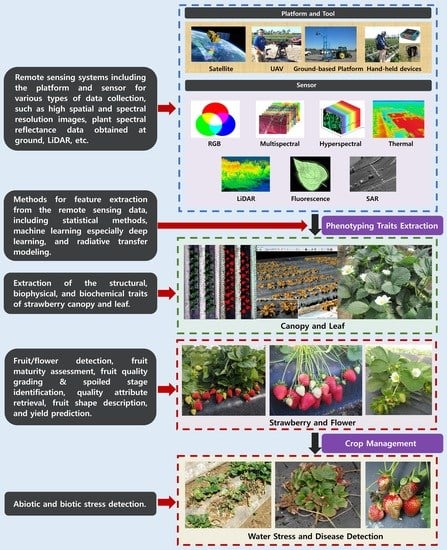
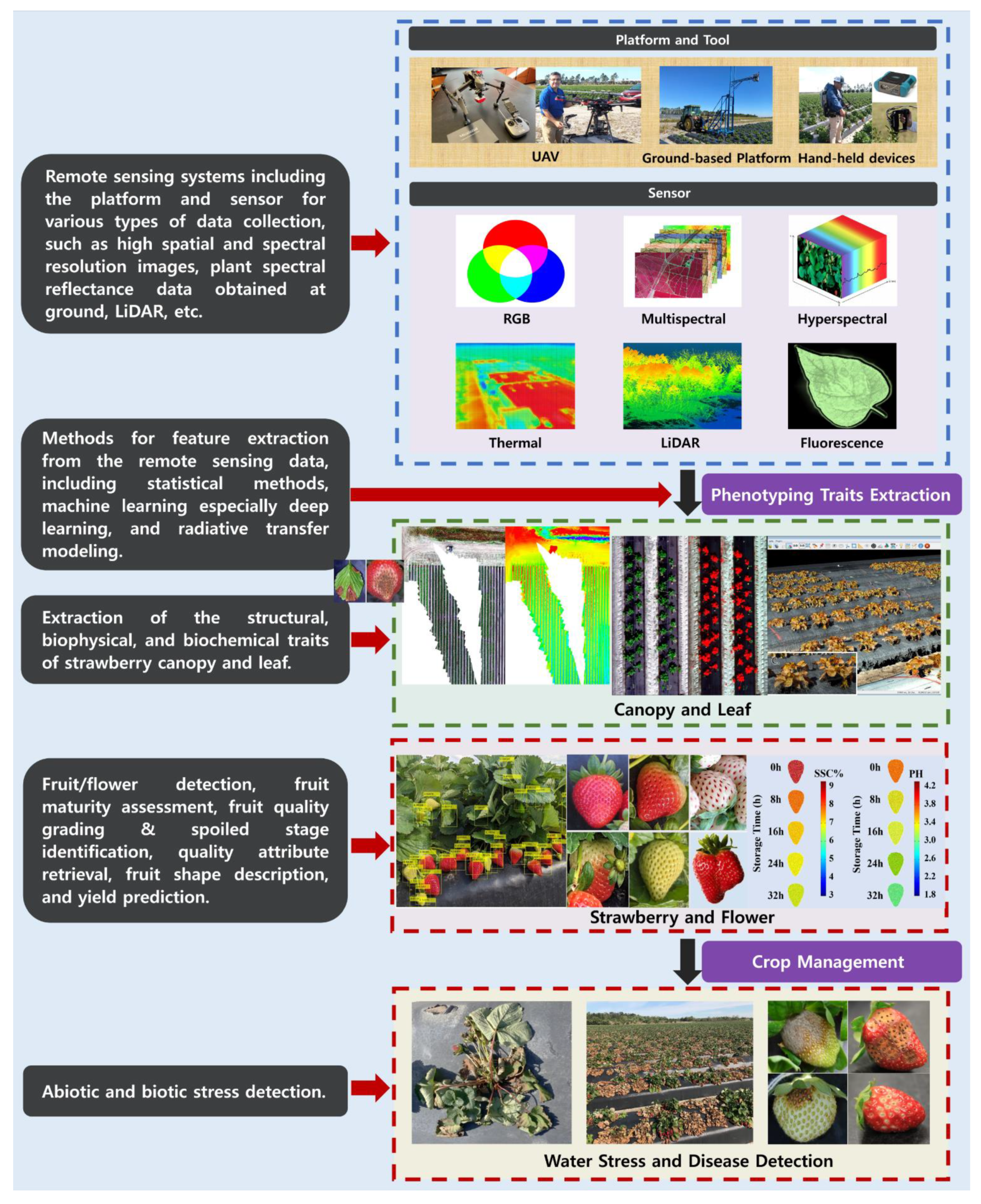
Remote Sensing and Machine Learning in Crop Phenotyping and Management, with an Emphasis on Applications in Strawberry Farming
Abstract
1. Introduction
2. Remote Sensing Platforms and Sensors
3. Machine and Deep Learning Analysis Methods
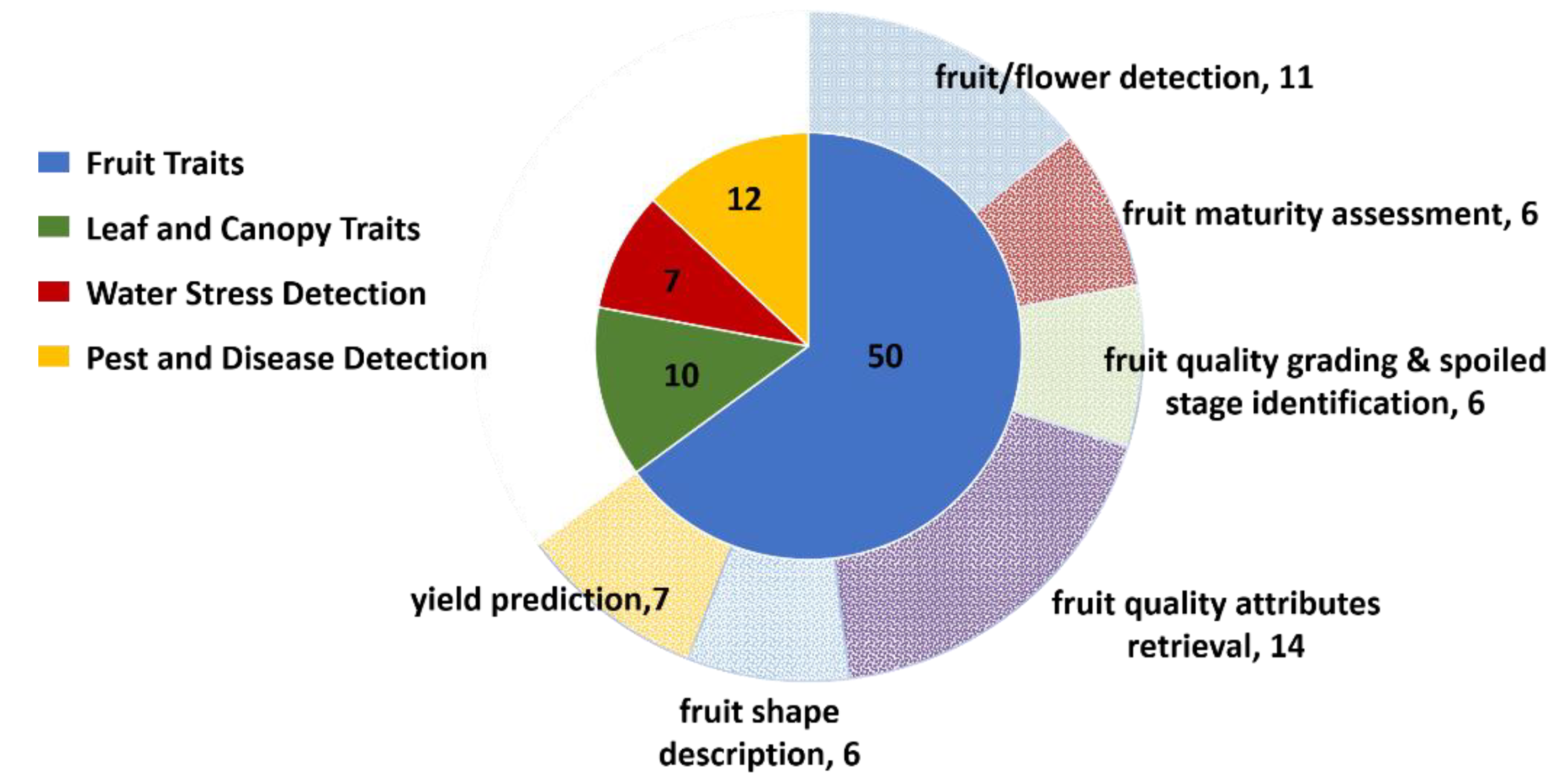
4. Fruit Traits
4.1. Fruit/Flower Detection
4.2. Fruit Maturity/Ripeness
4.3. Fruit Quality and Postharvest Monitoring
4.4. Internal Fruit Attributes
4.5. Fruit Shape
4.6. Strawberry Yield Prediction
5. Leaf and Canopy Traits
6. Abiotic/Biotic Stress Detection
6.1. Water Stress
6.2. Pest and Disease Detection
7. Discussion and Outlook
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix B
| Strawberry: Part of Interest | Phenotyping Traits | Data | Method and Model | Reference | |
|---|---|---|---|---|---|
| Fruit | Fruit/Flower detection | Mostly RGB images with high spatial resolution | Traditional morphological segmentation; CNNs (SSD, RCNN, Fast RCNN, Faster RCNN, Mask-RCNN, etc.) | [60,61,62,63,64,65] | |
| Ripeness and postharvest quality evaluation | RGB, multispectral, and hyperspectral images, especially for R, G, and NIR bands | 1) Feature extraction (spectral and textural indexes) + classifier (FLD, SVM, multivariate linear, multivariate nonlinear, SoftMax regression, etc.) 2) CNN classifier (AlexNet, CNN, etc.) | [12,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87] | ||
| Internal attributes’ retrieval (SSC, MC, pH, TA, vitamin C, TWSS, MPC, etc.) | NIR, multispectral, and hyperspectral spectroscopy and images | Feature extraction (spectral and textural features) + prediction model (PLSR, SVR, LWR, MLR, SVM, BPNN, etc.) | [73,90,91,92,93,94,95,96,97,98,99,100,101,102] | ||
| Shape description | Mostly RGB images | Shape descriptors extracted from 2D images; the SfM method was for generating 3D point clouds. | [103,104,105,106,107,108] | ||
| Yield prediction | RGB, multispectral, and hyperspectral images; weather parameters | 1) Feature extraction (fruit number, vegetation spectral indexes, LAI, weather condition parameters) + prediction model (MLP, GFNN, PPCR, NN, RF, etc.) for strawberry total weight 2) Strawberry detection and count of the number | [109,110,111,112,113,114] | ||
| Canopy and Leaf | Structural properties (planimetric canopy area, canopy surface area, canopy average height, standard deviation of canopy height, canopy volume, and canopy smoothness parameters) | RGB images with high spatial resolution | SfM and Arcgis analysis | [127,128,129,130,131] | |
| Biophysical features | Dry biomass and leaf area of canopy | RGB and NIR images | Feature extraction (canopy geometric parameters, including canopy area, canopy average height, etc.) + prediction model (MLR) | [129,130] | |
| Nitrogen content of leaves | RGB and NIR images | Feature extraction (green and red reflectance (550 and 680 nm), VI, and NDVI) + regression analysis | [126] | ||
| Leaf temperature | Thermal images | [132] | |||
| Water stress | Chlorophyll fluorescence, thermal, and hyperspectral images | Leaf temperature and spectral characteristics (CWSI, NDVI, REIP, PSSRb, PRI, MSI) were extracted for water stress detection. | [143,144,145,146,147] | ||
| Pest and disease stress | Powdery mildew, anthracnose crown rot, verticillium wilt, gray mold, etc. | RGB, multispectral, and hyperspectral images | Various types of color and texture features were imported to supervised classifiers for disease detection. | [41,149,152,153,154,155,156,158,160,161,164,165] | |
References
- FAO. The Future of Food and Agriculture—Alternative Pathways to 2050; Food and Agriculture Organization of the United Nations: Rome, Italy, 2018. [Google Scholar]
- Bongiovanni, R.; Lowenberg-DeBoer, J. Precision agriculture and sustainability. Precis. Agric. 2004, 5, 359–387. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, M.; Wang, N. Precision agriculture—A worldwide overview. Comput. Electron. Agric. 2002, 36, 113–132. [Google Scholar] [CrossRef]
- Liaghat, S.; Balasundram, S.K. A review: The role of remote sensing in precision agriculture. Am. J. Agric. Biol. Sci. 2010, 5, 50–55. [Google Scholar] [CrossRef]
- Say, S.M.; Keskin, M.; Sehri, M.; Sekerli, Y.E. Adoption of precision agriculture technologies in developed and developing countries. Online J. Sci. Technol. 2018, 8, 7–15. [Google Scholar]
- Costa, C.; Schurr, U.; Loreto, F.; Menesatti, P.; Carpentier, S. Plant phenotyping research trends, a science mapping approach. Front. Plant Sci. 2019, 9, 1933. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned aerial vehicle remote sensing for field-based crop phenotyping: Current status and perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef]
- Chawade, A.; van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-throughput field-phenotyping tools for plant breeding and precision agriculture. Agronomy 2019, 9, 258. [Google Scholar] [CrossRef]
- Pasala, R.; Pandey, B.B. Plant phenomics: High-throughput technology for accelerating genomics. J. Biosci. 2020, 45, 1–6. [Google Scholar] [CrossRef]
- Pauli, D.; Chapman, S.C.; Bart, R.; Topp, C.N.; Lawrence-Dill, C.J.; Poland, J.; Gore, M.A. The quest for understanding phenotypic variation via integrated approaches in the field environment. Plant Physiol. 2016, 172, 622–634. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop phenomics and high-throughput phenotyping: Past decades, current challenges, and future perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef]
- Weng, S.; Yu, S.; Dong, R.; Pan, F.; Liang, D. Nondestructive detection of storage time of strawberries using visible/near-infrared hyperspectral imaging. Int. J. Food Prop. 2020, 23, 269–281. [Google Scholar] [CrossRef]
- Mezzetti, B.; Giampieri, F.; Zhang, Y.-T.; Zhong, C.-F. Status of strawberry breeding programs and cultivation systems in Europe and the rest of the world. J. Berry Res. 2018, 8, 205–221. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. FAOSTAT Database; 2018. Available online: http://www.fao.org/faostat/en/?#data/QC (accessed on 20 November 2020).
- Huang, Y.; Chen, Z.-X.; Tao, Y.; Huang, X.-Z.; Gu, X.-F. Agricultural remote sensing big data: Management and applications. J. Integr. Agric. 2018, 17, 1915–1931. [Google Scholar] [CrossRef]
- Sicre, C.M.; Fieuzal, R.; Baup, F. Contribution of multispectral (optical and radar) satellite images to the classification of agricultural surfaces. Int. J. Appl. Earth Obs. Geoinf. 2020, 84, 101972. [Google Scholar] [CrossRef]
