Computational Screening and Experimental Evaluation of Wheat Proteases for Use in the Enzymatic Therapy of Gluten-Related Disorders
Abstract
1. Introduction
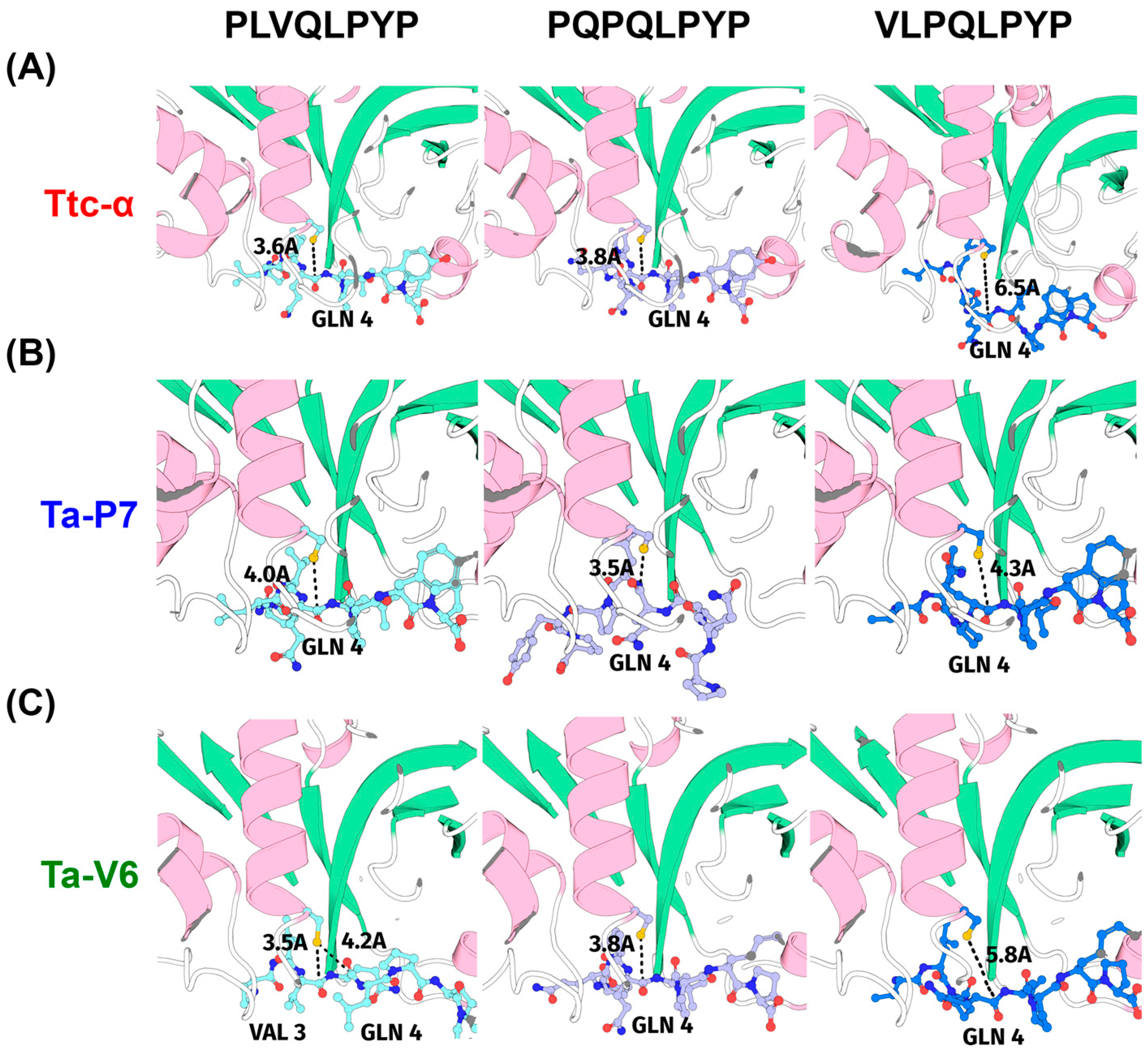
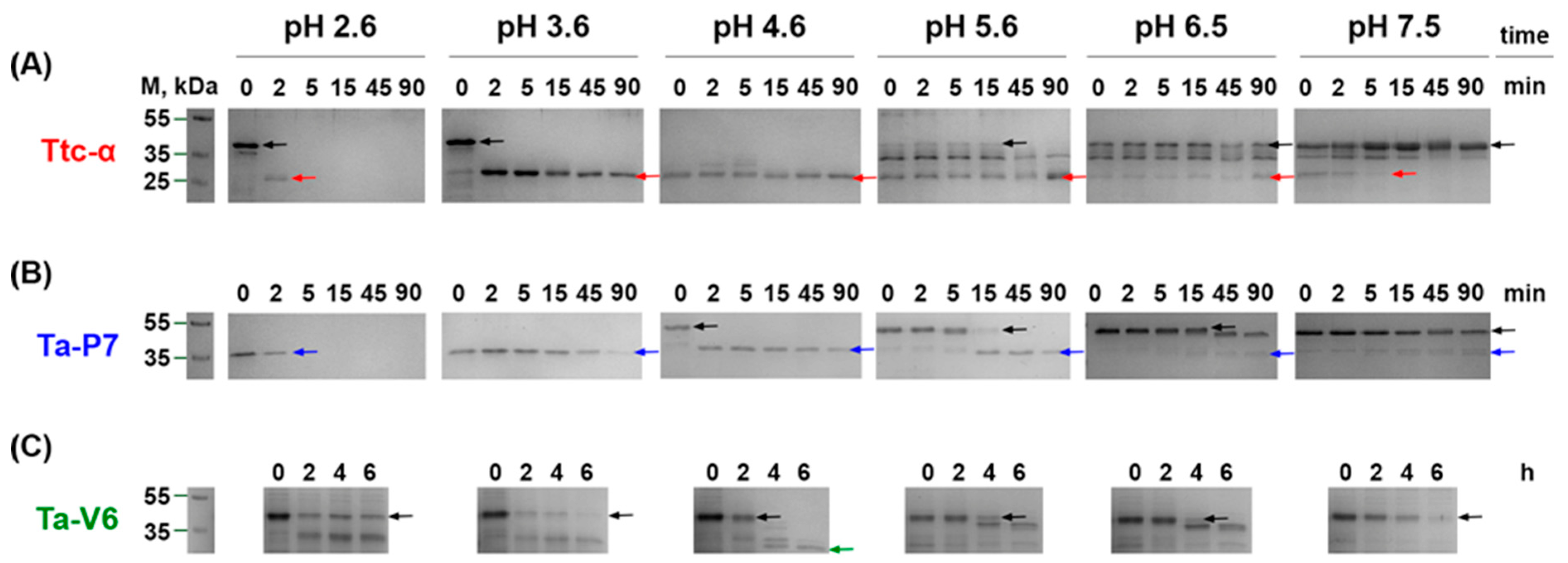
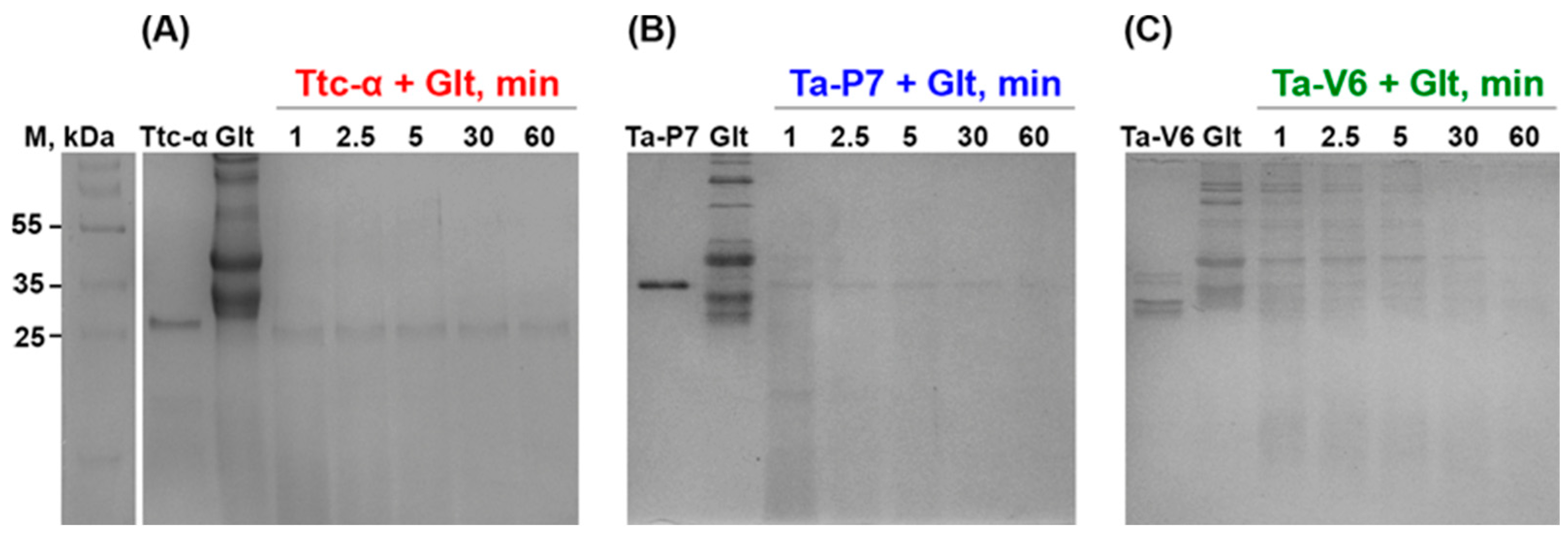
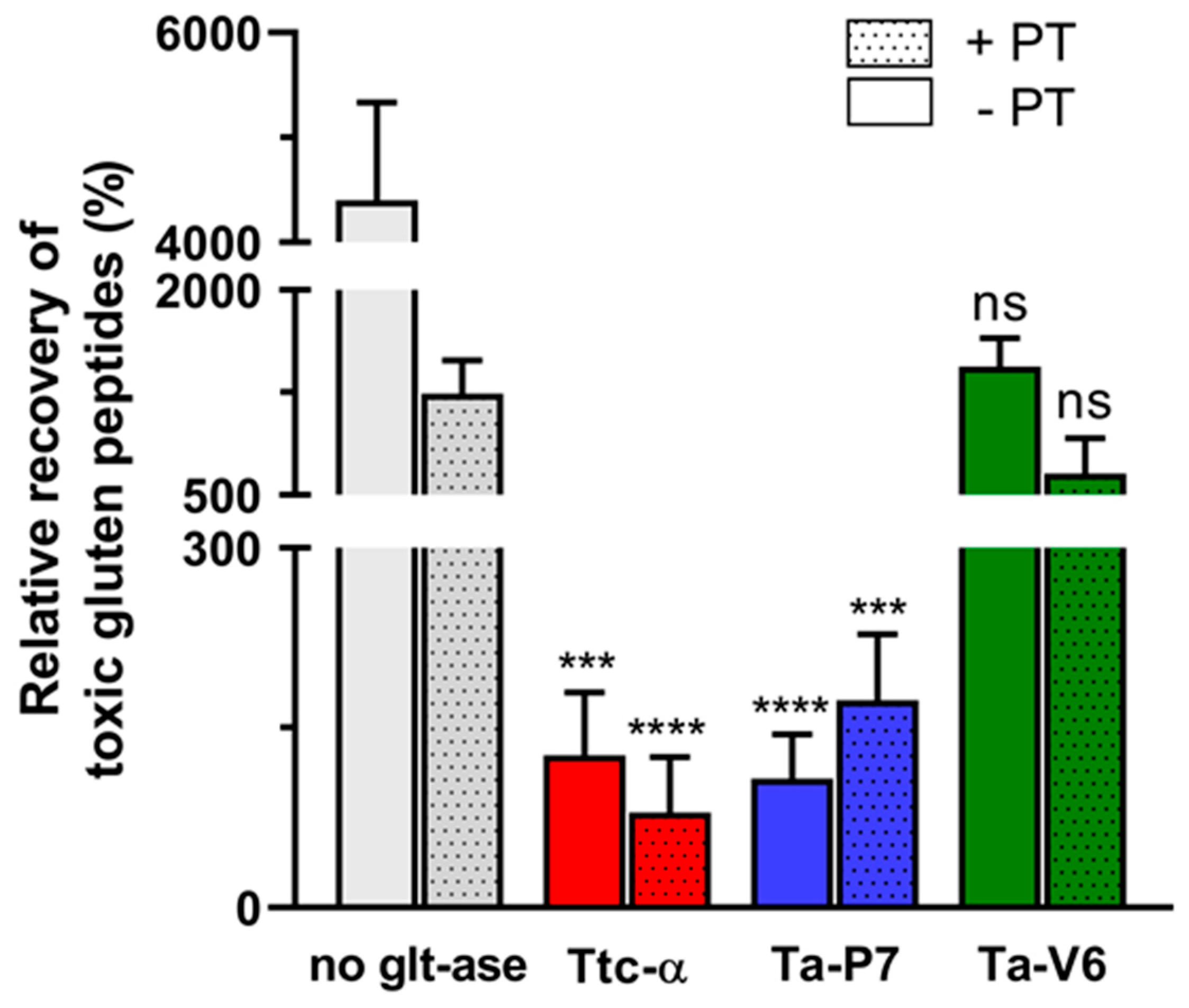
2. Results
3. Discussion
4. Materials and Methods
4.1. Computational Screening
4.2. Construction of Escherichia coli Expression Vectors
4.3. Protein Expression and Purification
4.4. Autocatalytic Activation
4.5. Protease Activity Assays
4.5.1. Protease Activity Assays with Fluorogenic Substrate
4.5.2. Gluten Cleavage
4.6. ELISA-Based Gluten Toxicity Test
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AFm | AlphaFold2-multimer |
| CD | celiac disease |
| Glt | gluten |
| Glt-ase | glutenase |
| GFD | gluten-free diet |
| PLCP | papain-like cysteine protease |
| PT | pepsin and trypsin |
| Ta-P7 | papain-like cysteine protease, Uniprot identifier: A0A3B6JDP7 |
| Ta-V6 | papain-like cysteine protease, Uniprot identifier: V6A0A3B6B7V6 |
| Ttc-α | papain-like cysteine protease Triticain-α |
| UTR | untranslated region |
References
- Gasbarrini, G.; Mangiola, F. Wheat-related disorders: A broad spectrum of ‘evolving’ diseases. United Eur. Gastroenterol. J. 2014, 2, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Kurppa, K.; Mulder, C.J.; Stordal, K.; Kaukinen, K. Celiac Disease Affects 1% of Global Population: Who Will Manage All These Patients? Gastroenterology 2024, 167, 148–158. [Google Scholar]
