Identification of Oil Tea (Camellia oleifera C.Abel) Cultivars Using EfficientNet-B4 CNN Model with Attention Mechanism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data Acquisition and Dataset Construction
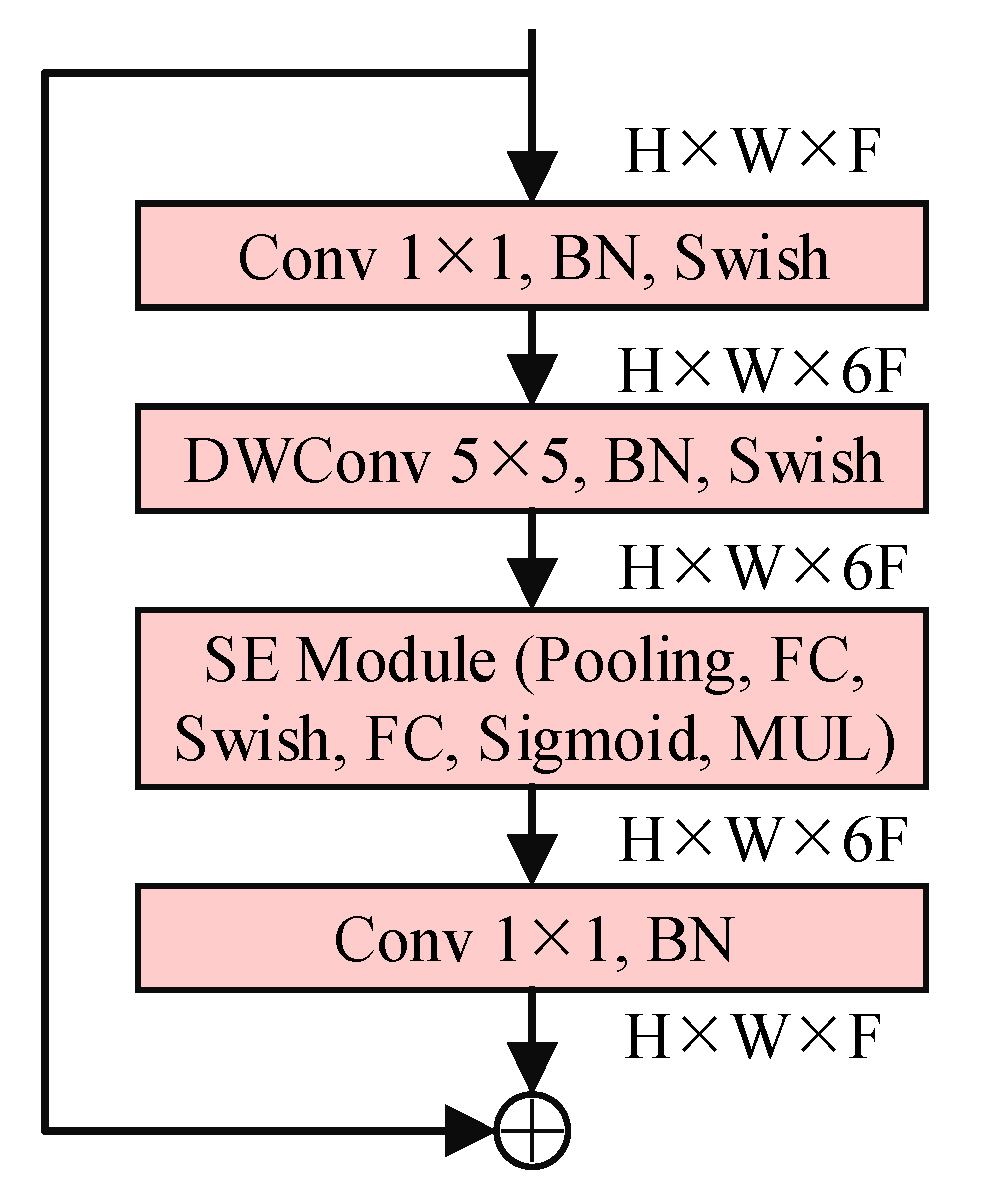
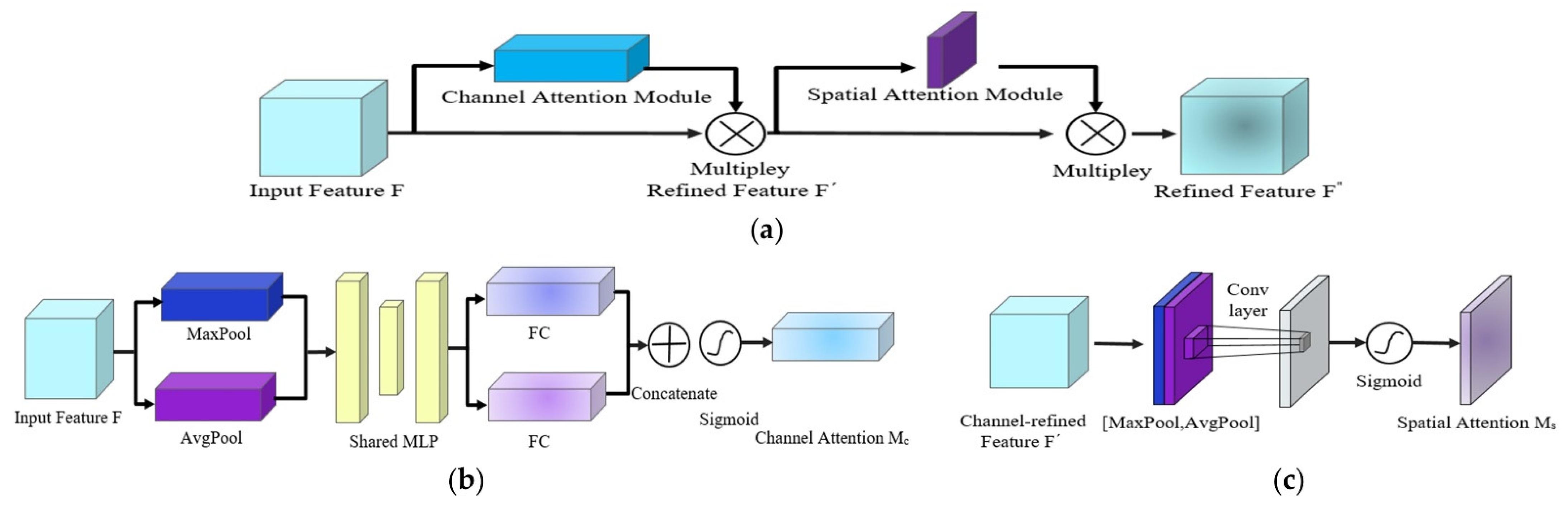
2.3. EfficientNet-B4-CBAM Model
2.4. Evaluation Indicators
2.5. Experimental Environment Configuration
3. Results and Discussion
3.1. Analysis Results of Cultivar Identification Using EfficientNet-B4-CBAM
3.2. Comparison of Cultivar Identification Results with Different Models
3.3. Visual Analysis of Cultivar Identification Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wu, L.; Li, J.; Li, Z.; Zhang, F.; Tan, X. Transcriptomic analyses of Camellia oleifera ‘Huaxin’ leaf reveal candidate genes related to long-term cold stress. Int. J. Mol. Sci. 2020, 21, 846. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Wang, A.; Qin, M.; Qin, X.; Yang, S.; Su, S.; Sun, Y.; Zhang, L. Direct and indirect somatic embryogenesis induction in Camellia oleifera Abel. Front. Plant Sci. 2021, 12, 644389. [Google Scholar] [CrossRef]
- Han, J.; Sun, R.; Zeng, X.; Zhang, J.; Xing, R.; Sun, C.; Chen, Y. Rapid classification and quantification of Camellia (Camellia oleifera Abel.) oil blended with rapeseed oil using FTIR-ATR spectroscopy. Molecules 2020, 25, 2036. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Li, J.; Gao, C.; Cheng, J.; Deng, X.; Jiang, D.; Li, L.; Yan, P. New perspective for evaluating the main Camellia oleifera cultivars in China. Sci. Rep. 2020, 10, 20676. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, B.; Chen, J.; Wang, X.; Wang, R.; Peng, S.; Chen, L.; Ma, L.; Luo, J. Identification of rubisco rbcL and rbcS in Camellia oleifera and their potential as molecular markers for selection of high tea oil cultivars. Front. Plant Sci. 2015, 6, 189. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.; Su, S.; Ma, L.; Yang, S.; Wang, Y.; Wang, X. Effects of canopy microclimate on fruit yield and quality of Camellia oleifera. Sci. Hortic. 2018, 235, 132–141. [Google Scholar] [CrossRef]
- Zeng, W.; Endo, Y. Effects of Cultivars and geography in China on the lipid characteristics of Camellia oleifera seeds. J. Oleo Sci. 2019, 68, 1051–1061. [Google Scholar] [CrossRef] [Green Version]
- Sahari, M.; Ataii, D.; Hamedi, M. Characteristics of tea seed oil in comparison with sunflower and olive oils and its effect as a natural antioxidant. J. Am. Oil Chem. Soc. 2004, 81, 585–588. [Google Scholar] [CrossRef]
