Predicting Leaf Nitrogen Content in Cotton with UAV RGB Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Data Acquisition
2.3. Image Preprocessing
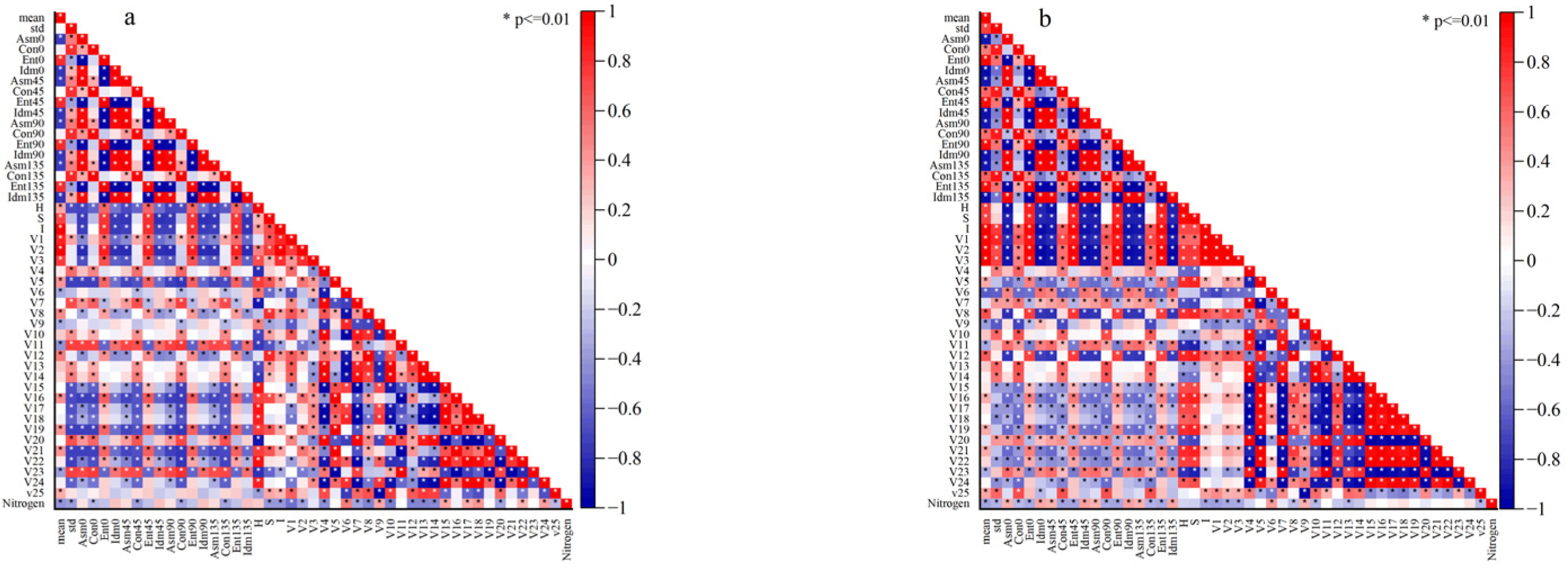
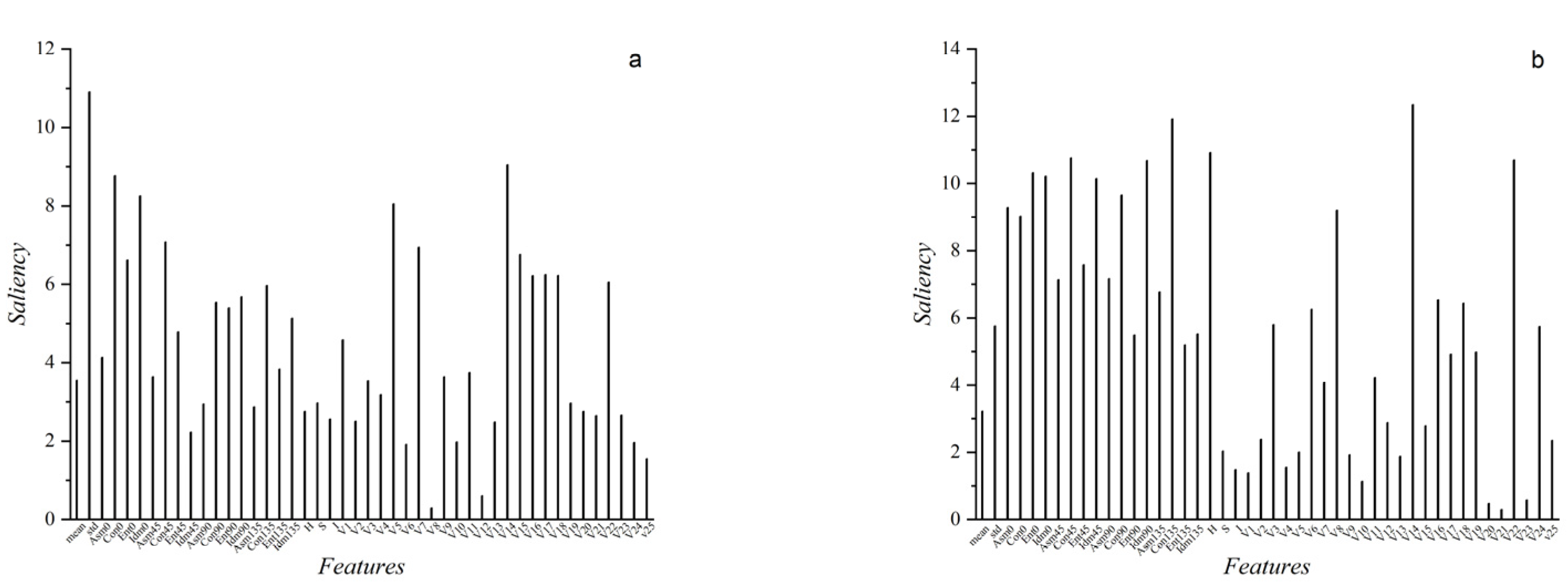
2.4. Feature Extraction
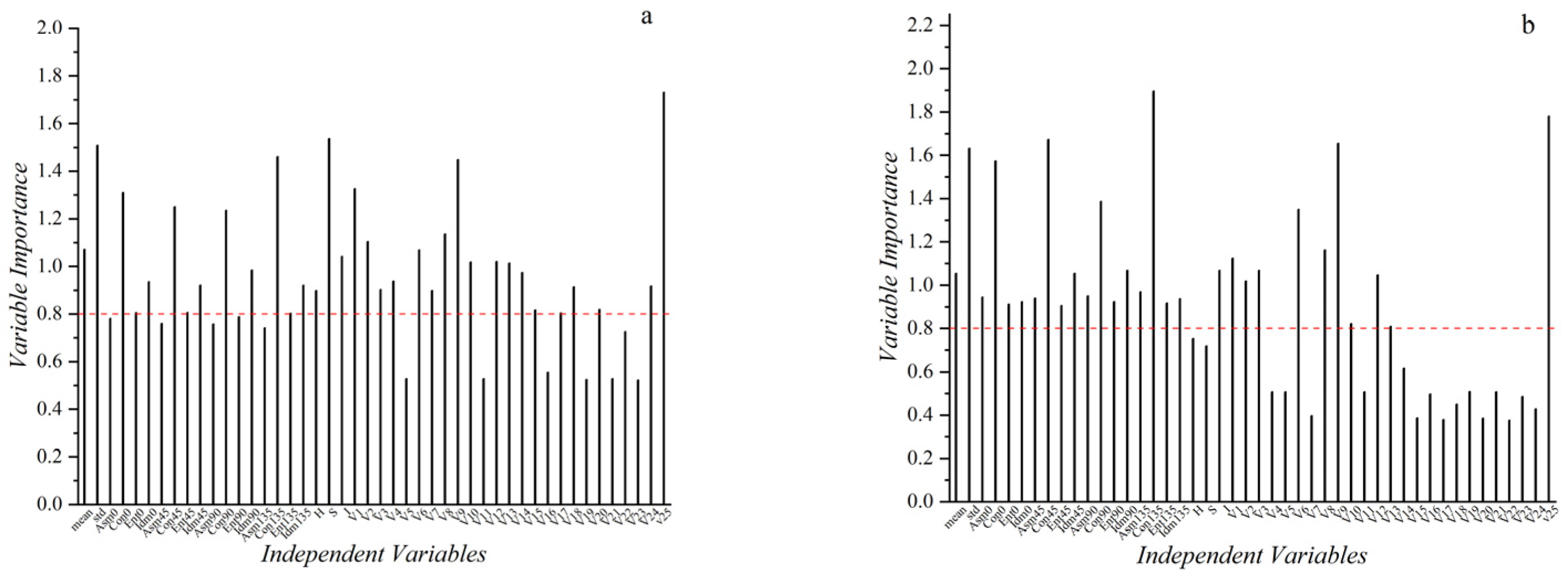
2.5. Feature Dimensionality Reduction
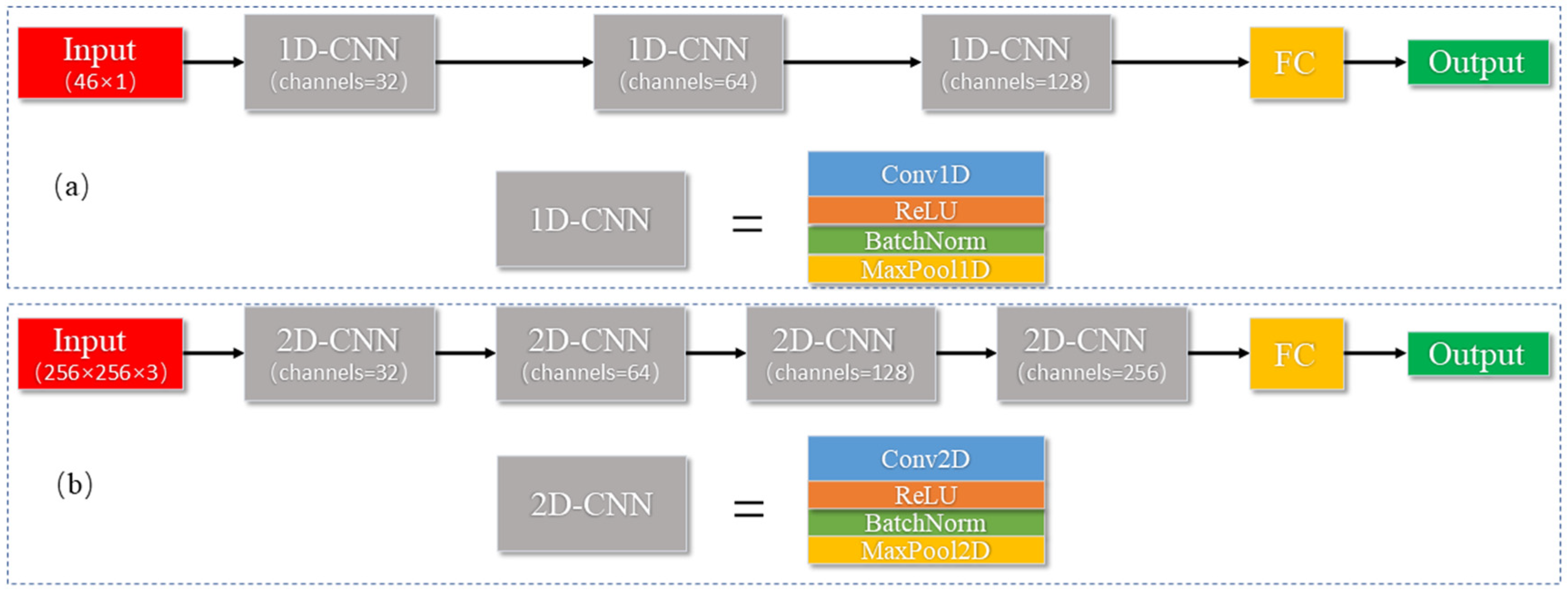
2.6. Model Construction
3. Results and Discussion
3.1. Analysis of Data Dimensionality Reduction Results
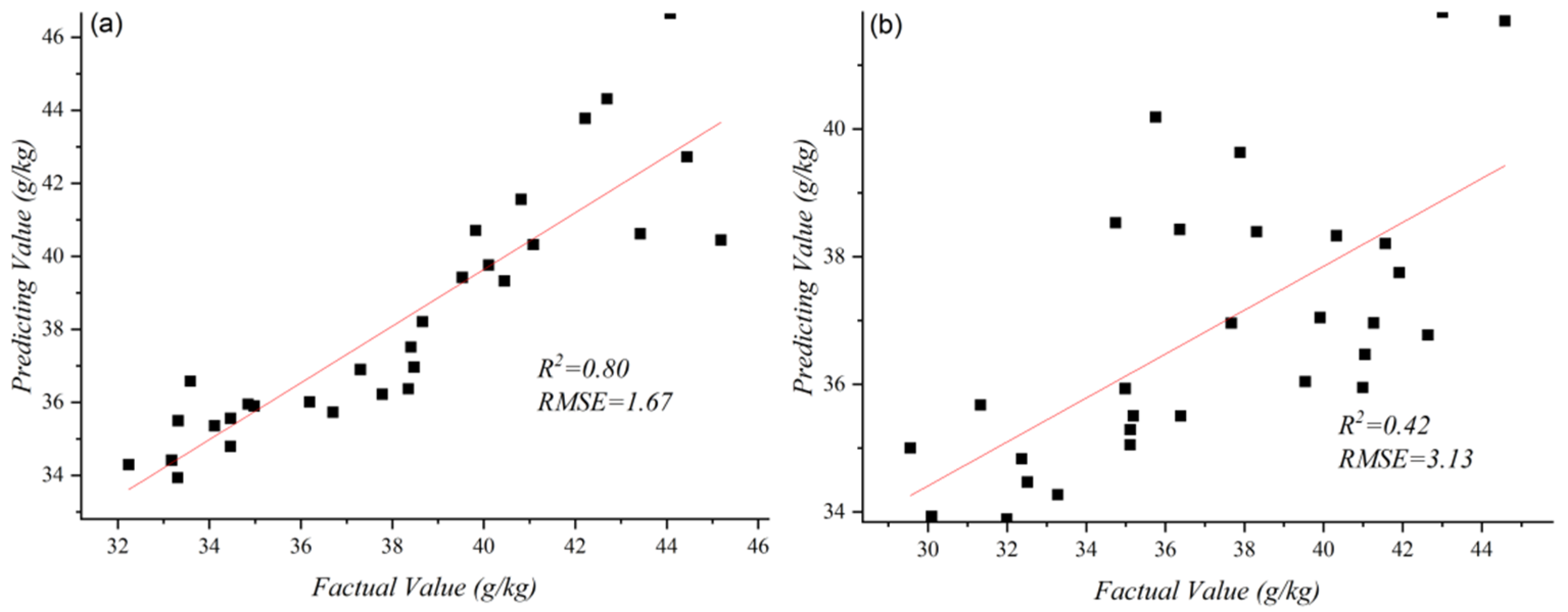
3.2. Manual Feature Regression Model
3.2.1. Results of Traditional Machine Learning Methods
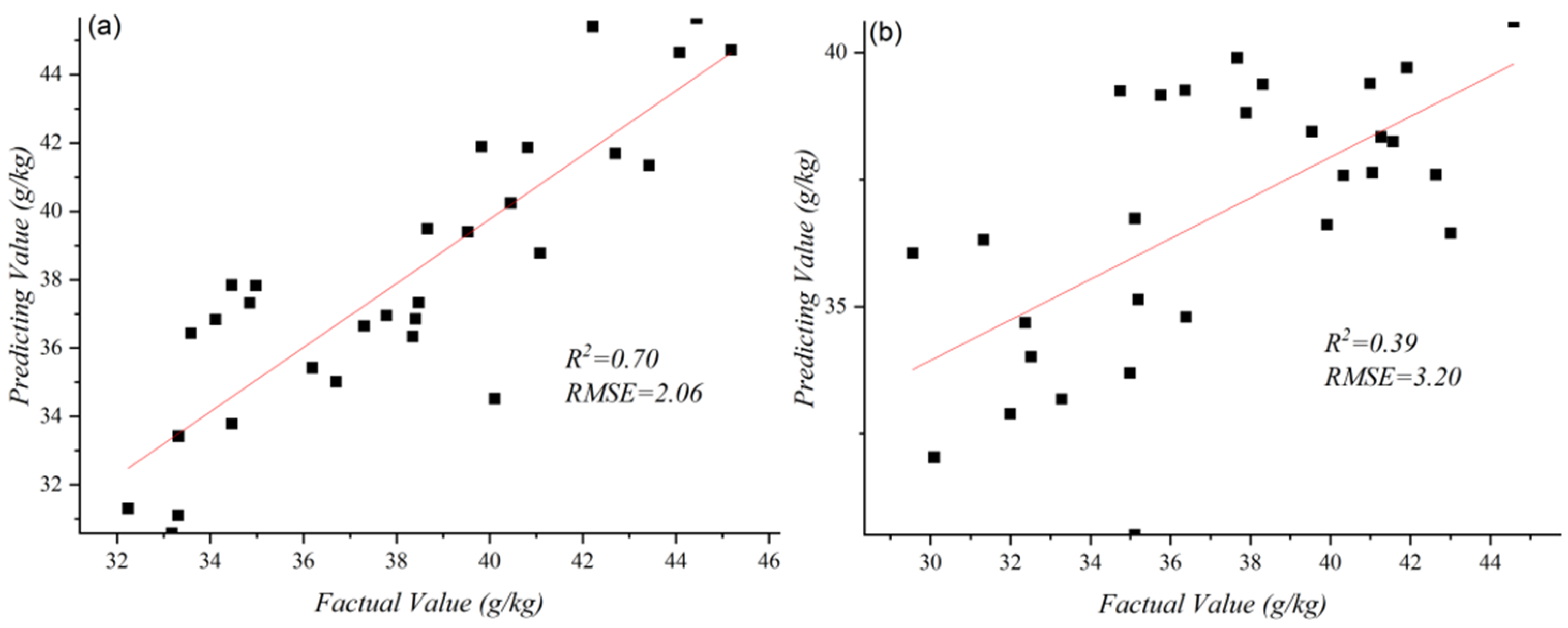
3.2.2. Results of 1D CNN
3.2.3. Results of Deep Feature Regression Model
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Snider, J.; Harris, G.; Roberts, P.; Meeks, C.; Chastain, D.; Bange, M.; Virk, G. Cotton physiological and agronomic response to nitrogen application rate. Field Crops Res. 2021, 270, 108194–108205. [Google Scholar] [CrossRef]
