato late blight disease detection (** means ).
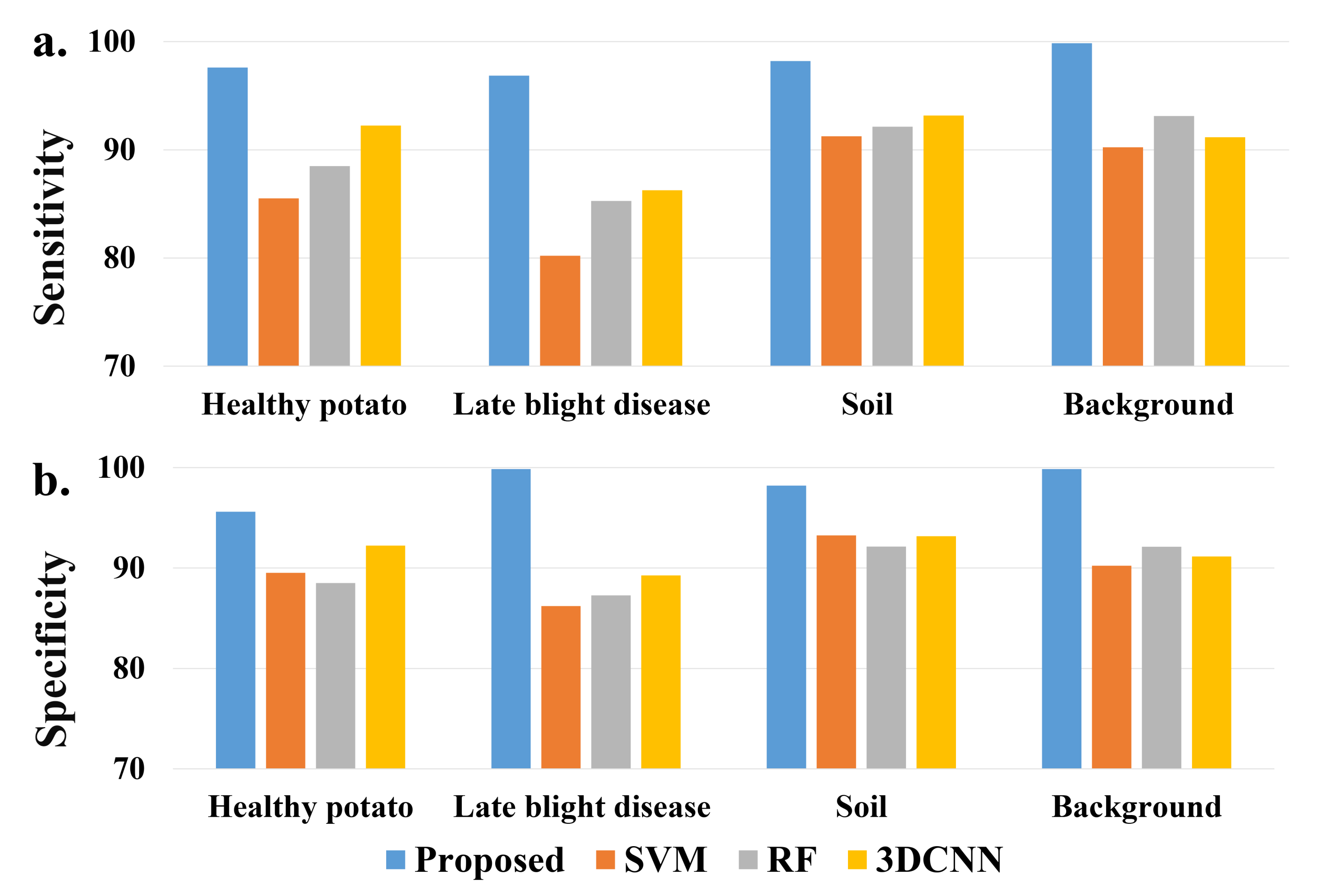
Moreover, a sensitivity and specificity comparison of detailed classes is shown in Figure 4. Similar to the classification evaluation results, the proposed model achieves the best sensitivity and specificity on all of the ground classes, especially for the class of potato late blight disease.
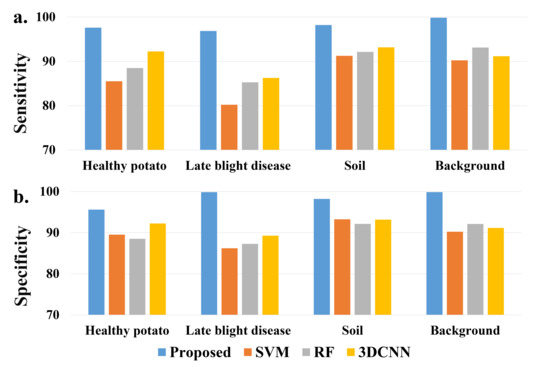
Figure 4.
A comparison of (a) sensitivity and (b) specificity of each class from different models.