- Xu, Y.; Smith, S.E.; Grunwald, S.; Abd-Elrahman, A.; Wani, S.P. Incorporation of satellite remote sensing pan-sharpened imagery into digital soil prediction and mapping models to characterize soil property variability in small agricultural fields. ISPRS J. Photogramm. Remote. Sens. 2017, 123, 1–19. [Google Scholar] [CrossRef]
- Zhu, Q.; Luo, Y.; Xu, Y.-P.; Tian, Y.; Yang, T. Satellite soil moisture for agricultural drought monitoring: Assessment of SMAP-derived soil water deficit index in Xiang River Basin, China. Remote. Sens. 2019, 11, 362. [Google Scholar] [CrossRef]
- Du, T.L.T.; Bui, D.D.; Nguyen, M.D.; Lee, H. Satellite-based, multi-indices for evaluation of agricultural droughts in a highly dynamic tropical catchment, Central Vietnam. Water 2018, 10, 659. [Google Scholar] [CrossRef]
- Estel, S.; Mader, S.; Levers, C.; Verburg, P.H.; Baumann, M.; Kuemmerle, T. Combining satellite data and agricultural statistics to map grassland management intensity in Europe. Environ. Res. Lett. 2018, 13, 074020. [Google Scholar] [CrossRef]
- Fieuzal, R.; Baup, F. Forecast of wheat yield throughout the agricultural season using optical and radar satellite images. Int. J. Appl. Earth Obs. Geoinf. 2017, 59, 147–156. [Google Scholar] [CrossRef]
- Sharma, A.K.; Hubert-Moy, L.; Buvaneshwari, S.; Sekhar, M.; Ruiz, L.; Bandyopadhyay, S.; Corgne, S. Irrigation history estimation using multitemporal landsat satellite images: Application to an intensive groundwater irrigated agricultural watershed in India. Remote. Sens. 2018, 10, 893. [Google Scholar] [CrossRef]
- Xie, Q.; Dash, J.; Huete, A.; Jiang, A.; Yin, G.; Ding, Y.; Peng, D.; Hall, C.C.; Brown, L.; Shi, Y. Retrieval of crop biophysical parameters from Sentinel-2 remote sensing imagery. Int. J. Appl. Earth Obs. Geoinf. 2019, 80, 187–195. [Google Scholar] [CrossRef]
- Mateo-Sanchis, A.; Piles, M.; Muñoz-Marí, J.; Adsuara, J.E.; Pérez-Suay, A.; Camps-Valls, G. Synergistic integration of optical and microwave satellite data for crop yield estimation. Remote. Sens. Environ. 2019, 234, 111460. [Google Scholar] [CrossRef] [PubMed]
- Radoglou-Grammatikis, P.; Sarigiannidis, P.; Lagkas, T.; Moscholios, I. A compilation of UAV applications for precision agriculture. Comput. Netw. 2020, 172, 107148. [Google Scholar] [CrossRef]
- Giles, D.; Billing, R. Deployment and Performance of a UAV for Crop Spraying. Chem. Eng. Trans. 2015, 44, 307–312. [Google Scholar]
- Faiçal, B.S.; Freitas, H.; Gomes, P.H.; Mano, L.Y.; Pessin, G.; de Carvalho, A.C.; Krishnamachari, B.; Ueyama, J. An adaptive approach for UAV-based pesticide spraying in dynamic environments. Comput. Electron. Agric. 2017, 138, 210–223. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Matese, A.; Gioli, B.; Toscano, P.; Zaldei, A.; Palliotti, A.; Genesio, L. Multisensor approach to assess vineyard thermal dynamics combining high-resolution unmanned aerial vehicle (UAV) remote sensing and wireless sensor network (WSN) proximal sensing. Sci. Hortic. 2017, 221, 83–87. [Google Scholar] [CrossRef]
- Popescu, D.; Stoican, F.; Stamatescu, G.; Ichim, L.; Dragana, C. Advanced UAV–WSN System for Intelligent Monitoring in Precision Agriculture. Sensors 2020, 20, 817. [Google Scholar] [CrossRef]
- Abd-Elrahman, A.; Pande-Chhetri, R.; Vallad, G. Design and development of a multi-purpose low-cost hyperspectral imaging system. Remote. Sens. 2011, 3, 570–586. [Google Scholar] [CrossRef]
- Jin, X.; Li, Z.; Atzberger, C. Editorial for the Special Issue “Estimation of Crop Phenotyping Traits using Unmanned Ground Vehicle and Unmanned Aerial Vehicle Imagery. Remote Sens. 2020, 12, 940. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote. Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Mishra, P.; Asaari, M.S.M.; Herrero-Langreo, A.; Lohumi, S.; Diezma, B.; Scheunders, P. Close range hyperspectral imaging of plants: A review. Biosyst. Eng. 2017, 164, 49–67. [Google Scholar] [CrossRef]
- Corp, L.A.; McMurtrey, J.E.; Middleton, E.M.; Mulchi, C.L.; Chappelle, E.W.; Daughtry, C.S. Fluorescence sensing systems: In vivo detection of biophysical variations in field corn due to nitrogen supply. Remote. Sens. Environ. 2003, 86, 470–479. [Google Scholar] [CrossRef]
- Wallace, L.; Lucieer, A.; Watson, C.; Turner, D. Development of a UAV-LiDAR system with application to forest inventory. Remote Sens. 2012, 4, 1519–1543. [Google Scholar] [CrossRef]
- Steele-Dunne, S.C.; McNairn, H.; Monsivais-Huertero, A.; Judge, J.; Liu, P.-W.; Papathanassiou, K. Radar remote sensing of agricultural canopies: A review. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2017, 10, 2249–2273. [Google Scholar] [CrossRef]
- McNairn, H.; Shang, J. A review of multitemporal synthetic aperture radar (SAR) for crop monitoring. In Multitemporal Remote Sensing. Remote Sensing and Digital Image Processing; Ban, Y., Ed.; Springer: Cham, Switzerland, 2016; Volume 20. [Google Scholar] [CrossRef]
- Liu, C.-A.; Chen, Z.-X.; Yun, S.; Chen, J.-S.; Hasi, T.; Pan, H.-Z. Research advances of SAR remote sensing for agriculture applications: A review. J. Integr. Agric. 2019, 18, 506–525. [Google Scholar] [CrossRef]
- Kuester, M.; Thome, K.; Krause, K.; Canham, K.; Whittington, E. Comparison of surface reflectance measurements from three ASD FieldSpec FR spectroradiometers and one ASD FieldSpec VNIR spectroradiometer. In Proceedings of the IGARSS 2001. Scanning the Present and Resolving the Future. Proceedings. IEEE 2001 International Geoscience and Remote Sensing Symposium (Cat. No.01CH37217), Sydney, NSW, Australia, 9–13 July 2001; pp. 72–74. [Google Scholar]
- Danner, M.; Locherer, M.; Hank, T.; Richter, K. Spectral Sampling with the ASD FieldSpec 4—Theory, Measurement, Problems, Interpretation; EnMAP Field Guides Technical Report; GFZ Data Services: Potsdam, Germany, 2015. [Google Scholar] [CrossRef]
- Mahmud, M.S.; Zaman, Q.U.; Esau, T.J.; Chang, Y.K.; Price, G.W.; Prithiviraj, B. Real-Time Detection of Strawberry Powdery Mildew Disease Using a Mobile Machine Vision System. Agronomy 2020, 10, 1027. [Google Scholar] [CrossRef]
- Liakos, K.G.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine learning in agriculture: A review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef]
- Mochida, K.; Koda, S.; Inoue, K.; Hirayama, T.; Tanaka, S.; Nishii, R.; Melgani, F. Computer vision-based phenotyping for improvement of plant productivity: A machine learning perspective. GigaScience 2019, 8, giy153. [Google Scholar] [CrossRef]
- Cai, J.; Luo, J.; Wang, S.; Yang, S. Feature selection in machine learning: A new perspective. Neurocomputing 2018, 300, 70–79. [Google Scholar] [CrossRef]