- Lexner, J.; Clarkson, S.; Sjöberg, K. Decreasing incidence of celiac disease in Southern Sweden. Scand. J. Gastroenterol. 2024, 59, 1039–1048. [Google Scholar] [CrossRef]
- Sollid, L.M.; Tye-Din, J.A.; Qiao, S.W.; Anderson, R.P.; Gianfrani, C.; Koning, F. Update 2020: Nomenclature and listing of celiac disease-relevant gluten epitopes recognized by CD4+ T cells. Immunogenetics 2020, 72, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Savvateeva, L.V.; Erdes, S.I.; Antishin, A.S.; Zamyatnin, A.A., Jr. Overview of Celiac Disease in Russia: Regional Data and Estimated Prevalence. J. Immunol. Res. 2017, 2017, 2314813. [Google Scholar] [CrossRef]
- Tamai, T.; Ihara, K. Celiac Disease Genetics, Pathogenesis, and Standard Therapy for Japanese Patients. Int. J. Mol. Sci. 2023, 24, 2075. [Google Scholar] [CrossRef]
- Bianchi, P.I.; Lenti, M.V.; Petrucci, C.; Gambini, G.; Aronico, N.; Varallo, M.; Rossi, C.M.; Pozzi, E.; Groppali, E.; Siccardo, F.; et al. Diagnostic Delay of Celiac Disease in Childhood. JAMA Netw. Open 2024, 7, e245671. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P. What Is Gluten-Why Is It Special? Front. Nutr. 2019, 6, 101. [Google Scholar] [CrossRef]
- Balakireva, A.V.; Zamyatnin, A.A. Properties of Gluten Intolerance: Gluten Structure, Evolution, Pathogenicity and Detoxification Capabilities. Nutrients 2016, 8, 644. [Google Scholar] [CrossRef]
- Mazzola, A.M.; Zammarchi, I.; Valerii, M.C.; Spisni, E.; Saracino, I.M.; Lanzarotto, F.; Ricci, C. Gluten-Free Diet and Other Celiac Disease Therapies: Current Understanding and Emerging Strategies. Nutrients 2024, 16, 1006. [Google Scholar] [CrossRef]