4.3. The Model’s Performance When Mapping Potato Late Blight Disease from UAV HSI Data
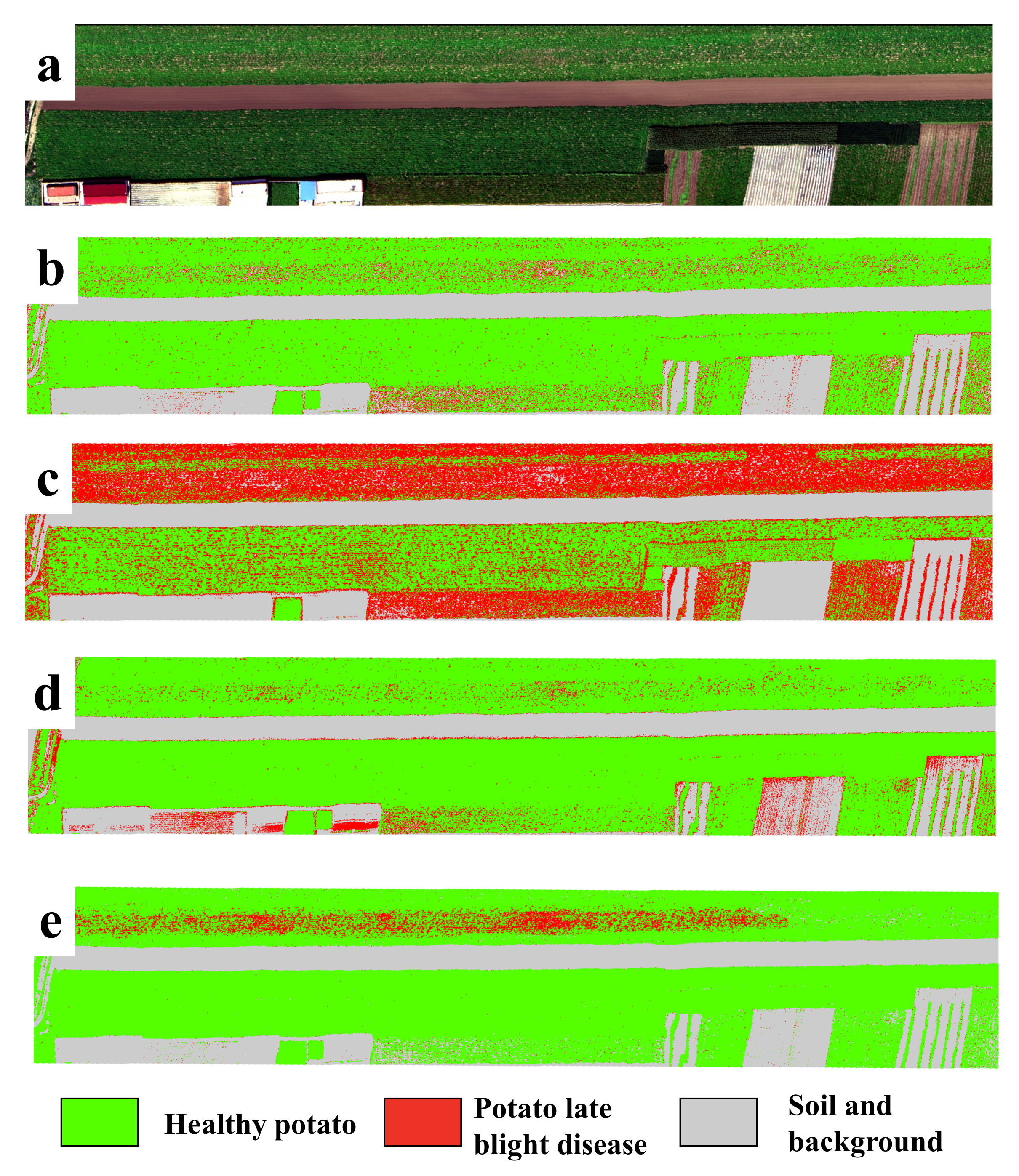
In order to show the model’s performance and generalizability for the detection of potato late blight disease, Figure 5 illustrates the classification maps of all four models for the independent testing dataset (collected under natural conditions). Here, to highlight the display of healthy potato and late blight, we show the classes of soil and background in the same color. We find that the potato late blight disease area produced by the proposed CropdocNet is located in a hot-spot area, which is consistent with our ground investigations. In comparison, there are noticeable “salt and pepper” noises found in the classification maps produced by SVM, RF and 3D-CNN. More importantly, the proposed CropdocNet method outperforms the competitors in the classification of the mixed pixels located in the potato field edge and low density area; thus, a clear boundary between the plant (i.e., the class of healthy potato) and bare soil (i.e., the class of background) can be observed in the classification map of CropdocNet (see Figure 5e), but the pixels in the potato field edge and low density area are misclassified as late blight disease in the maps of SVM, RF and 3D-CNN (see Figure 5b–d).
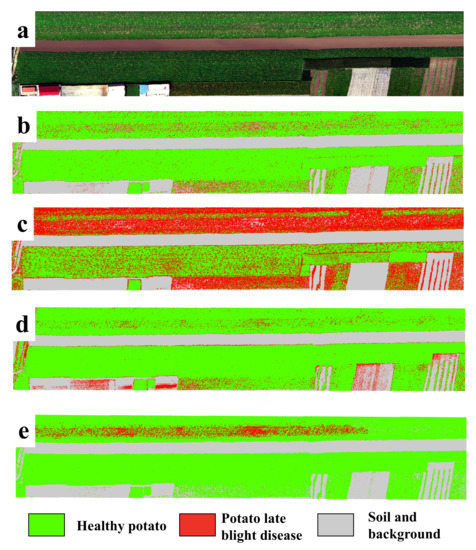
Figure 5.
A comparison of the classification maps for the independent testing dataset from four models. (a) RGB composition map of the raw data, (b–e) Classification maps of SVM, RF, 3D-CNN and the proposed CropdocNet model.
Table 5 shows the confusion matrix of the proposed model and the existing models for the pixel-scale disease classification by using the independent testing dataset from site 3. Our results demonstrate that, compared with the accuracies based on the test dataset mentioned in Section 4.2, the proposed model performs a robust classification on the evaluation dataset with an overall accuracy of and Kappa of 0.812. In comparison, the competitors that only considered spectral (i.e., SVM) or spatial information (i.e., RF) showed a significant degradation in terms of classification accuracy and robustness. The execution time of the proposed model is 721 ms, which is faster than the 3D-CNN but slower than SVM and RF. This findings suggest that the proposed model has better performance in terms of both accuracy and computing efficiency compared to 3D-CNN.

Table 5.
The confusion matrix of the proposed model and the existing models on the pixel-scale detection of potato late blight disease. Here, UA is the user’s accuracy, PA is the producer’s accuracy.