- Xu, Z.; Cao, Z.; Yao, H.; Li, C.; Zhao, Y.; Yuan, D.; Yang, G. The physicochemical properties and fatty acid composition of two new woody oil resources: Camellia hainanica seed oil and Camellia sinensis seed oil. CyTA-J. Food 2021, 19, 208–211. [Google Scholar] [CrossRef]
- He, Z.; Liu, C.; Wang, X.; Wang, R.; Tian, Y.; Chen, Y. Leaf transcriptome and weight gene co-expression network analysis uncovers genes associated with photosynthetic efficiency in Camellia oleifera. Biochem. Genet. 2021, 59, 398–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, L.; Chen, J.; Chen, Y.; Zhang, Z.; Wang, X.; Peng, Y.; Peng, S.; Li, A.; Wei, X. Different nitrate and ammonium ratios affect growth and physiological characteristics of Camellia oleifera Abel. seedlings. Forests 2018, 9, 784. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Chen, L.; Tang, W.; Peng, S.; Li, M.; Deng, N.; Chen, Y. Predicting potential distribution and evaluating suitable soil condition of oil tea Camellia in China. Forests 2018, 9, 487. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Jiang, D.; Cheng, H.; Zhou, X.; Fang, Y.; Zhang, X.; Xiao, X.; Deng, X.; Li, L. Determination of Camellia oleifera Abel. germplasm resources of genetic diversity in China using ISSR markers. Not. Bot. Horti Agrobot. Cluj-Napoca 2018, 46, 501–508. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Jung, J.; Shim, E.; Chung, S.; Park, Y.; Lee, G.; Sim, S. Genome-wide SNP discovery and core marker sets for DNA barcoding and variety identification in commercial tomato cultivars. Sci. Hortic. 2021, 276, 109734. [Google Scholar] [CrossRef]
- Shamustakimova, A.; Mavlyutov, Y.; Klimenko, I. Application of SRAP markers for DNA identification of Russian alfalfa Cultivars. Russ. J. Genet. 2021, 57, 540–547. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.; Zheng, D.; Wang, W. Nondestructive identification of millet varieties using hyperspectral imaging technology. J. Appl. Spectrosc. 2020, 87, 54–61. [Google Scholar] [CrossRef]
- Zhao, F.; Yan, Z.; Xue, J.; Xu, B. Identification of wild black and cultivated goji berries by hyperspectral image. Spectrosc. Spectr. Anal. 2021, 41, 201–205. [Google Scholar] [CrossRef]
- Sachar, S.; Kumar, A. Survey of feature extraction and classification techniques to identify plant through leaves. Expert Syst. Appl. 2021, 167, 114181. [Google Scholar] [CrossRef]
- Waldchen, J.; Mader, P. Machine learning for image based species identification. Methods Ecol. Evol. 2018, 9, 2216–2225. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, L. Traditional system versus DNA barcoding in identification of bamboo species: A systematic review. Mol. Biotechnol. 2021, 63, 651–675. [Google Scholar] [CrossRef]