- Simpson, D.J. Proteolytic degradation of cereal prolamins—The problem with proline. Plant Sci. 2001, 161, 825–838. [Google Scholar] [CrossRef]
- Zamyatnin, A.A., Jr. Plant Proteases Involved in Regulated Cell Death. Biochemistry 2015, 80, 1701–1715. [Google Scholar] [CrossRef]
- Marttila, S.; Porali, I.; Ho, T.-H.D.; Mikkonen, A. Expression of the 30 kD cysteine endoprotease B in germinating barley seeds. Cell Biol. Int. 1993, 17, 205–212. [Google Scholar] [CrossRef]
- Bottari, A.; Capocchi, A.; Fontanini, D.; Galleschi, L. Major proteinase hydrolysing gliadin during wheat germination. Phytochemistry 1996, 43, 39–44. [Google Scholar] [CrossRef]
- Terp, N.; Thomsen, K.K.; Svendsen, I.; Davy, A.; Simpson, D.J. Purification and Characterization of Hordolisin, a Subtilisin-like Serine Endoprotease from Barley. J. Plant Physiol. 2000, 156, 468–476. [Google Scholar] [CrossRef]
- Stenman, S.M.; Venäläinen, J.I.; Lindfors, K.; Auriola, S.; Mauriala, T.; Kaukovirta-Norja, A.; Jantunen, A.; Laurila, K.; Qiao, S.W.; Sollid, L.M.; et al. Enzymatic detoxification of gluten by germinating wheat proteases: Implications for new treatment of celiac disease. Ann. Med. 2009, 41, 390–400. [Google Scholar] [CrossRef]
- Hartmann, G.; Koehler, P.; Wieser, H. Rapid degradation of gliadin peptides toxic for coeliac disease patients by proteases from germinating cereals. J. Cereal Sci. 2006, 44, 368–371. [Google Scholar] [CrossRef]