- Miao, J.; Niu, L. A survey on feature selection. Procedia Comput. Sci. 2016, 91, 919–926. [Google Scholar] [CrossRef]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine learning approaches for crop yield prediction and nitrogen status estimation in precision agriculture: A review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- Sabanci, K.; Kayabasi, A.; Toktas, A. Computer vision-based method for classification of wheat grains using artificial neural network. J. Sci. Food Agric. 2017, 97, 2588–2593. [Google Scholar] [CrossRef] [PubMed]
- Koirala, A.; Walsh, K.B.; Wang, Z.; McCarthy, C. Deep learning–Method overview and review of use for fruit detection and yield estimation. Comput. Electron. Agric. 2019, 162, 219–234. [Google Scholar] [CrossRef]
- Jakhar, D.; Kaur, I. Artificial intelligence, machine learning and deep learning: Definitions and differences. Clin. Exp. Dermatol. 2020, 45, 131–132. [Google Scholar] [CrossRef]
- Miikkulainen, R.; Liang, J.; Meyerson, E.; Rawal, A.; Fink, D.; Francon, O.; Raju, B.; Shahrzad, H.; Navruzyan, A.; Duffy, N. Evolving deep neural networks. arXiv 2016, arXiv:1703.00548. [Google Scholar]
- Seifert, C.; Aamir, A.; Balagopalan, A.; Jain, D.; Sharma, A.; Grottel, S.; Gumhold, S. Visualizations of deep neural networks in computer vision: A survey. In Transparent Data Mining for Big and Small Data; Springer: Berlin/Heidelberg, Germany, 2017; pp. 123–144. [Google Scholar]
- Zhang, J.; Man, K.F. Time series prediction using RNN in multi-dimension embedding phase space. In Proceedings of the SMC’98 Conference Proceedings. 1998 IEEE International Conference on Systems, Man, and Cybernetics (Cat. No. 98CH36218), San Diego, CA, USA, 14 October 1998; pp. 1868–1873. [Google Scholar]
- Yu, S.; Jia, S.; Xu, C. Convolutional neural networks for hyperspectral image classification. Neurocomputing 2017, 219, 88–98. [Google Scholar] [CrossRef]
- Liu, T.; Abd-Elrahman, A. An object-based image analysis method for enhancing classification of land covers using fully convolutional networks and multi-view images of small unmanned aerial system. Remote. Sens. 2018, 10, 457. [Google Scholar] [CrossRef]
- Salakhutdinov, R. Learning deep generative models. Ann. Rev. Stat. Appl. 2015, 2, 361–385. [Google Scholar] [CrossRef]
- Pu, Y.; Gan, Z.; Henao, R.; Yuan, X.; Li, C.; Stevens, A.; Carin, L. Variational autoencoder for deep learning of images, labels and captions. arXiv 2016, arXiv:1609.08976. [Google Scholar]
- Bauer, A.; Bostrom, A.G.; Ball, J.; Applegate, C.; Cheng, T.; Laycock, S.; Rojas, S.M.; Kirwan, J.; Zhou, J. Combining computer vision and deep learning to enable ultra-scale aerial phenotyping and precision agriculture: A case study of lettuce production. Hortic. Res. 2019, 6, 1–12. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Du, B. Deep learning for remote sensing data: A technical tutorial on the state of the art. IEEE Geosci. Remote. Sens. Mag. 2016, 4, 22–40. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Puttemans, S.; Vanbrabant, Y.; Tits, L.; Goedemé, T. Automated visual fruit detection for harvest estimation and robotic harvesting. In Proceedings of the 2016 sixth international conference on image processing theory, tools and applications (IPTA), Oulu, Finland, 12–15 December 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Feng, G.; Qixin, C.; Masateru, N. Fruit detachment and classification method for strawberry harvesting robot. Int. J. Adv. Robot. Syst. 2008, 5, 4. [Google Scholar] [CrossRef]
- Lin, P.; Chen, Y. Detection of Strawberry Flowers in Outdoor Field by Deep Neural Network. In Proceedings of the 2018 IEEE 3rd International Conference on Image, Vision and Computing (ICIVC), Chongqing, China, 27–29 June 2018; pp. 482–486. [Google Scholar] [CrossRef]
- Lamb, N.; Chuah, M.C. A strawberry detection system using convolutional neural networks. In Proceedings of the 2018 IEEE International Conference on Big Data (Big Data), Seattle, WA, USA, 10–13 December 2018; pp. 2515–2520. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, K.; Yang, L.; Zhang, D. Fruit detection for strawberry harvesting robot in non-structural environment based on Mask-RCNN. Comput. Electron. Agric. 2019, 163, 104846. [Google Scholar] [CrossRef]
- Zhou, C.; Hu, J.; Xu, Z.; Yue, J.; Ye, H.; Yang, G. A novel greenhouse-based system for the detection and plumpness assessment of strawberry using an improved deep learning technique. Front. Plant Sci. 2020, 11, 559. [Google Scholar] [CrossRef]
- Kafkas, E.; Koşar, M.; Paydaş, S.; Kafkas, S.; Başer, K. Quality characteristics of strawberry genotypes at different maturation stages. Food Chem. 2007, 100, 1229–1236. [Google Scholar] [CrossRef]
- Azodanlou, R.; Darbellay, C.; Luisier, J.-L.; Villettaz, J.-C.; Amadò, R. Changes in flavour and texture during the ripening of strawberries. Eur. Food Res. Technol. 2004, 218, 167–172. [Google Scholar]
- Kader, A.A. Quality and its maintenance in relation to the postharvest physiology of strawberry. In The Strawberry into the 21st Century; Timber Press: Portland, OR, USA, 1991; pp. 145–152. [Google Scholar]
- Rahman, M.M.; Moniruzzaman, M.; Ahmad, M.R.; Sarker, B.; Alam, M.K. Maturity stages affect the postharvest quality and shelf-life of fruits of strawberry genotypes growing in subtropical regions. J. Saudi Soc. Agric. Sci. 2016, 15, 28–37. [Google Scholar] [CrossRef]
- Li, B.; Lecourt, J.; Bishop, G. Advances in non-destructive early assessment of fruit ripeness towards defining optimal time of harvest and yield prediction—A review. Plants 2018, 7, 3. [Google Scholar]
- Rico, D.; Martin-Diana, A.B.; Barat, J.; Barry-Ryan, C. Extending and measuring the quality of fresh-cut fruit and vegetables: A review. Trends Food Sci. Technol. 2007, 18, 373–386. [Google Scholar] [CrossRef]
- Kader, A.A. Quality parameters of fresh-cut fruit and vegetable products. In Fresh-Cut Fruits and Vegetables; CRC Press: Boca Raton, FL, USA, 2002; pp. 20–29. [Google Scholar]
- Liu, C.; Liu, W.; Lu, X.; Ma, F.; Chen, W.; Yang, J.; Zheng, L. Application of multispectral imaging to determine quality attributes and ripeness stage in strawberry fruit. PLoS ONE 2014, 9, e87818. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Plotto, A.; Baldwin, E.; Whitaker, V.; Rouseff, R. Electronic nose for detecting strawberry fruit maturity. In Proceedings of the Florida State Horticultural Society, Crystal River, FL, USA, 6–8 June 2010; Volume 123, pp. 259–263. [Google Scholar]
- Raut, K.D.; Bora, V. Assessment of Fruit Maturity using Direct Color Mapping. Int. Res. J. Eng. Technol. 2016, 3, 1540–1543. [Google Scholar]