- Banerjee, B.P.; Joshi, S.; Thoday-Kennedy, E.; Pasam, R.K.; Tibbits, J.; Hayden, M.; Spangenberg, G.; Kant, S. High-throughput phenotyping using digital and hyperspectral imaging-derived biomarkers for genotypic nitrogen response. J. Exp. Bot. 2020, 71, 4604–4615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Wang, J.; Leblon, B. Using Linear Regression, Random Forests, and Support Vector Machine with Unmanned Aerial Vehicle Multispectral Images to Predict Canopy Nitrogen Weight in Corn. Remote Sens. 2020, 12, 2071. [Google Scholar] [CrossRef]
- Zhao, W.; Guo, Z.; Yue, J.; Zhang, X.; Luo, L. On combining multiscale deep learning features for the classification of hyperspectral remote sensing imagery. Int. J. Remote Sens. 2015, 36, 3368–3379. [Google Scholar] [CrossRef]
- Liu, S.; Yang, G.; Jing, H.; Feng, H.; Li, H.; Chen, P.; Yang, W. Retrieval of winter wheat nitrogen content based on UAV digital image. Trans. Chin. Soc. Agric. Eng. 2019, 35, 75–85. (In Chinese) [Google Scholar]
- Xu, S.; Wang, M.; Shi, X.; Yu, Q.; Zhang, Z. Integrating hyperspectral imaging with machine learning techniques for the high-resolution mapping of soil nitrogen fractions in soil profiles. Sci. Total Environ. 2021, 754, 142135–142151. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Zhang, C.; Bai, X.; Du, X.; He, Y. Discrimination of Chrysanthemum Varieties Using Hyperspectral Imaging Combined with a Deep Convolutional Neural Network. Molecules 2018, 23, 2831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, T.; Xu, W.; Lin, J.; Duan, L.; Gao, P.; Zhang, C.; Lv, X. Combining Multi-Dimensional Convolutional Neural Network (CNN) With Visualization Method for Detection of Aphis gossypii Glover Infection in Cotton Leaves Using Hyperspectral Imaging. Front. Plant Sci. 2021, 12, 604510–604525. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Mounir, L.; Borman, M.; Johnson, D. Spatially Located Platform and Aerial Photography for Documentation of Grazing Impacts on Wheat. Geocarto Int. 2001, 16, 65–70. [Google Scholar]
- Meyer, G.E.; Neto, J.C. Verification of color vegetation indices for automated crop imaging applications. Comput. Electron. Agric. 2008, 63, 282–293. [Google Scholar]