- Li, H.; Zhang, L.; Sun, H.; Rao, Z.; Ji, H. Identification of soybean varieties based on hyperspectral imaging technology and one-dimensional convolutional neural network. J. Food Process Eng. 2021, 44, e13767. [Google Scholar] [CrossRef]
- Yang, C. Plant leaf recognition by integrating shape and texture features. Pattern Recognit. 2021, 112, 107809. [Google Scholar] [CrossRef]
- Alexandru Borz, S.; Păun, M. Integrating offline object tracking, signal processing, and artificial intelligence to classify relevant events in sawmilling operations. Forests 2020, 11, 1333. [Google Scholar] [CrossRef]
- Roshani, M.; Sattari, M.A.; Ali, P.J.M.; Roshani, G.H.; Nazemi, B.; Corniani, E.; Nazemi, E. Application of GMDH neural network technique to improve measuring precision of a simplified photon attenuation based two-phase flowmeter. Flow Meas. Instrum. 2020, 75, 101804. [Google Scholar] [CrossRef]
- Javanmardi, S.; Ashtiani, S.; Verbeek, F.; Martynenko, A. Computer-vision classification of corn seed varieties using deep convolutional neural network. J. Stored Prod. Res. 2021, 92, 101800. [Google Scholar] [CrossRef]
- Jin, X.; Che, J.; Chen, Y. Weed identification using deep learning and image processing in vegetable plantation. IEEE Access 2021, 9, 10940–10950. [Google Scholar] [CrossRef]
- Cheţa, M.; Marcu, M.V.; Iordache, E.; Borz, S.A. Testing the capability of low-cost tools and artificial intelligence techniques to automatically detect operations done by a small-sized manually driven bandsaw. Forests 2020, 11, 739. [Google Scholar] [CrossRef]
- Zhang, R.; Tian, Y.; Zhang, J.; Dai, S.; Hou, X.; Wang, J.; Guo, Q. Metric learning for image-based flower cultivars identification. Plant Methods 2021, 17, 65. [Google Scholar] [CrossRef]
- Yang, B.; Gao, Z.; Gao, Y.; Zhu, Y. Rapid detection and counting of wheat ears in the field using YOLOv4 with attention module. Agronomy 2021, 11, 1202. [Google Scholar] [CrossRef]
- Kaya, A.; Keceli, A.; Catal, C.; Yalic, H.; Temucin, H.; Tekinerdogan, B. Analysis of transfer learning for deep neural network based plant classification models. Comput. Electron. Agric. 2019, 158, 20–29. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, Y.; Yan, T.; Bai, X.; Xiao, Q.; Gao, P.; Li, M.; Huang, W.; Bao, Y.; He, Y.; et al. Application of near-infrared hyperspectral imaging for variety identification of coated maize kernels with deep learning. Infrared Phys. Technol. 