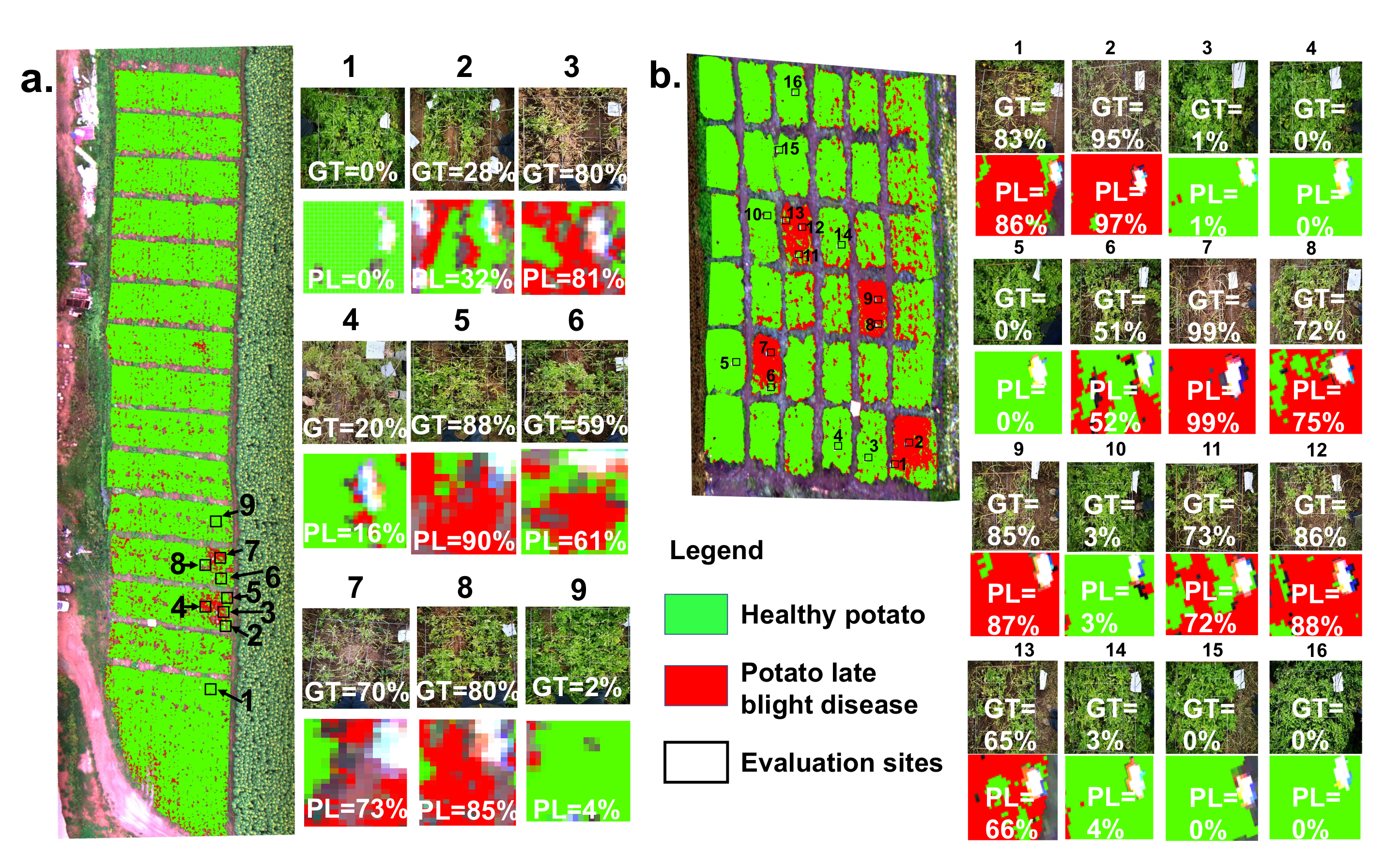
In addition, a patch-scale evaluation between the ground truth and classification result is significant for guiding agricultural management and control in practice. Figure 6 shows the patch-scale test for the classification maps of healthy potato and potato late blight disease overlaid on the UAV HSI in experimental site 1 and site 2, respectively. The percentage rate revealed in each patch is the ratio of the late blight disease pixels and the total pixels of the patch. For experimental site 1, nine patches with a size of 1 m × 1 m are ground truth data. Our results illustrate that the average difference in the disease ratio within the patches between the ground truth data and the classification map is . The maximum difference occurring in patch 8 is . For experimental site 2, there are 16 1 m × 1 m ground truth patches. Our findings suggest that the average difference in the disease ratio within the patches between the ground truth patches and the patches from the classification map is , and the maximum difference occurring in patch 1 is .
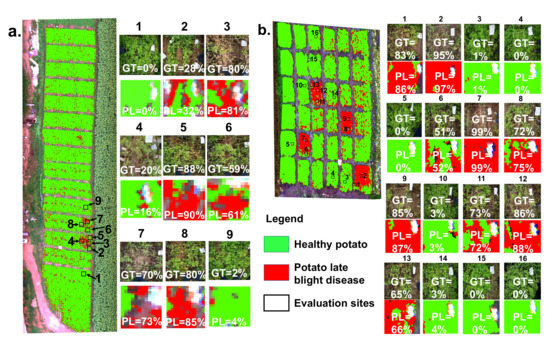
Figure 6.
The patch-scale test for the classification maps of healthy potato and potato late blight disease in (a) experimental site 1 and (b) experimental site 2. Here, the example patches on the right side illustrate the accuracy comparison between the ground truth (GT) investigations and the predicted levels (PL) of the late blight disease. Each value inside the patch represents the disease ratio (the late blight disease pixels/the total pixels).
5. Discussion
The hierarchical structure of the spectral–spatial information extracted from HSI data has been proven to be an effective way to represent the invariance of the target entities on HSI [36]. In this paper, we propose the CropdocNet model to learn the late blight disease-associated hierarchical structure information from the UAV HSI data, providing more accurate crop disease diagnosis at the farm scale. Unlike the traditional scalar features used in the existing machine learning/deep learning approaches, our proposed method introduces the capsule layers to learn the hierarchical structure of the late blight disease-associated spectral–spatial characteristics, which allows the capture of the rotation invariance of the late blight disease under complicated field conditions, leading to improvements in terms of the model’s accuracy, robustness and generalizability.