- Woebbecke, D.M.U.O.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color Indices for Weed Identification Under Various Soil, Residue, and Lighting Conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar]
- Kataoka, T.; Kaneko, T.; Okamoto, H.; Hata, S. Crop growth estimation system using machine vision. In Proceedings of the 2003 IEEE/ASME International Conference on Advanced Intelligent Mechatronics (AIM 2003), Kobe, Japan, 20–24 July 2003; Volume 2, pp. b1079–b1083. [Google Scholar]
- Gitelson, A.A.; Andrés Via Arkebauer, T.J.; Rundquist, D.C.; Keydan, G. Remote estimation of leaf area index and green leaf biomass in maize canopies. Geophys. Res. Lett. 2003, 30, 1248–1252. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; Lv, Y.; Zhu, H.; Zhao, Y.; He, Y. Identification and Classification of Different Producing Years of Dried Tangerine Using Hyperspectral Technique with Chemometrics Models. Spectrosc. Spectr. Anal. 2017, 37, 1866–1871. (In Chinese) [Google Scholar]
- Panahi, M.; Sadhasivam, N.; Pourghasemi, H.R.; Rezaie, F.; Lee, S. Spatial prediction of groundwater potential mapping based on convolutional neural network and support vector regression. J. Hydrol. 2020, 588, 125033–125045. [Google Scholar] [CrossRef]
- Sun, X.; Liu, L.; Li, C.; Yin, J.; Zhao, J.; Si, W. Classification for Remote Sensing Data with Improved CNN-SVM Method. IEEE Access 2019, 7, 164507–164516. [Google Scholar] [CrossRef]
- Chen, J.; Fan, Y.; Wang, T.; Zhang, C.; Qiu, Z.; He, Y. Automatic Segmentation and Counting of Aphid Nymphs on Leaves Using Convolutional Neural Networks. Agronomy 2018, 8, 129. [Google Scholar] [CrossRef] [Green Version]
- Pyo, J.; Duan, H.; Baek, S.; Kim, M.S.; Jeon, T.; Kwon, Y.S.; Lee, H.; Cho, K.H. A convolutional neural network regression for quantifying cyanobacteria using hyperspectral imagery. Remote Sens. Environ. 2019, 233, 111350. [Google Scholar]



| Variety | Data Set | June 28 | July 6 | July 16 | July 28 | August 7 | Total |
|---|---|---|---|---|---|---|---|