- Balakireva, A.V.; Kuznetsova, N.V.; Petushkova, A.I.; Savvateeva, L.V.; Zamyatnin, A.A., Jr. Trends and Prospects of Plant Proteases in Therapeutics. Curr. Med. Chem. 2019, 26, 465–486. [Google Scholar] [CrossRef]
- Segura, V.; Ruiz-Carnicer, Á.; Sousa, C.; Moreno, M.L. New Insights into Non-Dietary Treatment in Celiac Disease: Emerging Therapeutic Options. Nutrients 2021, 13, 2146. [Google Scholar] [CrossRef]
- Tye-Din, J.A. Evolution in coeliac disease diagnosis and management. JGH Open 2024, 8, e13107. [Google Scholar] [CrossRef]
- Dhali, A.; Maity, R.; Bharadwaj, H.R.; Ali, S.H.; Shah, M.H.; Sanders, D.S. Analyzing the landscape of coeliac crisis in adult and paediatric populations: A systematic review and meta-analysis. Dig. Liver Dis. 2025, 17, S1590-8658(25)00246-4. [Google Scholar] [CrossRef] [PubMed]
- Balakireva, A.V.; Deviatkin, A.A.; Zgoda, V.G.; Kartashov, M.I.; Zhemchuzhina, N.S.; Dzhavakhiya, V.G.; Golovin, A.V.; Zamyatnin, A.A., Jr. Proteomics Analysis Reveals That Caspase-Like and Metacaspase-Like Activities Are Dispensable for Activation of Proteases Involved in Early Response to Biotic Stress in Triticum aestivum L. Int. J. Mol. Sci. 2018, 19, 3991. [Google Scholar] [CrossRef] [PubMed]
- Savvateeva, L.V.; Gorokhovets, N.V.; Makarov, V.A.; Serebryakova, M.V.; Solovyev, A.G.; Morozov, S.Y.; Reddy, V.P.; Zernii, E.Y.; Zamyatnin, A.A., Jr.; Aliev, G. Glutenase and collagenase activities of wheat cysteine protease Triticain-α: Feasibility for enzymatic therapy assays. Int. J. Biochem. Cell Biol. 2015, 62, 115–124. [Google Scholar] [CrossRef]