2020, 111, 103550. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, J.; Tian, Y.; Dai, S. Deep learning for image-based large-flowered chrysanthemum cultivar recognition. Plant Methods 2019, 4, 146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dong, Z.; Chen, X.; Jia, W.; Du, S.; Muhammad, K.; Wang, S. Image based fruit category classification by 13-layer deep convolutional neural network and data augmentation. Multimed. Tools Appl. 2019, 78, 3613–3632. [Google Scholar] [CrossRef]
- Ponce, J.; Aquino, A.; Andújar, J. Olive-fruit variety classification by means of image processing and convolutional neural networks. IEEE Access 2019, 7, 147629–147641. [Google Scholar] [CrossRef]
- Rodríguez, F.; García, A.; Pardo, P.; Chávez, F.; Luque-Baena, R. Study and classification of plum varieties using image analysis and deep learning techniques. Prog. Artif. Intell. 2018, 7, 119–127. [Google Scholar] [CrossRef]
- Wei, X.; Xie, C.; Wu, J.; Shen, C. Mask-CNN: Localizing parts and selecting descriptors for fine-grained bird species categorization. Pattern Recogn. 2018, 76, 704–714. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar] [CrossRef] [Green Version]
- Nazemi, B.; Rafiean, M. Forecasting house prices in Iran using GMDH. Int. J. Hous. Mark. Anal. 2021, 14, 555–568. [Google Scholar] [CrossRef]
- Borz, S.A. Development of a modality-invariant multi-layer perceptron to predict operational events in motor-manual willow felling operations. Forests 2021, 12, 406. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.; Kweon, I. CBAM: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision ECCV 2018, Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Ni, J.; Gao, J.; Han, Z.; Luan, T. A novel method for peanut variety identification and classification by improved VGG16. Sci. Rep. 2021, 11, 15756. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, J.; Wu, X.; Shen, J.; Lu, B.; Tan, W. Identification of crop diseases using improved convolutional neural networks. IET Comput. Vis. 2020, 14, 538–545. [Google Scholar] [CrossRef]
- Spiesman, B.; Gratton, C.; Hatfield, R.; Hsu, W.; Jepsen, S.; McCornack, B.; Patel, K.; Wang, G. Assessing the potential for deep learning and computer vision to identify bumble bee species from images. Sci. Rep. 2021, 11, 7580. [Google Scholar] [CrossRef]
- Heidary-Sharifabad, A.; Zarchi, M.; Emadi, S.; Zarei, G. An efficient deep learning model for cultivars identification of a pistachio tree. Br. Food J. 2021, 123, 3592–3609. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.; Suzauddola, M.; Nanehkaran, Y.; Sun, Y. Identification of plant disease images via a squeeze-and-excitation MobileNet model and twice transfer learning. IET Image Process. 2021, 15, 1115–1127. [Google Scholar] [CrossRef]