To trade off between the accuracy and computing efficiency, the effects of the depth of the convolutional filters are investigated. Our findings suggest that there is no obvious improvement in accuracy when the depth of 1D convolutional kernels and the depth of 3D convolutional kernels . We also find that, by using the multi-scale capsule units (), the model’s performance on HSI-based potato late blight disease detection could be improved.
To investigate the effectiveness of using the hierarchical vector features for accurate disease detection, we have compared the proposed model with three typical machine learning models considering only the spectral or spatial scalar features. The results illustrate that the proposed model outperforms the traditional models in terms of overall accuracy, average accuracy, sensitivity and specificity on both the training dataset (collected under controlled field conditions) and the independent testing dataset (collected under natural conditions). In addition, the classification differences between the proposed model and the existing models are statistically significance based on the McNemar’s Chi-squared test.
5.1. The Assessment of the Hierarchical Vector Feature
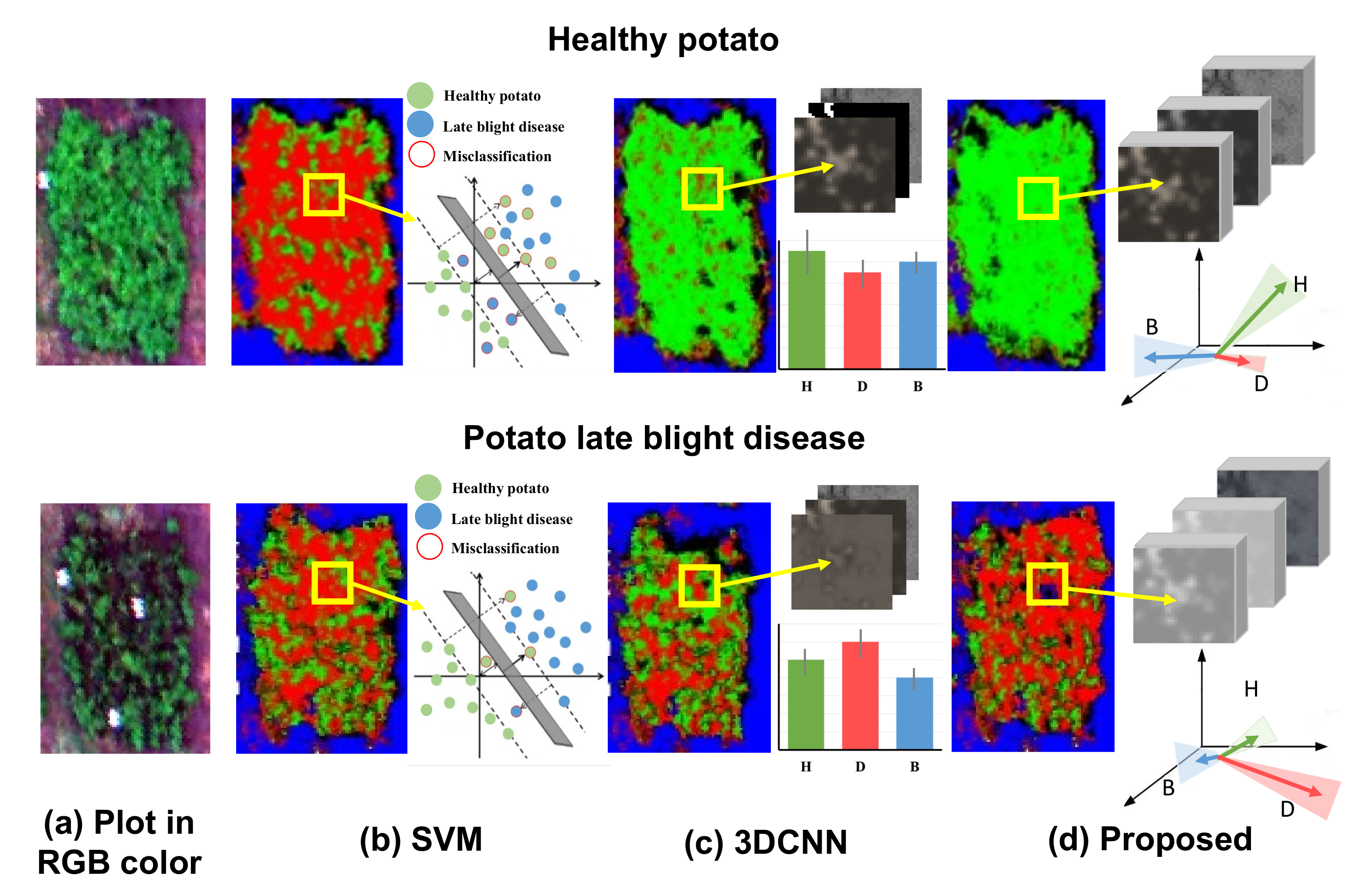
To further visually demonstrate the benefit of using hierarchical vector features in the proposed CropdocNet model, we have compared the visualized feature space and the mapping results of the healthy (see the first row of Figure 7) and diseased plots (see the second row of Figure 7) from three models: SVM, 3D-CNN and the proposed CropdocNet model. Our quantitative assessment reveals that the accuracy of the potato late blight disease plots is , and for SVM, 3D-CNN and CropdocNet, respectively. Specifically, for the SVM-based model, which only maps the spectral information into the feature space, a total of of the areas in the healthy plots are misclassified as potato late blight disease (see the left subgraph of Figure 7b), and the feature space of the samples in the yellow frame, as shown in the right subgraph of Figure 7b, explains the reason for these misclassifications. Thus, no cluster characteristics can be observed between the spectral features in the SVM-based feature space, indicating that the inter-class spectral variances are not significant in the SVM decision hyperplane.