| Xinluzao45 | Training set | 12 | 12 | 12 | 12 | 12 | 60 |
| Test set | 6 | 6 | 6 | 6 | 6 | 30 | |
| Xinluzao53 | Training set | 12 | 12 | 12 | 12 | 12 | 60 |
| Test set | 6 | 6 | 6 | 6 | 6 | 30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kou, J.; Duan, L.; Yin, C.; Ma, L.; Chen, X.; Gao, P.; Lv, X. Predicting Leaf Nitrogen Content in Cotton with UAV RGB Images. Sustainability 2022, 14, 9259. https://doi.org/10.3390/su14159259
Kou J, Duan L, Yin C, Ma L, Chen X, Gao P, Lv X. Predicting Leaf Nitrogen Content in Cotton with UAV RGB Images. Sustainability. 2022; 14(15):9259. https://doi.org/10.3390/su14159259
Chicago/Turabian StyleKou, Jinmei, Long Duan, Caixia Yin, Lulu Ma, Xiangyu Chen, Pan Gao, and Xin Lv. 2022. "Predicting Leaf Nitrogen Content in Cotton with UAV RGB Images" Sustainability 14, no. 15: 9259. https://doi.org/10.3390/su14159259
APA StyleKou, J., Duan, L., Yin, C., Ma, L., Chen, X., Gao, P., & Lv, X. (2022). Predicting Leaf Nitrogen Content in Cotton with UAV RGB Images. Sustainability, 14(15), 9259. https://doi.org/10.3390/su14159259