- Barré, P.; Stöver, B.; Müller, K.; Steinhage, V. LeafNet: A computer vision system for automatic plant species identification. Ecol. Inform. 2017, 40, 50–56. [Google Scholar] [CrossRef]
- Xu, R.; Lin, H.; Lu, K.; Cao, L.; Liu, Y. A forest fire detection system based on ensemble learning. Forests 2021, 12, 217. [Google Scholar] [CrossRef]
- Mi, Z.; Zhang, X.; Su, J.; Han, D.; Su, B. Wheat stripe rust grading by deep learning with attention mechanism and images from mobile devices. Front. Plant Sci. 2020, 11, 558126. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Zhang, Y.; Su, S.; Yang, S.; Ma, L.; Zhang, L.; Wang, X. Effects of tree shape on the microclimate and fruit quality parameters of Camellia oleifera Abel. Forests 2019, 10, 563. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Tian, X.; Chai, G.; Zhang, X.; Chen, E. A new CBAM-P-Net model for few-shot forest species classification using airborne hyperspectral images. Remote Sens. 2021, 13, 1269. [Google Scholar] [CrossRef]
- Selvaraju, R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Li, Z.; Zhou, J.; Gu, Y.; Tan, X. Comparative study on fruit development and oil synthesis in two cultivars of Camellia oleifera. BMC Plant Biol. 2021, 21, 348. [Google Scholar] [CrossRef]





| Dataset | Xinglin 210 | Huashuo | Huajin | Huaxin |
|---|---|---|---|---|
| Training dataset | 795 | 861 | 592 | 597 |
| Validation dataset | 264 | 285 | 195 | 196 |
| Testing dataset | 264 | 285 | 195 | 196 |
| Cultivars | TP | TN | FP | FN | ACC (%) | P (%) | R (%) | F1-score (%) | OA (%) | Kc |
|---|---|---|---|---|---|---|---|---|---|---|
| Xianglin 210 | 260 | 671 | 5 | 4 | 99.04 | 98.11 | 98.48 | 98.29 | 97.02 | 0.96 |
| Huashuo | 278 | 642 | 13 | 7 | 97.87 | 95.53 | 97.54 | 96.52 | ||
| Huajin | 184 | 738 | 7 | 11 | 98.09 | 96.34 | 94.36 | 95.34 | ||
| Huaxin | 190 | 741 | 3 | 6 | 99.04 | 98.44 | 96.94 | 97.68 |
| Model | Cultivars | TP | TN | FP | FN | ACC (%) | P (%) | R (%) | F1-score (%) | OA (%) | Kc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| InceptionV3 | Xianglin 210 | 151 | 624 | 52 | 113 | 82.45 | 74.38 | 57.20 | 64.67 | 69.04 | 0.59 |
| Huashuo | 190 | 589 | 66 | 95 | 82.87 | 74.22 | 66.67 | 70.24 | |||
| Huajin | 160 | 684 | 61 | 35 | 89.79 | 72.40 | 82.05 | 76.92 | |||
| Huaxin | 148 | 632 | 112 | 48 | 82.98 | 56.92 | 75.51 | 64.91 | |||