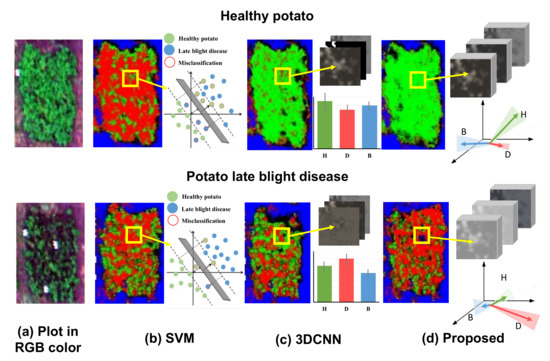
Figure 7.
The visualized feature space and the mapping results of the healthy and diseased plots based on the different machine learning/deep learning methods: (a) the original RGB image for the healthy potato (H), diseased potato (D) and background (B). (b) The classification results and the visualized spectral feature space of SVM, (c) the classification results and the averages and the standard deviations of the activated high-level spectral–spatial features of 3D-CNN, and (d) the classification results and the visualized hierarchical capsule feature space of the proposed CropdocNet.
In contrast, the spectral–spatial information based on 3D-CNN (Figure 7c) performs better than the SVM-based model. However, looking at the edge of the plots, there are obvious misclassifications. The right subgraph of Figure 7c reveals the averages and the standard deviations of the activated high-level features of the samples within the yellow frame. It is worth noting that, for the healthy potato (the first row of Figure 7c), the average values of the activated joint spectral–spatial features for different classes are quite close, and the standard deviations are relatively high, illustrating that the inter-class distances between the healthy potato and potato late blight disease are not significant in the features space. Similar results can be found in the late blight disease (see second row of Figure 7c). Thus, no significant inter-class separability can be represented in the joint spectral–spatial feature space owning to the mixed spectral–spatial signatures of plants and the background.
In comparison, the hierarchical vector features-based CropdocNet model provides more accurate classification because the hierarchical structural capsule features can express the various spectral-spatial characteristics of the target entities. For example, the white panels in the diseased plot (see the second row of of Figure 7d) are successfully classified as the background. The right subgraphs of Figure 7d demonstrate the average, direction and standard deviations of the activated hierarchical capsule features of the samples within the yellow frame. It is noteworthy that the average length and direction of the activated features for different classes are quite different, and the standard deviations (see the shadow under the arrows) do not overlap with each other. These results fully demonstrate the significant clustering of each class in the hierarchical capsule feature space; thus, the hierarchical vector features are capable of capturing most of the spectral–spatial variability found in practice.
5.2. The General Comparison of CropdocNet and the Existing Models
For an indirect comparison between the proposed CropdocNet model and the existing case studies, we have drawn Table 6 and present the accuracy and computing efficiency. As shown in Table 6, our proposed CropdocNet model has the best accuracy () compared to the existing works. For computing efficiency, due to the deep-layered network architecture and large scale samples, the deep learning models (3D-CNN and CropdocNet) require more computing time compared to traditional machine learning methods (such as SVM, RF) which only use fewer samples.

Table 6.
The performance comparison of the proposed CropdocNet model with the existing study cases. Note: “-” means no record found in the relevant literature.
5.3. Limitations and Future Works
Benefiting from the hierarchical capsule features, the proposed CropdocNet model performs better for potato late blight disease detection than the existing spectral-based or spectral–spatial based deep/machine learning models, and the generalizability of the network architecture is better than the existing models. The previous experimental evaluation has demonstrated the robustness and generalizability of our proposed model. Our model can be adapted to the detection of other crop diseases since our proposed method introduces the capsule layers to learn the hierarchical structure of the disease-associated spectral–spatial characteristics, which allows for the capture of the rotation invariance of diseases under complicated conditions. However, it is worth mentioning that our current input data for model training are mainly based on the full bloom period of potato growth, when the canopy closure reaches maximum and the field microclimate is mostly suitable for the occurrence of late blight disease; thus, the direct use of the pre-trained model may lead to limited performance. The reason is that the hyperspectral imagery is generally influenced by the mixed pixel effect, which depends on the crop growth and stress types. Therefore, in future studies, we will validate the proposed model on more UAV-based HSI data with various potato growth stages and various diseases. Specifically, we will further test the receptive field of CropdocNet and fine-tune the model on HSI data for performance enhancement under various field conditions.
6. Conclusions
In this study, a novel end-to-end deep learning model (CropdocNet) is proposed to extract the spectral–spatial hierarchical structure of late blight disease and automatically detect the disease from UAV HSI data. The innovation of CropdocNet is the deep-layered network architecture, which integrates the spectral–spatial scalar features into the hierarchical vector features to represent the rotation invariance of potato late blight disease in complicated field conditions. The model has been tested and evaluated on controlled and natural field data and compared with the existing machine/deep learning models. The average accuracies for the training dataset and independent testing dataset are and , respectively. The experimental findings demonstrate that the proposed model is able to significantly improve the accuracy of potato late blight disease detection with HSI data.
Since the proposed model is mainly based on data collected from the limited potato growth stage and one type of potato disease, to further enhance the proposed model, future work will include two aspects: (1) we will validate the proposed model on more UAV-based HSI data with various potato growth stages and various diseases under various field conditions. This is important for UAV-based crop disease detection and monitoring at the canopy and regional levels since the hyperspectral imaging is generally influenced by the mixed pixel effect, which is highly dependent on the canopy geometry associated with the crop growth and stresses. (2) We will also investigate whether the size of the receptive field of CropdocNet is able to characterize the spectral–spatial hierarchical features of different crop diseases.