- Gorokhovets, N.V.; Makarov, V.A.; Petushkova, A.I.; Prokopets, O.S.; Rubtsov, M.A.; Savvateeva, L.V.; Zernii, E.Y.; Zamyatnin, A.A., Jr. Rational Design of Recombinant Papain-Like Cysteine Protease: Optimal Domain Structure and Expression Conditions for Wheat-Derived Enzyme Triticain-α. Int. J. Mol. Sci. 2017, 18, 1395. [Google Scholar] [CrossRef]
- Tatham, A.S.; Gilbert, S.M.; Fido, R.J.; Shewry, P.R. Extraction, separation, and purification of wheat gluten proteins and related proteins of barley, rye, and oats. Methods Mol. Med. 2000, 41, 55–73. [Google Scholar] [PubMed]
- Scherf, K.A.; Wieser, H.; Koehler, P. Novel approaches for enzymatic gluten degradation to create high-quality gluten-free products. Food Res. Int. 2018, 110, 62–72. [Google Scholar] [CrossRef]
- Gray, G.M. Dietary Protein Processing: Intraluminal and Enterocyte Surface Events. In Supplement 19. Handbook of Physiology, the Gastrointestinal System, Intestinal Absorption and Secretion; Terjung, R., Ed.; Wiley: New York, NY, USA, 2011. [Google Scholar]
- Akeroyd, M.; Zandycke, S.; Hartog, J.; Mutsaers, J.; Edens, L.; Berg, M.; Christis, C. AN-PEP, proline-specific endopeptidase, degrades all known immunostimulatory gluten peptides in beer made from barley malt. J. Am. Soc. Brew. Chem. 2016, 74, 91–99. [Google Scholar] [CrossRef]
- Ehren, J.; Morón, B.; Martin, E.; Bethune, M.T.; Gray, G.M.; Khosla, C. A food-grade enzyme preparation with modest gluten detoxification properties. PLoS ONE 2009, 4, e6313. [Google Scholar] [CrossRef]