- Jiang, H.; Zhang, C.; Liu, F.; Zhu, H.; He, Y. Identification of strawberry ripeness based on multispectral indexes extracted from hyperspectral images. Guang Pu Xue Yu Guang Pu Fen Xi = Guang Pu 2016, 36, 1423–1427. [Google Scholar] [PubMed]
- Guo, C.; Liu, F.; Kong, W.; He, Y.; Lou, B. Hyperspectral imaging analysis for ripeness evaluation of strawberry with support vector machine. J. Food Eng. 2016, 179, 11–18. [Google Scholar]
- Yue, X.-Q.; Shang, Z.-Y.; Yang, J.-Y.; Huang, L.; Wang, Y.-Q. A smart data-driven rapid method to recognize the strawberry maturity. Inf. Proc. Agric. 2019. [Google Scholar] [CrossRef]
- Gao, Z.; Shao, Y.; Xuan, G.; Wang, Y.; Liu, Y.; Han, X. Real-time hyperspectral imaging for the in-field estimation of strawberry ripeness with deep learning. Artif. Intell. Agric. 2020, 4, 31–38. [Google Scholar] [CrossRef]
- Xiong, Y.; Peng, C.; Grimstad, L.; From, P.J.; Isler, V. Development and field evaluation of a strawberry harvesting robot with a cable-driven gripper. Comput. Electron. Agric. 2019, 157, 392–402. [Google Scholar] [CrossRef]
- Sustika, R.; Subekti, A.; Pardede, H.F.; Suryawati, E.; Mahendra, O.; Yuwana, S. Evaluation of deep convolutional neural network architectures for strawberry quality inspection. Int. J. Eng. Technol. 2018, 7, 75–80. [Google Scholar]
- Usha, S.; Karthik, M.; Jenifer, R.; Scholar, P. Automated Sorting and Grading of Vegetables Using Image Processing. Int. J. Eng. Res. Gen. Sci. 2017, 5, 53–61. [Google Scholar]
- Shen, J.; Qi, H.-F.; Li, C.; Zeng, S.-M.; Deng, C. Experimental on storage and preservation of strawberry. Food Sci. Tech 2011, 36, 48–51. [Google Scholar]
- Liming, X.; Yanchao, Z. Automated strawberry grading system based on image processing. Comput. Electron. Agric. 2010, 71, S32–S39. [Google Scholar] [CrossRef]
- Mahendra, O.; Pardede, H.F.; Sustika, R.; Kusumo, R.B.S. Comparison of Features for Strawberry Grading Classification with Novel Dataset. In Proceedings of the 2018 International Conference on Computer, Control, Informatics and Its Applications (IC3INA), Tangerang, Indonesia, 1–2 November 2018; pp. 7–12. [Google Scholar] [CrossRef]
- Péneau, S.; Brockhoff, P.B.; Escher, F.; Nuessli, J. A comprehensive approach to evaluate the freshness of strawberries and carrots. Postharvest Biol. Technol. 2007, 45, 20–29. [Google Scholar] [CrossRef]
- Dong, D.; Zhao, C.; Zheng, W.; Wang, W.; Zhao, X.; Jiao, L. Analyzing strawberry spoilage via its volatile compounds using longpath fourier transform infrared spectroscopy. Sci. Rep. 2013, 3, 2585. [Google Scholar] [CrossRef] [PubMed]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Wang, H.; Peng, J.; Xie, C.; Bao, Y.; He, Y. Fruit quality evaluation using spectroscopy technology: A review. Sensors 2015, 15, 11889–11927. [Google Scholar] [CrossRef] [PubMed]
- ElMasry, G.; Wang, N.; ElSayed, A.; Ngadi, M. Hyperspectral imaging for nondestructive determination of some quality attributes for strawberry. J. Food Eng. 2007, 81, 98–107. [Google Scholar] [CrossRef]
- Weng, S.; Yu, S.; Guo, B.; Tang, P.; Liang, D. Non-Destructive Detection of Strawberry Quality Using Multi-Features of Hyperspectral Imaging and Multivariate Methods. Sensors 2020, 20, 3074. [Google Scholar] [CrossRef]
- Liu, Q.; Wei, K.; Xiao, H.; Tu, S.; Sun, K.; Sun, Y.; Pan, L.; Tu, K. Near-infrared hyperspectral imaging rapidly detects the decay of postharvest strawberry based on water-soluble sugar analysis. Food Anal. Methods 2019, 12, 936–946. [Google Scholar] [CrossRef]
- Liu, S.; Xu, H.; Wen, J.; Zhong, W.; Zhou, J. Prediction and analysis of strawberry sugar content based on partial least squares prediction model. J. Anim. Plant Sci. 2019, 29, 1390–1395. [Google Scholar]
- Amodio, M.L.; Ceglie, F.; Chaudhry, M.M.A.; Piazzolla, F.; Colelli, G. Potential of NIR spectroscopy for predicting internal quality and discriminating among strawberry fruits from different production systems. Postharvest Biol. Technol. 2017, 125, 112–121. [Google Scholar] [CrossRef]
- LI, J.-B.; GUO, Z.-M.; HUANG, W.-Q.; ZHANG, B.-H.; ZHAO, C.-J. Near-infrared spectra combining with CARS and SPA algorithms to screen the variables and samples for quantitatively determining the soluble solids content in strawberry. Spectrosc. Spectr. Anal. 2015, 35, 372–378. [Google Scholar]
- Ding, X.; Zhang, C.; Liu, F.; Song, X.; Kong, W.; He, Y. Determination of soluble solid content in strawberry using hyperspectral imaging combined with feature extraction methods. Guang Pu Xue Yu Guang Pu Fen Xi = Guang Pu 2015, 35, 1020–1024. [Google Scholar] [PubMed]
- Sánchez, M.-T.; De la Haba, M.J.; Benítez-López, M.; Fernández-Novales, J.; Garrido-Varo, A.; Pérez-Marín, D. Non-destructive characterization and quality control of intact strawberries based on NIR spectral data. J. Food Eng. 2012, 110, 102–108. [Google Scholar] [CrossRef]
- Nishizawa, T.; Mori, Y.; Fukushima, S.; Natsuga, M.; Maruyama, Y. Non-destructive analysis of soluble sugar components in strawberry fruits using near-infrared spectroscopy. Nippon Shokuhin Kagaku Kogaku Kaishi = J. Jpn. Soc. Food Sci. Technol. 2009, 56, 229–235. [Google Scholar] [CrossRef]
- Wulf, J.; Rühmann, S.; Rego, I.; Puhl, I.; Treutter, D.; Zude, M. Nondestructive application of laser-induced fluorescence spectroscopy for quantitative analyses of phenolic compounds in strawberry fruits (Fragaria × ananassa). J. Agric. Food Chem. 2008, 56, 2875–2882. [Google Scholar] [CrossRef]
- Tallada, J.G.; Nagata, M.; Kobayashi, T. Non-destructive estimation of firmness of strawberries (Fragaria × ananassa Duch.) using NIR hyperspectral imaging. Environ. Control. Biol. 2006, 44, 245–255. [Google Scholar] [CrossRef]
- Nagata, M.; Tallada, J.G.; Kobayashi, T.; Toyoda, H. NIR hyperspectral imaging for measurement of internal quality in strawberries. In Proceedings of the 2005 ASAE Annual Meeting, Tampa, FL, USA, 17–20 July 2005. ASAE Paper No. 053131. [Google Scholar]
- Nagata, M.; Tallada, J.G.; Kobayashi, T.; Cui, Y.; Gejima, Y. Predicting maturity quality parameters of strawberries using hyperspectral imaging. In Proceedings of the ASAE/CSAE Annual International Meeting, Ottawa, ON, Canada, 1–4 August 2004. Paper No. 043033. [Google Scholar]
- Ishikawa, T.; Hayashi, A.; Nagamatsu, S.; Kyutoku, Y.; Dan, I.; Wada, T.; Oku, K.; Saeki, Y.; Uto, T.; Tanabata, T.; et al. Classification of strawberry fruit shape by machine learning. Int. Arch. Photogramm. Remote. Sens. Spat. Inf. Sci. 2018, 42. [Google Scholar] [CrossRef]
- Oo, L.M.; Aung, N.Z. A simple and efficient method for automatic strawberry shape and size estimation and classification. Biosyst. Eng. 2018, 170, 96–107. [Google Scholar] [CrossRef]