| VGG16 | Xianglin 210 | 235 | 585 | 91 | 29 | 87.23 | 72.09 | 89.02 | 79.67 | 72.02 | 0.62 |
| Huashuo | 213 | 557 | 98 | 72 | 81.91 | 68.49 | 74.74 | 71.48 | |||
| Huajin | 112 | 732 | 13 | 83 | 89.79 | 89.60 | 57.44 | 70.00 | |||
| Huaxin | 117 | 683 | 61 | 79 | 85.11 | 65.73 | 59.69 | 62.56 | |||
| ResNet50 | Xianglin 210 | 235 | 647 | 29 | 29 | 93.83 | 89.02 | 89.02 | 89.02 | 85.53 | 0.81 |
| Huashuo | 245 | 621 | 34 | 40 | 92.13 | 87.81 | 85.96 | 86.88 | |||
| Huajin | 169 | 713 | 32 | 26 | 93.83 | 84.08 | 86.67 | 85.36 | |||
| Huaxin | 155 | 703 | 41 | 41 | 91.28 | 79.08 | 79.08 | 79.08 | |||
| EfficientNet-B4 | Xianglin 210 | 249 | 670 | 6 | 15 | 97.77 | 97.65 | 94.32 | 95.96 | 91.38 | 0.88 |
| Huashuo | 277 | 607 | 48 | 8 | 94.04 | 85.23 | 97.19 | 90.82 | |||
| Huajin | 178 | 735 | 10 | 17 | 97.13 | 94.68 | 91.28 | 92.95 | |||
| Huaxin | 155 | 727 | 17 | 41 | 93.83 | 90.12 | 79.08 | 84.24 | |||
| EfficientNet-B4-SE | Xianglin 210 | 261 | 665 | 11 | 3 | 98.51 | 95.96 | 98.86 | 97.39 | 92.98 | 0.90 |
| Huashuo | 281 | 614 | 48 | 4 | 94.51 | 85.41 | 98.60 | 91.53 | |||
| Huajin | 188 | 740 | 5 | 7 | 98.72 | 97.41 | 96.41 | 96.91 | |||
| Huaxin | 144 | 745 | 2 | 52 | 94.27 | 98.63 | 73.47 | 84.21 | |||
| EfficientNet-B4-CBAM | Xianglin 210 | 260 | 671 | 5 | 4 | 99.04 | 98.11 | 98.48 | 98.29 | 97.02 | 0.96 |
| Huashuo | 278 | 642 | 13 | 7 | 97.87 | 95.53 | 97.54 | 96.52 | |||
| Huajin | 184 | 738 | 7 | 11 | 98.09 | 96.34 | 94.36 | 95.34 | |||
| Huaxin | 190 | 741 | 3 | 6 | 99.04 | 98.44 | 96.94 | 97.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Zhang, X.; Sun, Z.; Zheng, Y.; Su, S.; Chen, F. Identification of Oil Tea (Camellia oleifera C.Abel) Cultivars Using EfficientNet-B4 CNN Model with Attention Mechanism. Forests 2022, 13, 1. https://doi.org/10.3390/f13010001
Zhu X, Zhang X, Sun Z, Zheng Y, Su S, Chen F. Identification of Oil Tea (Camellia oleifera C.Abel) Cultivars Using EfficientNet-B4 CNN Model with Attention Mechanism. Forests. 2022; 13(1):1. https://doi.org/10.3390/f13010001
Chicago/Turabian StyleZhu, Xueyan, Xinwei Zhang, Zhao Sun, Yili Zheng, Shuchai Su, and Fengjun Chen. 2022. "Identification of Oil Tea (Camellia oleifera C.Abel) Cultivars Using EfficientNet-B4 CNN Model with Attention Mechanism" Forests 13, no. 1: 1. https://doi.org/10.3390/f13010001
APA StyleZhu, X., Zhang, X., Sun, Z., Zheng, Y., Su, S., & Chen, F. (2022). Identification of Oil Tea (Camellia oleifera C.Abel) Cultivars Using EfficientNet-B4 CNN Model with Attention Mechanism. Forests, 13(1), 1. https://doi.org/10.3390/f13010001