- Cavaletti, L.; Taravella, A.; Carrano, L.; Carenzi, G.; Sigurtà, A.; Solinas, N.; Caro, S.; Stasio, L.D.; Picascia, S.; Laezza, M.; et al. E40, a novel microbial protease efficiently detoxifying gluten proteins, for the dietary management of gluten intolerance. Sci. Rep. 2019, 9, 13147. [Google Scholar] [CrossRef]
- Mamone, G.; Comelli, M.C.; Vitale, S.; Di Stasio, L.; Kessler, K.; Mottola, I.; Siano, F.; Cavaletti, L.; Gianfrani, C. E40 glutenase detoxification capabilities of residual gluten immunogenic peptides in in vitro gastrointestinal digesta of food matrices made of soft and durum wheat. Front. Nutr. 2022, 9, 974771. [Google Scholar] [CrossRef]
- Wolf, C.; Siegel, J.B.; Tinberg, C.; Camarca, A.; Gianfrani, C.; Paski, S.; Guan, R.; Montelione, G.; Baker, D.; Pultz, I.S. Engineering of Kuma030: A Gliadin Peptidase That Rapidly Degrades Immunogenic Gliadin Peptides in Gastric Conditions. J. Am. Chem. Soc. 2015, 137, 13106–13113. [Google Scholar] [CrossRef] [PubMed]
- Rey, M.; Yang, M.; Lee, L.; Zhang, Y.; Sheff, J.G.; Sensen, C.W.; Mrazek, H.; Halada, P.; Man, P.; McCarville, J.L.; et al. Addressing proteolytic efficiency in enzymatic degradation therapy for celiac disease. Sci. Rep. 2016, 6, 30980. [Google Scholar] [CrossRef]
- Bethune, M.T.; Strop, P.; Tang, Y.; Sollid, L.M.; Khosla, C. Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chem. Biol. 2006, 13, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Mackie, A.; Rigby, N. InfoGest Consensus Method. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; pp. 13–22. [Google Scholar]