- Feldmann, M.J.; Hardigan, M.A.; Famula, R.A.; López, C.M.; Tabb, A.; Cole, G.S.; Knapp, S.J. Multi-dimensional machine learning approaches for fruit shape phenotyping in strawberry. GigaScience 2020, 9, giaa030. [Google Scholar] [CrossRef]
- He, J.Q.; Harrison, R.J.; Li, B. A novel 3D imaging system for strawberry phenotyping. Plant Methods 2017, 13, 1–8. [Google Scholar] [CrossRef]
- Kochi, N.; Tanabata, T.; Hayashi, A.; Isobe, S. A 3D shape-measuring system for assessing strawberry fruits. Int. J. Autom. Technol. 2018, 12, 395–404. [Google Scholar] [CrossRef]
- Li, B.; Cockerton, H.M.; Johnson, A.W.; Karlström, A.; Stavridou, E.; Deakin, G.; Harrison, R.J. Defining Strawberry Uniformity using 3D Imaging and Genetic Mapping. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Pathak, T.B.; Dara, S.K.; Biscaro, A. Evaluating correlations and development of meteorology based yield forecasting model for strawberry. Adv. Meteorol. 2016, 2016, 1–7. [Google Scholar] [CrossRef]
- Misaghi, F.; Dayyanidardashti, S.; Mohammadi, K.; Ehsani, M. Application of Artificial Neural Network and Geostatistical Methods in Analyzing Strawberry Yield Data; American Society of Agricultural and Biological Engineers: Minneapolis, MN, USA, 2004; p. 1. [Google Scholar]
- MacKenzie, S.J.; Chandler, C.K. A method to predict weekly strawberry fruit yields from extended season production systems. Agron. J. 2009, 101, 278–287. [Google Scholar] [CrossRef]
- Hassan, H.A.; Taha, S.S.; Aboelghar, M.A.; Morsy, N.A. Comparative the impact of organic and conventional strawberry cultivation on growth and productivity using remote sensing techniques under Egypt climate conditions. Asian J. Agric. Biol. 2018, 6, 228–244. [Google Scholar]
- Maskey, M.L.; Pathak, T.B.; Dara, S.K. Weather Based Strawberry Yield Forecasts at Field Scale Using Statistical and Machine Learning Models. Atmosphere 2019, 10, 378. [Google Scholar] [CrossRef]
- Chen, Y.; Lee, W.S.; Gan, H.; Peres, N.; Fraisse, C.; Zhang, Y.; He, Y. Strawberry yield prediction based on a deep neural network using high-resolution aerial orthoimages. Remote. Sens. 2019, 11, 1584. [Google Scholar] [CrossRef]
- Fonstad, M.A.; Dietrich, J.T.; Courville, B.C.; Jensen, J.L.; Carbonneau, P.E. Topographic structure from motion: A new development in photogrammetric measurement. Earth Surf. Proc. Landf. 2013, 38, 421–430. [Google Scholar] [CrossRef]
- Ozyesil, O.; Voroninski, V.; Basri, R.; Singer, A. A survey of structure from motion. arXiv 2017, arXiv:1701.08493. [Google Scholar]
- Patrick, A.; Li, C. High throughput phenotyping of blueberry bush morphological traits using unmanned aerial systems. Remote. Sens. 2017, 9, 1250. [Google Scholar] [CrossRef]
- Makanza, R.; Zaman-Allah, M.; Cairns, J.E.; Magorokosho, C.; Tarekegne, A.; Olsen, M.; Prasanna, B.M. High-throughput phenotyping of canopy cover and senescence in maize field trials using aerial digital canopy imaging. Remote. Sens. 2018, 10, 330. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Yang, G.; Dai, H.; Yang, H.; Xu, B.; Feng, H.; Li, Z.; Yang, X. Fuzzy Clustering of Maize Plant-Height Patterns Using Time Series of UAV Remote-Sensing Images and Variety Traits. Front. Plant Sci. 2019, 10, 926. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1–17. [Google Scholar] [CrossRef]
- Hunt, E.R., Jr.; Doraiswamy, P.C.; McMurtrey, J.E.; Daughtry, C.S.; Perry, E.M.; Akhmedov, B. A visible band index for remote sensing leaf chlorophyll content at the canopy scale. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 103–112. [Google Scholar] [CrossRef]
- Clevers, J.G.; Kooistra, L. Using hyperspectral remote sensing data for retrieving canopy chlorophyll and nitrogen content. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2011, 5, 574–583. [Google Scholar] [CrossRef]
- Kattenborn, T.; Schmidtlein, S. Radiative transfer modelling reveals why canopy reflectance follows function. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Yuan, H.; Yang, G.; Li, C.; Wang, Y.; Liu, J.; Yu, H.; Feng, H.; Xu, B.; Zhao, X.; Yang, X. Retrieving soybean leaf area index from unmanned aerial vehicle hyperspectral remote sensing: Analysis of RF, ANN, and SVM regression models. Remote. Sens. 2017, 9, 309. [Google Scholar] [CrossRef]
- Wolanin, A.; Camps-Valls, G.; Gómez-Chova, L.; Mateo-García, G.; van der Tol, C.; Zhang, Y.; Guanter, L. Estimating crop primary productivity with Sentinel-2 and Landsat 8 using machine learning methods trained with radiative transfer simulations. Remote. Sens. Environ. 2019, 225, 441–457. [Google Scholar] [CrossRef]
- Luisa España-Boquera, M.; Cárdenas-Navarro, R.; López-Pérez, L.; Castellanos-Morales, V.; Lobit, P. Estimating the nitrogen concentration of strawberry plants from its spectral response. Commun. Soil Sci. Plant Anal. 2006, 37, 2447–2459. [Google Scholar] [CrossRef]
- Sandino, J.D.; Ramos-Sandoval, O.L.; Amaya-Hurtado, D. Method for estimating leaf coverage in strawberry plants using digital image processing. Rev. Bras. Eng. Agrícola Ambient. 2016, 20, 716–721. [Google Scholar] [CrossRef][Green Version]
- Jianlun, W.; Yu, H.; Shuangshuang, Z.; Hongxu, Z.; Can, H.; Xiaoying, C.; Yun, X.; Jianshu, C.; Shuting, W. A new multi-scale analytic algorithm for edge extraction of strawberry leaf images in natural light. Int. J. Agric. Biol. Eng. 2016, 9, 99–108. [Google Scholar]
- Guan, Z.; Abd-Elrahman, A.; Fan, Z.; Whitaker, V.M.; Wilkinson, B. Modeling strawberry biomass and leaf area using object-based analysis of high-resolution images. J. Photogramm. Remote. Sens. 2020, 163, 171–186. [Google Scholar] [CrossRef]
- Abd-Elrahman, A.; Guan, Z.; Dalid, C.; Whitaker, V.; Britt, K.; Wilkinson, B.; Gonzalez, A. Automated Canopy Delineation and Size Metrics Extraction for Strawberry Dry Weight Modeling Using Raster Analysis of High-Resolution Imagery. Remote. Sens. 2020, 12, 3632. [Google Scholar] [CrossRef]
- Takahashi, M.; Takayama, S.; Umeda, H.; Yoshida, C.; Koike, O.; Iwasaki, Y.; Sugeno, W. Quantification of Strawberry Plant Growth and Amount of Light Received Using a Depth Sensor. Environ. Control. Biol. 2020, 58, 31–36. [Google Scholar] [CrossRef]
- Kokin, E.; Palge, V.; Pennar, M.; Jürjenson, K. Strawberry leaf surface temperature dynamics measured by thermal camera in night frost conditions. Agron. Res. 2018, 16. [Google Scholar] [CrossRef]
- Touati, F.; Al-Hitmi, M.; Benhmed, K.; Tabish, R. A fuzzy logic based irrigation system enhanced with wireless data logging applied to the state of Qatar. Comput. Electron. Agric. 2013, 98, 233–241. [Google Scholar] [CrossRef]
