RT-dPCR in Mosquito Samples for ZIKV Detection: Effects of RNA Extraction and Reverse Transcription in Target Concentration
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. RNA Extraction and cDNA Synthesis
2.3. RT-dPCR
2.4. Data Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moureau, G.; Cook, S.; Lemey, P.; Nougairede, A.; Forrester, N.; Khasnatinov, M.; Charrel, R.N.; Firth, A.E.; Gould, E.A.; Lamballerie, X. New insights into Flavivirus evolution, taxonomy and biogeographic history, extended by analysis of canonical and alternative coding sequences. PLoS ONE 2015, 10, e0117849. [Google Scholar] [CrossRef] [PubMed]
- Fauver, J.R.; Grubaugh, N.D.; Krajacich, B.J.; Weger-Lucarelli, J.; Lakin, S.M.; Fakoli, L.S.; Bolay, F.K.; Diclaro, J.W.; Dabiré, K.R.; Foy, B.D.; et al. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology 2015, 498, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.L.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Inf. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Mlakar, J.; Korva, M.; Tul, N.; Popovic, M.; Pljsak-Prijatelj, M.; Mraz, J.; Kolenk, M.; Rus, K.R.; Vipotnik, T.V.; Vodusek, V.F.; et al. Zika virus associated with microcephaly. N. Eng. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, N.; Wessel, T.; Marks, J. Digital pcr modeling for maximal sensitivity, dynamic range and measurement precision. PLoS ONE 2015, 10, e0118833. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Liu, Y.; Liu, J.; Zhao, J.; Champagne, C.; Tong, L.; Zhang, R.; Zhang, F.; Qin, C.; Ma, P.; et al. Aedes mosquitoes acquire and transmit Zika virus by breeding in contaminated aquatic environments. Nat. Commun. 