Author Contributions
Conceptualization, all authors; methodology, Y.S. and L.H.; software, Y.S. and A.K.; data acquisition and validation, T.H., S.C. and A.K.; analysis: Y.S., L.H. and T.H.; writing—original draft preparation, Y.S. and L.H.; writing—review and editing, all authors; supervision, L.H.; funding acquisition, L.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research is supported by BBSRC (BB/S020969/1), BBSRC (BB/R019983/1), the Open Research Fund of Key Laboratory of Digital Earth Science, Chinese Academy of Sciences (No.2019LDE003) and National key R&D Program of China (2017YFE01227000).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All processed data and methodology in this research are available on request from the corresponding author for research purpose.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Demissie, Y.T. Integrated potato (Solanum tuberosum L.) late blight (Phytophthora infestans) disease management in Ethiopia. Am. J. BioSci. 2019, 7, 123–130. [Google Scholar] [CrossRef]
- Patil, P.; Yaligar, N.; Meena, S. Comparision of performance of classifiers-svm, rf and ann in potato blight disease detection using leaf images. In Proceedings of the 2017 IEEE International Conference on Computational Intelligence and Computing Research (ICCIC), Coimbatore, India, 4–16 December 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–5. [Google Scholar]
- Hirut, B.G.; Shimelis, H.A.; Melis, R.; Fentahun, M.; De Jong, W. Yield, Yield-related Traits and Response of Potato Clones to Late Blight Disease, in North-Western Highlands of Ethiopia. J. Phytopathol. 2017, 165, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Namugga, P.; Sibiya, J.; Melis, R.; Barekye, A. Yield Response of Potato (Solanum tuberosum L.) Genotypes to late blight caused by Phytophthora infestans in Uganda. Am. J. Potato Res. 2018, 95, 423–434. [Google Scholar] [CrossRef]
- Zhang, X.; Li, X.; Zhang, Y.; Chen, Y.; Tan, X.; Su, P.; Zhang, D.; Liu, Y. Integrated control of potato late blight with a combination of the photosynthetic bacterium Rhodopseudomonas palustris strain GJ-22 and fungicides. BioControl 2020, 65, 635–645. [Google Scholar] [CrossRef]
- Gao, J.; Westergaard, J.C.; Sundmark, E.H.R.; Bagge, M.; Liljeroth, E.; Alexandersson, E. Automatic late blight lesion recognition and severity quantification based on field imagery of diverse potato genotypes by deep learning. Knowl.-Based Syst. 2021, 214, 106723. [Google Scholar] [CrossRef]
- Lehsten, V.; Wiik, L.; Hannukkala, A.; Andreasson, E.; Chen, D.; Ou, T.; Liljeroth, E.; Lankinen, Å.; Grenville-Briggs, L. Earlier occurrence and increased explanatory power of climate for the first incidence of potato late blight caused by Phytophthora infestans in Fennoscandia. PLoS ONE 2017, 12, e0177580. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Singh, A.; Dutta, M.K.; Riha, K.; Kriz, P. Image processing based automated identification of late blight disease from leaf images of potato crops. In Proceedings of the 2017 40th International Conference on Telecommunications and Signal Processing (TSP), Barcelona, Spain, 5–7 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 758–762. [Google Scholar]
- Tung, P.X.; Vander Zaag, P.; Li, C.; Tang, W. Combining Ability for Foliar Resistance to Late Blight [Phytophthora infestans (Mont.) de Bary] of Potato Cultivars with Different Levels of Resistance. Am. J. Potato Res. 2018, 95, 670–678. [Google Scholar] [CrossRef]
- Franceschini, M.H.D.; Bartholomeus, H.; Van Apeldoorn, D.F.; Suomalainen, J.; Kooistra, L. Feasibility of unmanned aerial vehicle optical imagery for early detection and severity assessment of late blight in potato. Remote Sens. 2019, 11, 224. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.; Dinh, A.; Wahid, K.; Bhowmik, P. Detection of potato diseases using image segmentation and multiclass support vector machine. In Proceedings of the 2017 IEEE 30th canadian conference on electrical and computer engineering (CCECE), Windsor, ON, Canada, 30 April–3 May 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–4. [Google Scholar]
- Shi, Y.; Huang, W.; González-Moreno, P.; Luke, B.; Dong, Y.; Zheng, Q.; Ma, H.; Liu, L. Wavelet-based rust spectral feature set (WRSFs): A novel spectral feature set based on continuous wavelet transformation for tracking progressive host–pathogen interaction of yellow rust on wheat. Remote Sens. 2018, 10, 525. [Google Scholar] [CrossRef] [Green Version]
- Parra-Boronat, L.; Parra-Boronat, M.; Torices, V.; Marín, J.; Mauri, P.V.; Lloret, J. Comparison of single image processing techniques and their combination for detection of weed in Lawns. Int. J. Adv. Intell. Syst. 2019, 12, 177–190. [Google Scholar]
- Shin, J.; Chang, Y.K.; Heung, B.; Nguyen-Quang, T.; Price, G.W.; Al-Mallahi, A. A deep learning approach for RGB image-based powdery mildew disease detection on strawberry leaves. Comput. Electron. Agric. 2021, 183, 106042. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, W.; Luo, J.; Huang, L.; Zhou, X. Detection and discrimination of pests and diseases in winter wheat based on spectral indices and kernel discriminant analysis. Comput. Electron. Agric. 2017, 141, 171–180. [Google Scholar] [CrossRef]
- Yang, N.; Yuan, M.; Wang, P.; Zhang, R.; Sun, J.; Mao, H. Tea diseases detection based on fast infrared thermal image processing technology. J. Sci. Food Agric. 2019, 99, 3459–3466. [Google Scholar] [CrossRef] [PubMed]