- Pak, M.A.; Markhieva, K.A.; Novikova, M.S.; Petrov, D.S.; Vorobyev, I.S.; Maksimova, E.S.; Kondrashov, F.A.; Ivankov, D.N. Using AlphaFold to predict the impact of single mutations on protein stability and function. PLoS ONE 2023, 18, e0282689. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- McGibbon, R.T.; Beauchamp, K.A.; Harrigan, M.P.; Klein, C.; Swails, J.M.; Hernández, C.X.; Schwantes, C.R.; Wang, L.P.; Lane, T.J.; Pande, V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109, 1528–1532. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Makarov, V.A.; Tikhomirova, N.K.; Savvateeva, L.V.; Petushkova, A.I.; Serebryakova, M.V.; Baksheeva, V.E.; Gorokhovets, N.V.; Zernii, E.Y.; Zamyatnin, A.A., Jr. Novel applications of modification of thiol enzymes and redox-regulated proteins using S-methyl methanethiosulfonate (MMTS). Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 140259. [Google Scholar] [CrossRef]
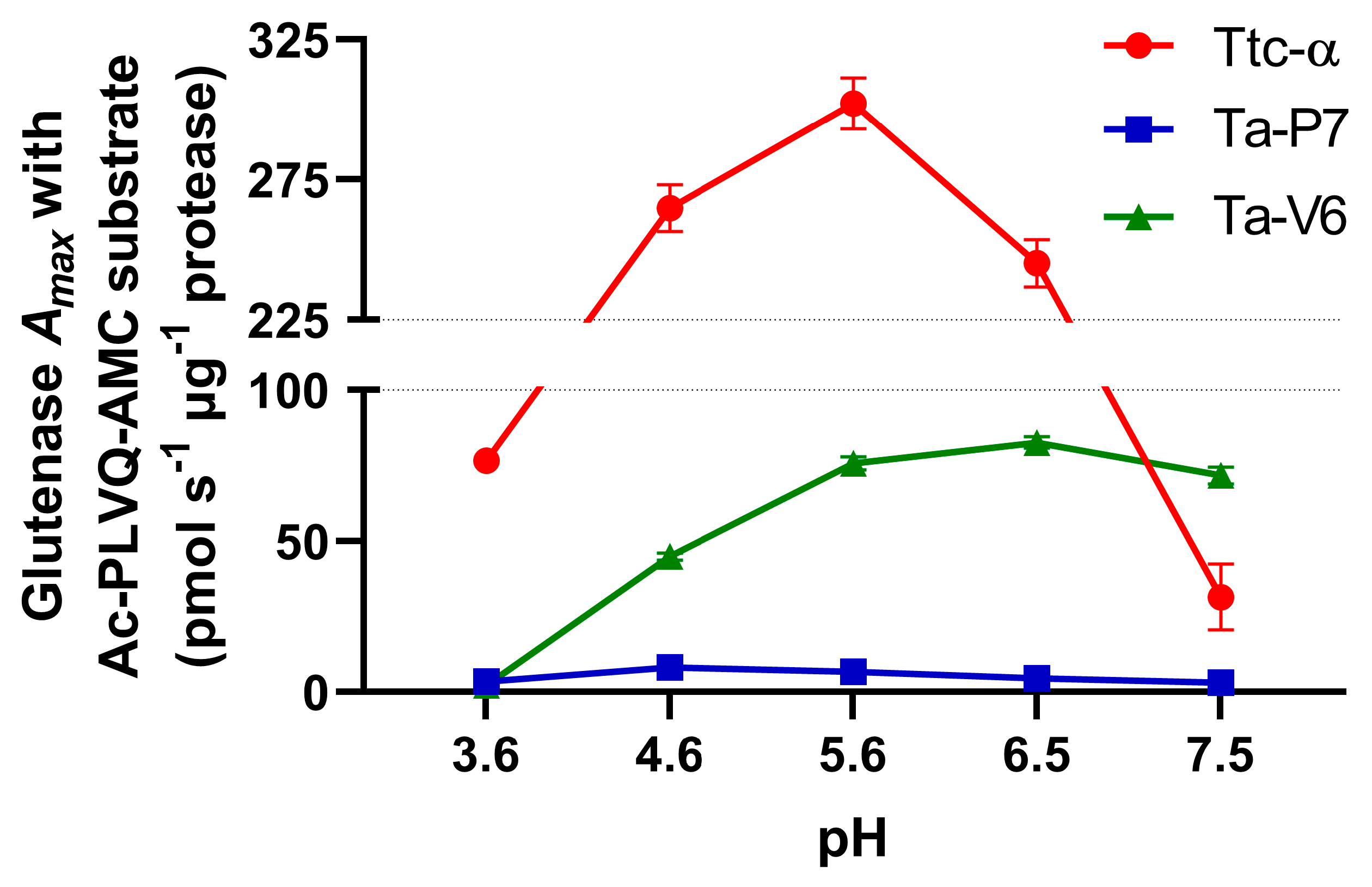
| Proteases | Kinetic Constants § | pH 3.6 | pH 4.6 | pH 5.6 | pH 6.5 | pH 7.5 |
|---|---|---|---|---|---|---|
| Ttc-α | KM, µM | 21.5 ± 1.8 | 34.6 ± 3.1 | 46.0 ± 3.4 | 49.4 ± 4.2 | 582.7 ± 259.9 |
| kcat, s−1 | 2.45 ± 0.06 | 8.48 ± 0.27 | 9.67 ± 0.29 | 7.85 ± 0.27 | 1.01 ± 0.35 | |
| Ta-P7 | KM, µM | 44.7 ± 6.6 ns | 32.2 ± 3.0 ns | 24.5 ± 1.9 ns | 31.4 ± 2.6 ns | 35.3 ± 7.7 *** |
| kcat, s−1 | 0.12 ± 0.01 **** | 0.30 ± 0.01 **** | 0.24 ± 0.01 **** | 0.16 ± 0.00 **** | 0.12 ± 0.01 *** | |
| Ta-V6 | KM, µM | 16.4 ± 7.3 ns | 20.5 ± 1.7 ns | 22.6 ± 1.8 ns | 26.9 ± 1.8 ns | 29.2 ± 3.7 *** |