2019, 10, 1324. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Yeo, J.; Kim, B.; Ha, M.; Kim, V.N. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol. Cell. 2012, 46, 893–895. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.; Dhillon, H.S.; Kirvell, S.; Greenwood, C.; Parker, M.; Shipley, G.L.; Nolan, T. Variability of the reverse transcription step: Practical implications. Clin. Chem. 2015, 61, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Minshal, N.; Git, A. Enzyme- and gene-specific biases in reverse transcription of RNA raise concerns for evaluating gene expression. Sci. Rep. 2020, 10, 8151. [Google Scholar] [CrossRef] [PubMed]
- Schwaber, J.; Andersen, S.; Nielsen, L. Shedding light: The importance of reverse transcription efficiency standards in data interpretation. Biomol. Detect. Quantif. 2019, 17, 100077. [Google Scholar] [CrossRef] [PubMed]
- Christopherson, C.; John Sninsky, J.; Kwok, S. The Effects of Internal Primer-Template Mismatches on RT-PCR: HIV-1 Model Studies. Nucleic Acids Res. 1997, 25, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Beerntsen, B.T.; James, A.A.; Christensen, B.M. Genetics of Mosquito Vector Competence. Microbiol. Mol. Biol. Rev. 2000, 64, 115–137. [Google Scholar] [CrossRef]
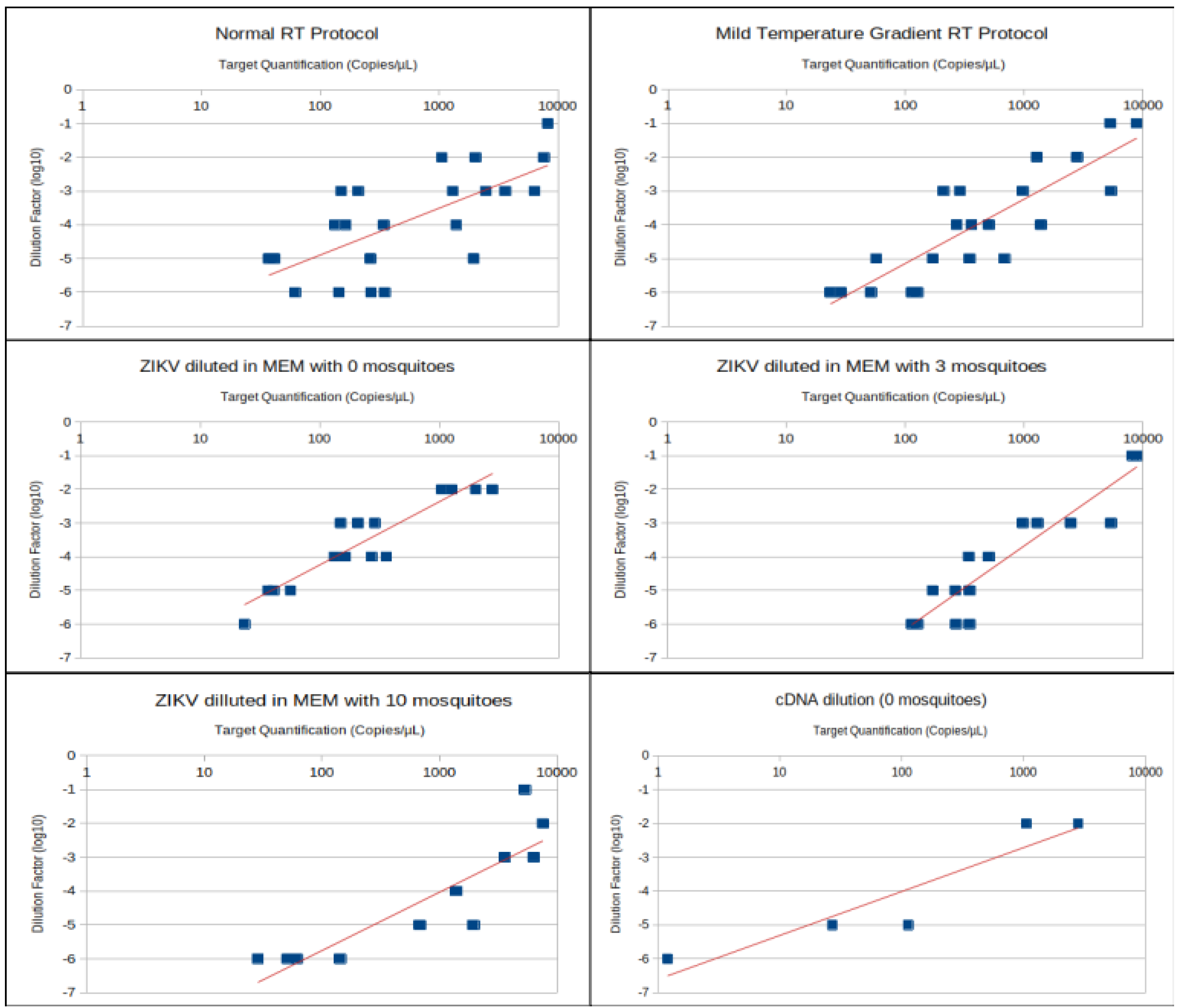
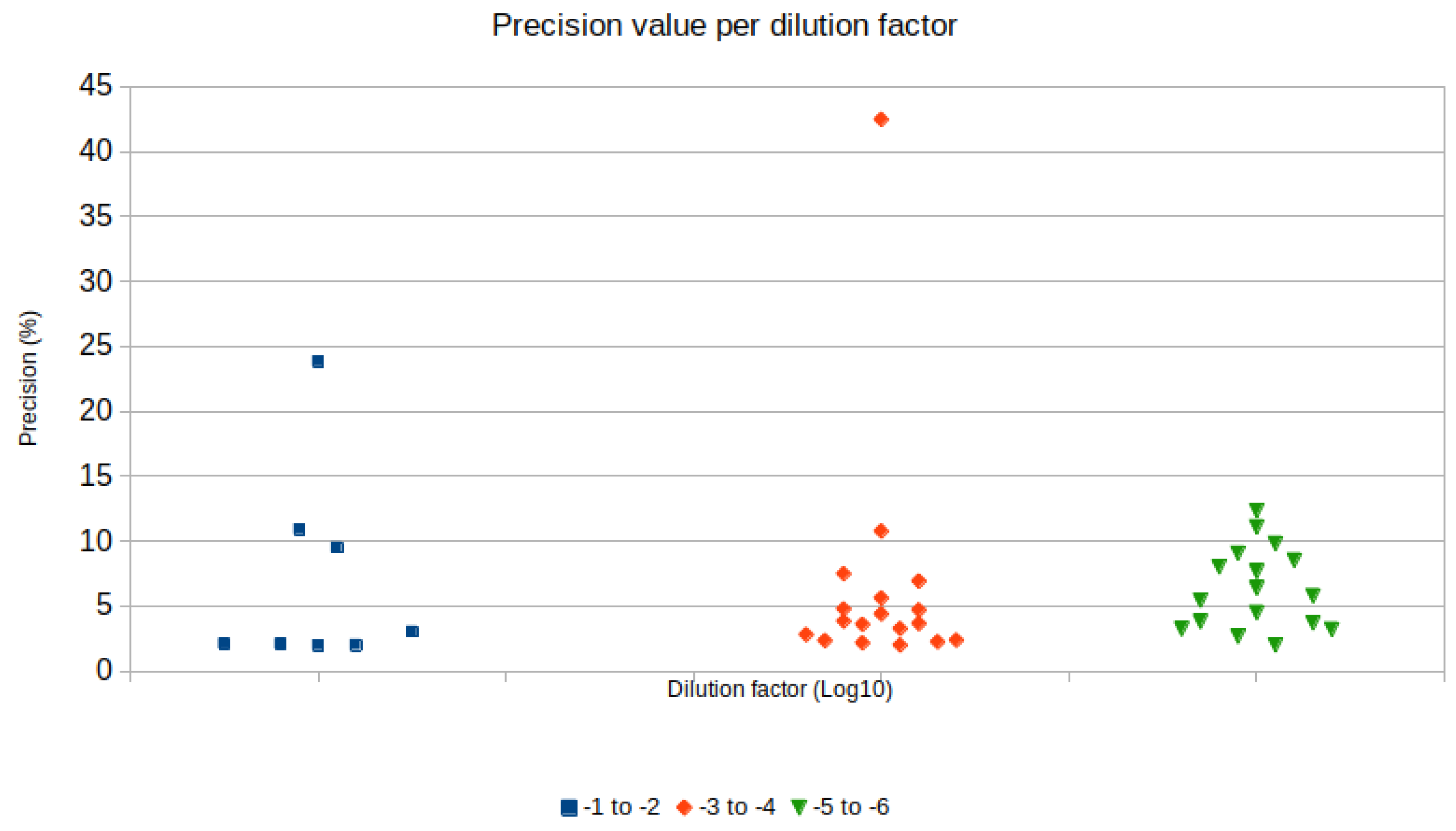
| Treatment | Number of Samples Used for r Calculation | Correlation (r) |
|---|---|---|
| 0 mosquitoes (normal RT) | 8 | 0.765 |
| 0 mosquitoes (MTG RT) | 8 | 0.702 |
| 3 mosquitoes (normal RT) | 7 | 0.845 |
| 3 mosquitoes (MTG RT) | 8 | 0.86 |
| 10 mosquitoes (normal RT) | 7 | 0.943 |
| 10 mosquitoes (MTG RT) | 5 | 0.988 |
| 0 mosquitoes (total) | 16 | 0.713 |