- Høye, T.T.; Ärje, J.; Bjerge, K.; Hansen, O.L.; Iosifidis, A.; Leese, F.; Mann, H.M.; Meissner, K.; Melvad, C.; Raitoharju, J. Deep learning and computer vision will transform entomology. Proc. Natl. Acad. Sci. USA 2021, 118, 2002545117. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Chen, M.; Wang, C.; Luo, L.; Li, J.; Lian, G.; Zou, X. Recognition and localization methods for vision-based fruit picking robots: A review. Front. Plant Sci. 2020, 11, 510. [Google Scholar] [CrossRef]
- Iqbal, U.; Perez, P.; Li, W.; Barthelemy, J. How computer vision can facilitate flood management: A systematic review. Int. J. Disaster Risk Reduct. 2021, 2021, 102030. [Google Scholar] [CrossRef]
- Wu, F.; Duan, J.; Chen, S.; Ye, Y.; Ai, P.; Yang, Z. Multi-target recognition of bananas and automatic positioning for the inflorescence axis cutting point. Front. Plant Sci. 2021, 12, 705021. [Google Scholar] [CrossRef]
- Cao, X.; Yan, H.; Huang, Z.; Ai, S.; Xu, Y.; Fu, R.; Zou, X. A Multi-Objective Particle Swarm Optimization for Trajectory Planning of Fruit Picking Manipulator. Agronomy 2021, 11, 2286. [Google Scholar] [CrossRef]
- Dhingra, G.; Kumar, V.; Joshi, H.D. Study of digital image processing techniques for leaf disease detection and classification. Multimed. Tools Appl. 2018, 77, 19951–20000. [Google Scholar] [CrossRef]
- Zhu, W.; Chen, H.; Ciechanowska, I.; Spaner, D. Application of infrared thermal imaging for the rapid diagnosis of crop disease. IFAC-PapersOnLine 2018, 51, 424–430. [Google Scholar] [CrossRef]
- Li, B.; Xu, X.; Zhang, L.; Han, J.; Bian, C.; Li, G.; Liu, J.; Jin, L. Above-ground biomass estimation and yield prediction in potato by using UAV-based RGB and hyperspectral imaging. ISPRS J. Photogramm. Remote Sens. 2020, 162, 161–172. [Google Scholar] [CrossRef]
- Wan, L.; Cen, H.; Zhu, J.; Zhang, J.; Zhu, Y.; Sun, D.; Du, X.; Zhai, L.; Weng, H.; Li, Y.; et al. Grain yield prediction of rice using multi-temporal UAV-based RGB and multispectral images and model transfer—A case study of small farmlands in the South of China. Agric. For. Meteorol. 2020, 291, 108096. [Google Scholar] [CrossRef]
- Moghadam, P.; Ward, D.; Goan, E.; Jayawardena, S.; Sikka, P.; Hernandez, E. Plant disease detection using hyperspectral imaging. In Proceedings of the 2017 International Conference on Digital Image Computing: Techniques and Applications (DICTA), Sydney, NSW, Australia, 29 November–1 December 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–8. [Google Scholar]
- Golhani, K.; Balasundram, S.K.; Vadamalai, G.; Pradhan, B. A review of neural networks in plant disease detection using hyperspectral data. Inf. Process. Agric. 2018, 5, 354–371. [Google Scholar] [CrossRef]
- Zhang, N.; Pan, Y.; Feng, H.; Zhao, X.; Yang, X.; Ding, C.; Yang, G. Development of Fusarium head blight classification index using hyperspectral microscopy images of winter wheat spikelets. Biosyst. Eng. 2019, 186, 83–99. [Google Scholar] [CrossRef]
- Khan, I.H.; Liu, H.; Cheng, T.; Tian, Y.; Cao, Q.; Zhu, Y.; Cao, W.; Yao, X. Detection of wheat powdery mildew based on hyperspectral reflectance through SPA and PLS-LDA. Int. J. Precis. Agric. Aviat. 2020, 3, 13–22. [Google Scholar]
- Abdulridha, J.; Batuman, O.; Ampatzidis, Y. UAV-based remote sensing technique to detect citrus canker disease utilizing hyperspectral imaging and machine learning. Remote Sens. 2019, 11, 1373. [Google Scholar] [CrossRef] [Green Version]
- Gogoi, N.; Deka, B.; Bora, L. Remote sensing and its use in detection and monitoring plant diseases: A review. Agric. Rev. 2018, 39, 307–313. [Google Scholar] [CrossRef]
- Arivazhagan, S.; Shebiah, R.N.; Ananthi, S.; Varthini, S.V. Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features. Agric. Eng. Int. CIGR J. 2013, 15, 211–217. [Google Scholar]
- Behmann, J.; Bohnenkamp, D.; Paulus, S.; Mahlein, A.K. Spatial referencing of hyperspectral images for tracing of plant disease symptoms. J. Imaging 2018, 4, 143. [Google Scholar] [CrossRef] [Green Version]
- De Castro, A.I.; Torres-Sánchez, J.; Peña, J.M.; Jiménez-Brenes, F.M.; Csillik, O.; López-Granados, F. An automatic random forest-OBIA algorithm for early weed mapping between and within crop rows using UAV imagery. Remote Sens. 2018, 10, 285. [Google Scholar] [CrossRef] [Green Version]
- Qiao, X.; Jiang, J.; Qi, X.; Guo, H.; Yuan, D. Utilization of spectral-spatial characteristics in shortwave infrared hyperspectral images to classify and identify fungi-contaminated peanuts. Food Chem. 2017, 220, 393–399. [Google Scholar] [CrossRef]
- Shi, Y.; Han, L.; Huang, W.; Chang, S.; Dong, Y.; Dancey, D.; Han, L. A Biologically Interpretable Two-Stage Deep Neural Network (BIT-DNN) for Vegetation Recognition From Hyperspectral Imagery. IEEE Trans. Geosci. Remote Sens. 2021, 60, 4401320. [Google Scholar] [CrossRef]
- Behmann, J.; Mahlein, A.K.; Rumpf, T.; Römer, C.; Plümer, L. A review of advanced machine learning methods for the detection of biotic stress in precision crop protection. Precis. Agric. 2015, 16, 239–260. [Google Scholar] [CrossRef]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Plant disease detection and classification by deep learning. Plants 2019, 8, 468. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Han, L.; Dong, Y.; Shi, Y.; Huang, W.; Han, L.; González-Moreno, P.; Ma, H.; Ye, H.; Sobeih, T. A deep learning-based approach for automated yellow rust disease detection from high-resolution hyperspectral UAV images. Remote Sens. 2019, 11, 1554. [Google Scholar] [CrossRef] [Green Version]