| kcat, s−1 | 0.07 ± 0.01 **** | 1.44 ± 0.04 **** | 2.42 ± 0.07 **** | 2.64 ± 0.06 **** | 2.42 ± 0.11 **** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savvateeva, L.V.; Chepikova, O.E.; Solonkina, A.D.; Sakharov, A.A.; Gorokhovets, N.V.; Golovin, A.V.; Zamyatnin, A.A., Jr. Computational Screening and Experimental Evaluation of Wheat Proteases for Use in the Enzymatic Therapy of Gluten-Related Disorders. Pharmaceuticals 2025, 18, 592. https://doi.org/10.3390/ph18040592
Savvateeva LV, Chepikova OE, Solonkina AD, Sakharov AA, Gorokhovets NV, Golovin AV, Zamyatnin AA Jr. Computational Screening and Experimental Evaluation of Wheat Proteases for Use in the Enzymatic Therapy of Gluten-Related Disorders. Pharmaceuticals. 2025; 18(4):592. https://doi.org/10.3390/ph18040592
Chicago/Turabian StyleSavvateeva, Lyudmila V., Olga E. Chepikova, Alena D. Solonkina, Artemiy A. Sakharov, Neonila V. Gorokhovets, Andrey V. Golovin, and Andrey A. Zamyatnin, Jr. 2025. "Computational Screening and Experimental Evaluation of Wheat Proteases for Use in the Enzymatic Therapy of Gluten-Related Disorders" Pharmaceuticals 18, no. 4: 592. https://doi.org/10.3390/ph18040592
APA StyleSavvateeva, L. V., Chepikova, O. E., Solonkina, A. D., Sakharov, A. A., Gorokhovets, N. V., Golovin, A. V., & Zamyatnin, A. A., Jr. (2025). Computational Screening and Experimental Evaluation of Wheat Proteases for Use in the Enzymatic Therapy of Gluten-Related Disorders. Pharmaceuticals, 18(4), 592. https://doi.org/10.3390/ph18040592