- Avşar, E.; Buluş, K.; Saridaş, M.A.; Kapur, B. Development of a cloud-based automatic irrigation system: A case study on strawberry cultivation. In Proceedings of the 2018 7th International Conference on Modern Circuits and Systems Technologies (MOCAST), Thessaloniki, Greece, 7–9 May 2018; pp. 1–4. [Google Scholar]
- Gutiérrez, J.; Villa-Medina, J.F.; Nieto-Garibay, A.; Porta-Gándara, M.Á. Automated irrigation system using a wireless sensor network and GPRS module. IEEE Trans. Instrum. Meas. 2013, 63, 166–176. [Google Scholar] [CrossRef]
- Morillo, J.G.; Martín, M.; Camacho, E.; Díaz, J.R.; Montesinos, P. Toward precision irrigation for intensive strawberry cultivation. Agric. Water Manag. 2015, 151, 43–51. [Google Scholar] [CrossRef]
- Gerhards, M.; Schlerf, M.; Mallick, K.; Udelhoven, T. Challenges and future perspectives of multi-/Hyperspectral thermal infrared remote sensing for crop water-stress detection: A review. Remote. Sens. 2019, 11, 1240. [Google Scholar] [CrossRef]
- Grant, O.M.; Davies, M.J.; Johnson, A.W.; Simpson, D.W. Physiological and growth responses to water deficits in cultivated strawberry (Fragaria× ananassa) and in one of its progenitors, Fragaria chiloensis. Environ. Exp. Bot. 2012, 83, 23–32. [Google Scholar] [CrossRef]
- Nezhadahmadi, A.; Faruq, G.; Rashid, K. The impact of drought stress on morphological and physiological parameters of three strawberry varieties in different growing conditions. Pak. J. Agric. Sci. 2015, 52, 79–92. [Google Scholar]
- Grant, O.M.; Johnson, A.W.; Davies, M.J.; James, C.M.; Simpson, D.W. Physiological and morphological diversity of cultivated strawberry (Fragaria× ananassa) in response to water deficit. Environ. Exp. Bot. 2010, 68, 264–272. [Google Scholar] [CrossRef]
- Klamkowski, K.; Treder, W. Response to drought stress of three strawberry cultivars grown under greenhouse conditions. J. Fruit Ornam. Plant Res. 2008, 16, 179–188. [Google Scholar]
- Adak, N.; Gubbuk, H.; Tetik, N. Yield, quality and biochemical properties of various strawberry cultivars under water stress. J. Sci. Food Agric. 2018, 98, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Peñuelas, J.; Savé, R.; Marfà, O.; Serrano, L. Remotely measured canopy temperature of greenhouse strawberries as indicator of water status and yield under mild and very mild water stress conditions. Agric. For. Meteorol. 1992, 58, 63–77. [Google Scholar] [CrossRef]
- Razavi, F.; Pollet, B.; Steppe, K.; Van Labeke, M.-C. Chlorophyll fluorescence as a tool for evaluation of drought stress in strawberry. Photosynthetica 2008, 46, 631–633. [Google Scholar] [CrossRef]
- Delalieux, S.; Delauré, B.; Tits, L.; Boonen, M.; Sima, A.; Baeck, P. High resolution strawberry field monitoring using the compact hyperspectral imaging solution COSI. Adv. Anim. Biosci. 2017, 8, 156. [Google Scholar] [CrossRef]
- Li, H.; Yin, J.; Zhang, M.; Sigrimis, N.; Gao, Y.; Zheng, W. Automatic diagnosis of strawberry water stress status based on machine vision. Int. J. Agric. Biol. Eng. 2019, 12, 159–164. [Google Scholar] [CrossRef]
- Gerhards, M.; Schlerf, M.; Rascher, U.; Udelhoven, T.; Juszczak, R.; Alberti, G.; Miglietta, F.; Inoue, Y. Analysis of airborne optical and thermal imagery for detection of water stress symptoms. Remote. Sens. 2018, 10, 1139. [Google Scholar] [CrossRef]
- Oliveira, M.S.; Peres, N.A. Common Strawberry Diseases in Florida. EDIS 2020, 2020. [Google Scholar] [CrossRef]
- Chang, Y.K.; Mahmud, M.; Shin, J.; Nguyen-Quang, T.; Price, G.W.; Prithiviraj, B. Comparison of Image Texture Based Supervised Learning Classifiers for Strawberry Powdery Mildew Detection. AgriEngineering 2019, 1, 434–452. [Google Scholar] [CrossRef]
- Mahlein, A.-K. Plant Disease detection by imaging sensors–parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Eun, J.-S.; Kim, S.-H. Image-based disease diagnosing and predicting of the crops through the deep learning mechanism. In Proceedings of the 2017 International Conference on Information and Communication Technology Convergence (ICTC), Jeju, Korea, 18–20 October 2017; pp. 129–131. [Google Scholar] [CrossRef]
- Shin, J.; Chang, Y.K.; Heung, B.; Nguyen-Quang, T.; Price, G.W.; Al-Mallahi, A. Effect of directional augmentation using supervised machine learning technologies: A case study of strawberry powdery mildew detection. Biosyst. Eng. 2020, 194, 49–60. [Google Scholar] [CrossRef]
- De Lange, E.S.; Nansen, C. Early detection of arthropod-induced stress in strawberry using innovative remote sensing technology. In Proceedings of the GeoVet 2019. Novel Spatio-Temporal Approaches in the Era of Big Data, Davis, CA, USA, 8–10 October 2019. [Google Scholar] [CrossRef]
- Liu, Q.; Sun, K.; Zhao, N.; Yang, J.; Zhang, Y.; Ma, C.; Pan, L.; Tu, K. Information fusion of hyperspectral imaging and electronic nose for evaluation of fungal contamination in strawberries during decay. Postharvest Biol. Technol. 2019, 153, 152–160. [Google Scholar] [CrossRef]
- Cockerton, H.M.; Li, B.; Vickerstaff, R.; Eyre, C.A.; Sargent, D.J.; Armitage, A.D.; Marina-Montes, C.; Garcia, A.; Passey, A.J.; Simpson, D.W. Image-based Phenotyping and Disease Screening of Multiple Populations for resistance to Verticillium dahliae in cultivated strawberry Fragaria x ananassa. bioRxiv 2018, 497107. [Google Scholar] [CrossRef]
- Altıparmak, H.; Al Shahadat, M.; Kiani, E.; Dimililer, K. Fuzzy classification for strawberry diseases-infection using machine vision and soft-computing techniques. In Proceedings of the Tenth International Conference on Machine Vision (ICMV 2017), Vienna, Austria, 13–15 November 2017; p. 106961N. [Google Scholar] [CrossRef]
- Hecht-Nielsen, R. Theory of the backpropagation neural network. In Proceedings of the International 1989 Joint Conference on Neural Networks, Washington, DC, USA, 1989; Volume 1, pp. 593–605. [Google Scholar] [CrossRef]
- Siedliska, A.; Baranowski, P.; Zubik, M.; Mazurek, W.; Sosnowska, B. Detection of fungal infections in strawberry fruit by VNIR/SWIR hyperspectral imaging. Postharvest Biol. Technol. 2018, 139, 115–126. [Google Scholar] [CrossRef]
- Thompson, B. Stepwise Regression and Stepwise Discriminant Analysis Need Not Apply Here: A Guidelines; Sage Publications: Thousand Oaks, CA, USA, 1995. [Google Scholar]
- Lu, J.; Ehsani, R.; Shi, Y.; Abdulridha, J.; de Castro, A.I.; Xu, Y. Field detection of anthracnose crown rot in strawberry using spectroscopy technology. Comput. Electron. Agric. 2017, 135, 289–299. [Google Scholar] [CrossRef]
- Abdel Wahab, H.; Aboelghar, M.; Ali, A.; Yones, M. Spectral and molecular studies on gray mold in strawberry. Asian J. Plant Pathol. 2017, 11, 167–173. [Google Scholar] [CrossRef][Green Version]