| 3 mosquitoes (total) | 15 | 0.848 |
| 10 mosquitoes (total) | 12 | 0.89 |
| Normal RT (total) | 22 | 0.65 |
| MTG RT (total) | 21 | 0.726 |
| Sample/RT Protocol | Copies/µL | CI | Precision | Sample Dilution | cDNA Dilution |
|---|---|---|---|---|---|
| 1:100 MTG | 2794.8 | 2737.1–2853.7 | 2.11% | 10−2 | pure |
| 1:100 2T | 1052.1 | 1031.2–1073.4 | 2.02% | 10−2 | pure |
| 1:10 MTG | 112.74 | 106.65–119.17 | 5.71% | 10−1 | 10−4 |
| 1:10 2T RT | 26.858 | 24.105–29.927 | 11.42% | 10−1 | 10−4 |
| 1:100 MTG | 1.197 | 0.695–2.061 | 72.22% | 10−2 | 10−4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, P.R.d.; Eisen, A.K.A.; Demoliner, M.; Spilki, F.R. RT-dPCR in Mosquito Samples for ZIKV Detection: Effects of RNA Extraction and Reverse Transcription in Target Concentration. Viruses 2020, 12, 827. https://doi.org/10.3390/v12080827
Almeida PRd, Eisen AKA, Demoliner M, Spilki FR. RT-dPCR in Mosquito Samples for ZIKV Detection: Effects of RNA Extraction and Reverse Transcription in Target Concentration. Viruses. 2020; 12(8):827. https://doi.org/10.3390/v12080827
Chicago/Turabian StyleAlmeida, Paula Rodrigues de, Ana Karolina Antunes Eisen, Meriane Demoliner, and Fernando Rosado Spilki. 2020. "RT-dPCR in Mosquito Samples for ZIKV Detection: Effects of RNA Extraction and Reverse Transcription in Target Concentration" Viruses 12, no. 8: 827. https://doi.org/10.3390/v12080827
APA StyleAlmeida, P. R. d., Eisen, A. K. A., Demoliner, M., & Spilki, F. R. (2020). RT-dPCR in Mosquito Samples for ZIKV Detection: Effects of RNA Extraction and Reverse Transcription in Target Concentration. Viruses, 12(8), 827. https://doi.org/10.3390/v12080827