- Rumpf, T.; Mahlein, A.K.; Steiner, U.; Oerke, E.C.; Dehne, H.W.; Plümer, L. Early detection and classification of plant diseases with support vector machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Li, J.; Tang, Y.; Zou, X.; Lin, G.; Wang, H. Detection of fruit-bearing branches and localization of litchi clusters for vision-based harvesting robots. IEEE Access 2020, 8, 117746–117758. [Google Scholar] [CrossRef]
- Chen, M.; Tang, Y.; Zou, X.; Huang, Z.; Zhou, H.; Chen, S. 3D global mapping of large-scale unstructured orchard integrating eye-in-hand stereo vision and SLAM. Comput. Electron. Agric. 2021, 187, 106237. [Google Scholar] [CrossRef]
- Nagasubramanian, K.; Jones, S.; Sarkar, S.; Singh, A.K.; Singh, A.; Ganapathysubramanian, B. Hyperspectral band selection using genetic algorithm and support vector machines for early identification of charcoal rot disease in soybean stems. Plant Methods 2018, 14, 1–13. [Google Scholar] [CrossRef]
- Huang, L.; Li, T.; Ding, C.; Zhao, J.; Zhang, D.; Yang, G. Diagnosis of the severity of fusarium head blight of wheat ears on the basis of image and spectral feature fusion. Sensors 2020, 20, 2887. [Google Scholar] [CrossRef]
- Mahlein, A.K. Plant disease detection by imaging sensors–parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Pu, R.; Zhang, J.; Wang, J.; Yang, H. Using high spatial resolution satellite imagery for mapping powdery mildew at a regional scale. Precis. Agric. 2016, 17, 332–348. [Google Scholar] [CrossRef]
- Xie, C.; He, Y. Spectrum and image texture features analysis for early blight disease detection on eggplant leaves. Sensors 2016, 16, 676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Wang, Y.; Zhang, X. Extraction of tree crowns damaged by Dendrolimus tabulaeformis Tsai et Liu via spectral-spatial classification using UAV-based hyperspectral images. Plant Methods 2020, 16, 1–19. [Google Scholar] [CrossRef]
- Karthik, R.; Hariharan, M.; Anand, S.; Mathikshara, P.; Johnson, A.; Menaka, R. Attention embedded residual CNN for disease detection in tomato leaves. Appl. Soft Comput. 2020, 86, 105933. [Google Scholar]
- Francis, M.; Deisy, C. Disease detection and classification in agricultural plants using convolutional neural networks—A visual understanding. In Proceedings of the 2019 6th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 7–8 March 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1063–1068. [Google Scholar]
- Nguyen, C.; Sagan, V.; Maimaitiyiming, M.; Maimaitijiang, M.; Bhadra, S.; Kwasniewski, M.T. Early Detection of Plant Viral Disease Using Hyperspectral Imaging and Deep Learning. Sensors 2021, 21, 742. [Google Scholar] [CrossRef]
- Suryawati, E.; Sustika, R.; Yuwana, R.S.; Subekti, A.; Pardede, H.F. Deep structured convolutional neural network for tomato diseases detection. In Proceedings of the 2018 International Conference on Advanced Computer Science and Information Systems (ICACSIS), Yogyakarta, India, 27–28 October 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 385–390. [Google Scholar]
- Nagasubramanian, K.; Jones, S.; Singh, A.K.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B. Plant disease identification using explainable 3D deep learning on hyperspectral images. Plant Methods 2019, 15, 98. [Google Scholar] [CrossRef]
- Kumar, K.V.; Jayasankar, T. An identification of crop disease using image segmentation. Int. J. Pharm. Sci. Res. 2019, 10, 1054–1064. [Google Scholar]
- La Rosa, L.E.C.; Sothe, C.; Feitosa, R.Q.; de Almeida, C.M.; Schimalski, M.B.; Oliveira, D.A.B. Multi-task fully convolutional network for tree species mapping in dense forests using small training hyperspectral data. arXiv 2021, arXiv:2106.00799. [Google Scholar] [CrossRef]
- Cai, S.; Shu, Y.; Wang, W. Dynamic Routing Networks. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikola, HI, USA, 5–9 January 2021; pp. 3588–3597. [Google Scholar]
- Zhao, J.; Fang, Y.; Chu, G.; Yan, H.; Hu, L.; Huang, L. Identification of leaf-scale wheat powdery mildew (Blumeria graminis f. sp. Tritici) combining hyperspectral imaging and an SVM classifier. Plants 2020, 9, 936. [Google Scholar] [CrossRef]
- Padol, P.B.; Yadav, A.A. SVM classifier based grape leaf disease detection. In Proceedings of the 2016 Conference on Advances in Signal Processing (CASP), Pune, India, 9–11 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 175–179. [Google Scholar]
- Semary, N.A.; Tharwat, A.; Elhariri, E.; Hassanien, A.E. Fruit-based tomato grading system using features fusion and support vector machine. In Intelligent Systems’ 2014; Springer: Berlin/Heidelberg, Germany, 2015; pp. 401–410. [Google Scholar]
- Govardhan, M.; Veena, M. Diagnosis of Tomato Plant Diseases using Random Forest. In Proceedings of the 2019 Global Conference for Advancement in Technology (GCAT), Bangalore, India, 18–20 October 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–5. [Google Scholar]
- Saha, S.; Ahsan, S.M.M. Rice Disease Detection using Intensity Moments and Random Forest. In Proceedings of the 2021 International Conference on Information and Communication Technology for Sustainable Development (ICICT4SD), Dhaka, Bangladesh, 27–28 February 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 166–170. [Google Scholar]
- Hu, G.; Wu, H.; Zhang, Y.; Wan, M. A low shot learning method for tea leafs disease identification. Comput. Electron. Agric. 2019, 163, 104852. [Google Scholar] [CrossRef]
- Elhassouny, A.; Smarandache, F. Smart mobile application to recognize tomato leaf diseases using Convolutional Neural Networks. In Proceedings of the 2019 International Conference of Computer Science and Renewable Energies (ICCSRE), Agadir, Morocco, 22–24 July 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–4. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).