- Yuhas, R.H.; Goetz, A.F.H.; Boardman, J.W. Discrimination among semi-arid landscape endmembers using the Spectral AngleMapper (SAM) algorithm. In Summaries of the Third Annual JPL Airborne Geoscience Workshop; AVIRIS Workshop: Pasadena, CA, USA, 1992; pp. 147–149. [Google Scholar]
- Levine, M.F. Self-developed QWL measures. J. Occup. Behav. 1983, 4, 35–46. [Google Scholar]
- Yeh, Y.-H.; Chung, W.-C.; Liao, J.-Y.; Chung, C.-L.; Kuo, Y.-F.; Lin, T.-T. Strawberry foliar anthracnose assessment by hyperspectral imaging. Comput. Electron. Agric. 2016, 122, 1–9. [Google Scholar] [CrossRef]
- Yeh, Y.-H.F.; Chung, W.-C.; Liao, J.-Y.; Chung, C.-L.; Kuo, Y.-F.; Lin, T.-T. A comparison of machine learning methods on hyperspectral plant disease assessments. IFAC Proc. Vol. 2013, 46, 361–365. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C.; Takeda, F.; Kramer, E.A.; Ashrafi, H.; Hunter, J. 3D point cloud data to quantitatively characterize size and shape of shrub crops. Hortic. Res. 2019, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Poliyapram, V.; İmamoğlu, N.; Uto, K.; Nakamura, R.; Kumar, D.N. Canopy Averaged Chlorophyll Content Prediction of Pear Trees Using Convolutional Autoencoder on Hyperspectral Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2020, 13, 1426–1437. [Google Scholar] [CrossRef]
- Li, D.; Li, C.; Yao, Y.; Li, M.; Liu, L. Modern imaging techniques in plant nutrition analysis: A review. Comput. Electron. Agric. 2020, 174, 105459. [Google Scholar] [CrossRef]
- Lu, X.; Liu, Z.; Zhao, F.; Tang, J. Comparison of total emitted solar-induced chlorophyll fluorescence (SIF) and top-of-canopy (TOC) SIF in estimating photosynthesis. Remote. Sens. Environ. 2020, 251, 112083. [Google Scholar] [CrossRef]
- Dechant, B.; Ryu, Y.; Badgley, G.; Zeng, Y.; Berry, J.A.; Zhang, Y.; Goulas, Y.; Li, Z.; Zhang, Q.; Kang, M.; et al. Canopy structure explains the relationship between photosynthesis and sun-induced chlorophyll fluorescence in crops. Remote. Sens. Environ. 2020, 241, 111733. [Google Scholar] [CrossRef]


| Author | Year | Parameters * | Data | Feature Extraction | Optimal Waveband Selection Method ** | Regression Model *** | Prediction Accuracy (R or R2) | Reference |
|---|---|---|---|---|---|---|---|---|
| Weng et al. | 2020 | SSC, pH, and vitamin C | Hyperspectral imaging (range: 374–1020 nm; spectral resolution: 2.31 nm) | All spectral information, 9 color features, and 36 textural features | CARS, UVE | PLSR, SVR, LWR | R2: 0.9370 for SSC, 0.8493 for PH, and 0.8769 for vitamin C | [91] |
| Liu et al. | 2019 | TWSS, glucose, fructose, and sucrose concentrations | Near-infrared hyperspectral imaging (range: 1000–2500 nm; spectral resolution: 6.8 nm) | Spectral information (range: 1085–1780 nm; 5 wavelengths for fructose, glucose, and sucrose; 7 wavelengths for TWSS) | SPA | SVR | R2: 0.589 for fructose, 0.503 for glucose, 0.724 for sucrose, and 0.807 for TWSS | [92] |
| Liu et al. | 2019 | Sugar content | Hyperspectral imaging (range: 391–1043 nm; spectral resolution: 2.8 nm) | Spectral information (range: 420–1007 nm; 76 wavelengths) | CCSD | PLS | R: 0.7708–0.8053 | [93] |
| Amodio et al. | 2017 | SSC, pH, TA, ascorbic acid content, and phenolic content | Fourier-transform (FT)-NIR spectrometer (range: 12,500–3600 cm−1; spectral interval: 8 cm−1) | Spectral information (range: 9401–4597 cm−1, 7507–6094 cm−1, 5454–4597 cm−1, 6103–5446 cm−1, and 4428–4242 cm−1; 219 spectral) | Bruker’s OPUS software | PLSR | R2: 0.85 for TSS, 0.86 for pH, and 0.58 for TA | [94] |
| Li et al. | 2015 | SSC | Near-infrared spectrometer (range: 12,000–3800 cm−1; spectral interval: 1.928 cm−1) | Spectral information (25 wavelengths) | CARS, SPA, MC-UVE | PLSR, MLR | R2: 0.9097 | [95] |
| Ding et al. | 2015 | SSC | Hyperspectral imaging (range: 874–1734 nm; spectral resolution: 5 nm) | Spectral information (range: 941–1612 nm; 14, 17, 24, and 25 wavelengths selected by four methods); 20 spectral features by PCA; 58 spectral features by wavelet transform (WT) | SPA, GAPLS & SPA, Bw, CARS | PLSR | R: >0.9 for SSC | [96] |
| Liu et al. | 2014 | Firmness and SSC | Multispectral imaging system (range: 405–970 nm; 19 wavelengths) | Spectral information (range: 405, 435, 450, 470, 505, 525, 570, 590, 630, 645, 660, 700, 780, 850, 870, 890, 910, 940, and 970 nm; 19 wavelengths) | None | PLSR, SVM, BPNN | R: 0.94 for firmness and 0.83 for SSC | [73] |
| Sánchez et al. | 2012 | SSC and TA | Handheld MEMS-based NIR spectrophotometer (range: 1600–2400 nm; spectral intervals: 12 nm) | Spectral information | MPLS, local algorithm | MPLS, local algorithm | R2: 0.48 for firmness, 0.62 for MPC, 0.69 for SSC, 0.65 for TA, and 0.40 for PH | [97] |
| Nishizawa et al. | 2009 | SSC and glucose, fructose, and sucrose concentrations | Near-infrared (NIR) spectroscopy | Spectral information (range: 700–925 nm) | SMLR | SMLR | R2: 0.86 for SSC, 0.74 for glucose, 0.50 for fructose, and 0.51 for sucrose | [98] |
| Wulf et al. | 2008 | Phenolic compound content | Laser-induced fluorescence spectroscopy (LIFS) (EX: 337 nm; EM: 400–820 nm; spectral interval: 2 nm) | Spectral information | None | PLSR | R2: 0.99 for p-coumaroyl-glucose and cinnamoyl-glucose | [99] |
| ElMasry et al. | 2007 | MC, SSC, and pH | Hyperspectral imaging in visible and near-infrared regions (range: 400–1000 nm; 826 wavelengths) | Spectral information (8, 6, and 8 wavelengths for MC, TSS, and pH, respectively) | β-coefficients from PLS models | MLR | R: 0.87 for MC, 0.80 for SSC, and 0.92 for pH | [90] |
| Tallada et al. | 2006 | Firmness | Near-infrared hyperspectral imaging (range: 650–1000 nm; spectral resolution: 5 nm) | Spectral information | SMLR | SMLR | R: 0.786 for firmness | [100] |
| Nagata et al. | 2005 | Firmness and SSC | Near-infrared hyperspectral imaging (range: 650–1000 nm; spectral resolution: 5 nm) | Spectral information (3 and 5 wavelengths for firmness and SSC, respectively) | SMLR | SMLR | R: 0.786 for firmness, and 0.87 for SSC | [101] |
| Nagata et al. | 2004 | Firmness and SSC | Hyperspectral imaging in visible regions (range: 400–650 nm; spectral resolution: 2 nm) | Spectral information (5 wavelengths for firmness) | SMLR | SMLR | R: 0.784 for firmness | [102] |
| Author | Year | Disease | Description | Reference |
|---|---|---|---|---|
| Mahmud et al. | 2020 | Powdery mildew | Mahmud et al. (2020) designed a mobile machine vision system for strawberry powdery mildew disease detection. The system contains GPS, two cameras, a custom image processing program integrated with color co-occurrence matrix-based texture analysis and ANN classifier, and a ruggedized laptop computer. The highest detection accuracy can reach 98.49%. | [41] |
| Shin et al. | 2020 | Powdery mildew | Shin et al. (2020) used three feature extraction methods (histogram of oriented gradients (HOG), speeded-up robust features (SURF), and gray level co-occurrence matrix (GLCM)) and two supervised learning classifiers (ANNs and SVMs) for the detection of strawberry powdery mildew disease. The classification accuracy was the highest, with 94.34% for ANNs and SURF and 88.98% for SVMs and the GLCM. | [152] |
| Chang et al. | 2019 | Powdery mildew | Chang et al. (2019) extracted 40 textural indices from high-resolution RGB images and compared the performance of three supervised learning classifiers, ANNs, SVMs, and KNNs, in the detection of powdery mildew disease in strawberry. The overall classification accuracy was 93.81%, 91.66%, and 78.80% for the ANN, SVM, and KNN classifiers, respectively. | [149] |
| De Lange, E. S., and Nansen C | 2019 | Arthropod pest influence | De Lange and Nansen (2019) used hyperspectral imaging instruments to detect the spectral response of three stress-induced changes on the strawberry leaves from the influence of three arthropod pests. Large differences were observed from the reflectance data. | [153] |
| Liu et al. | 2019 | Fungal contamination | Liu et al. (2019) combined spatial-spectral information from hyperspectral imaging and aroma information from an electronic nose (E-nose) to estimate external and internal compositions (total soluble solids, titratable acidity) of fungi-infected strawberries during various storage times. PCA was used to extract the features from the hyperspectral images and aroma information. These parameters were highly correlated with microbial content. | [154] |
| Cockerton et al. | 2018 | Verticillium wilt | Cockerton. et al. (2018) collected the high-resolution RGB and multispectral images of strawberry based on the UAV platform to study verticillium wilt resistance of multiple strawberry populations. The NDVI was linked to the disease susceptibility. | [155] |
| Altiparmak et al. | 2018 | Iron deficiency or fungal infection | Altiparmak et al. (2018) proposed a new strawberry leaf disease infection detection and classification method based on only the RGB spectral response value. First, a color-processing detection algorithm (CPDA) was applied to calculate the red and green indices to extract the strawberry leaf from the background and determine the infected area based on the threshold segmentation. Secondly, the fuzzy logic classification algorithm (FLCA) was used to determine the disease type and differentiate iron deficiency from fungal infection. | [156] |
| Siedliska et al. | 2018 | Fungal infection | Siedliska et al. (2018) tried to detect whether strawberry fruits were infected by the fungus using the VNIR/SWIR hyperspectral imaging technology. Nineteen optimal wavelengths were selected by the second derivative of the original spectra, and then the back propagation neural network (BPNN) [157] model was used to differentiate between good and infected fruits, with an accuracy of higher than 97%. The multiple linear regression model was used to estimate the total anthocyanin content (AC) and soluble solid content (SSC). The AC (681 and 1292 nm) and SSC (705, 842, 1162, and 2239 nm) prediction models were tested and produced R2 = 0.65 and R2 = 0.85, respectively. | [158] |
| Lu et al. | 2017 | Anthracnose crown rot | Lu et al. (2017) collected in-field hyperspectral data using a mobile platform on three types of strawberry plants: infected but asymptomatic, infected and symptomatic, and healthy. Thirty-two spectral vegetation indices were used to train the model using stepwise discriminant analysis (SDA) [159], Fisher discriminant analysis (FDA), and KNN algorithms. The achieved classification accuracies were 71.3%, 70.5%, and 73.6% for these three models, respectively. | [160] |
| Wahab et al. | 2017 | Gray mold | Wahab et al. (2017) compared two systems of qPCR and spectroradiometer to detect the gray mold pathogen Botrytis cinerea for infected and healthy strawberry fruits. The results indicated that spectral analysis can effectively detect the gray mold infection and VNIR spectra can distinguish healthy fruits from infected strawberry fruits based on the difference of cellular pigments, while the SWIR can classify infection degrees caused by the cellular structure and water content. | [161] |
| Yeh et al. | 2016 | Foliar anthracnose | Yeh et al. (2016) classified the strawberry leaf images into healthy, incubation, and symptomatic stages of the foliar anthracnose disease based on hyperspectral imaging. Three methods, spectral angle mapper (SAM) [162], SDA, and self-developed correlation measure (CM) [163], were used to carry out the classification. Meanwhile, partial least-squares regression (PLSR) [88], SDA, and CM were also used to select the optimal wavelengths. Wavelengths of 551, 706, 750, and 914 nm were chosen, and the classification accuracy was 80%. | [164] |
| Yeh et al. | 2013 | Foliar anthracnose | Yeh et al. (2013) applied three hyperspectral image analysis methods to determine whether strawberry plants were affected by foliar anthracnose: SDA, SAM, and the proposed simple slope measure (SSM) method. The classified statuses of the strawberry plants were healthy, incubation, and symptomatic. The classification accuracies were 82.0%, 80.7%, and 72.7%, respectively. | [165] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, C.; Abd-Elrahman, A.; Whitaker, V. Remote Sensing and Machine Learning in Crop Phenotyping and Management, with an Emphasis on Applications in Strawberry Farming. Remote Sens. 2021, 13, 531. https://doi.org/10.3390/rs13030531
Zheng C, Abd-Elrahman A, Whitaker V. Remote Sensing and Machine Learning in Crop Phenotyping and Management, with an Emphasis on Applications in Strawberry Farming. Remote Sensing. 2021; 13(3):531. https://doi.org/10.3390/rs13030531
Chicago/Turabian StyleZheng, Caiwang, Amr Abd-Elrahman, and Vance Whitaker. 2021. "Remote Sensing and Machine Learning in Crop Phenotyping and Management, with an Emphasis on Applications in Strawberry Farming" Remote Sensing 13, no. 3: 531. https://doi.org/10.3390/rs13030531
APA StyleZheng, C., Abd-Elrahman, A., & Whitaker, V. (2021). Remote Sensing and Machine Learning in Crop Phenotyping and Management, with an Emphasis on Applications in Strawberry Farming. Remote Sensing, 13(3), 531. https://doi.org/10.3390/rs13030531