N-Derivatives of (Z)-Methyl 3-(4-Oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylates as Antimicrobial Agents—In Silico and In Vitro Evaluation
Abstract
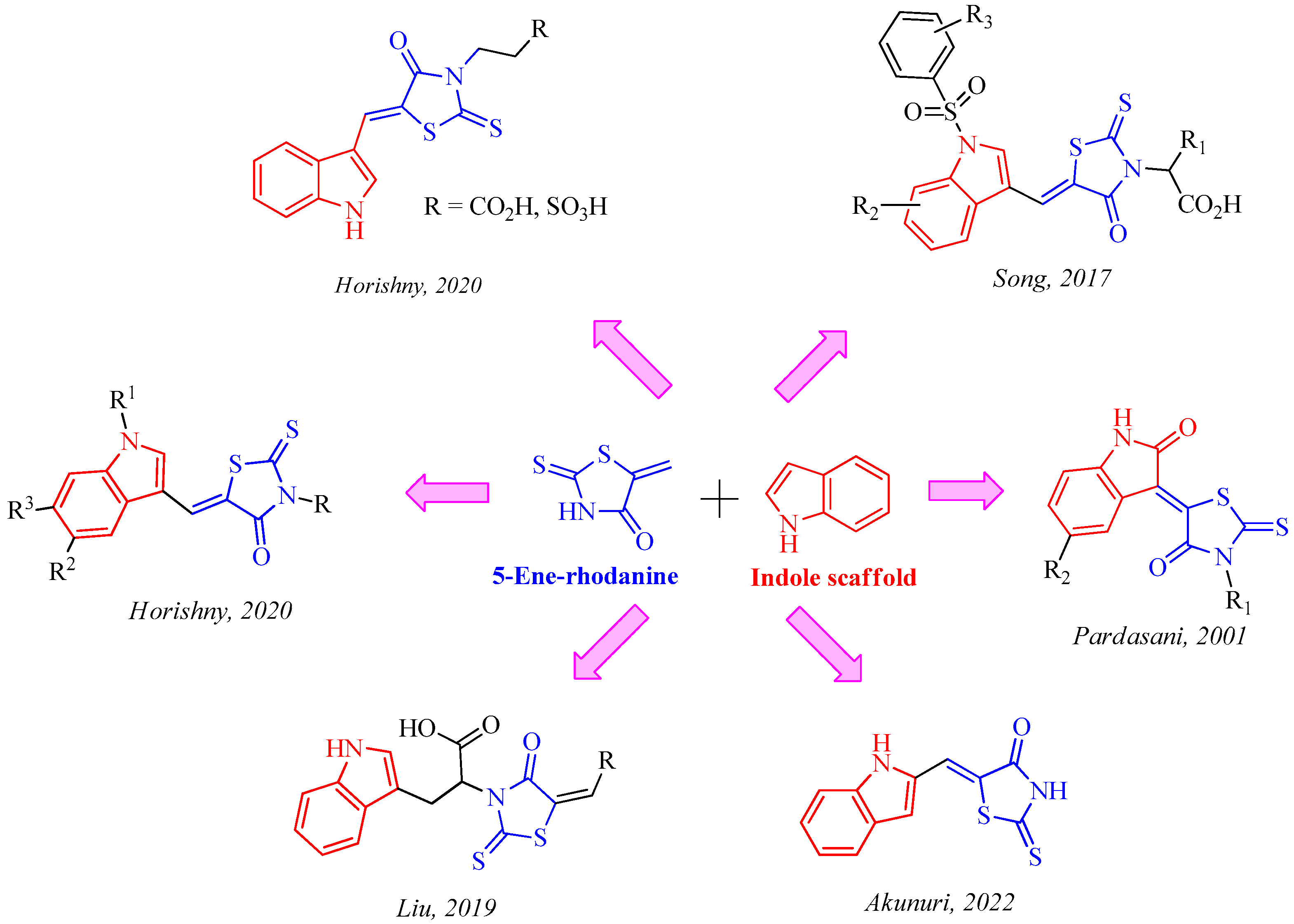
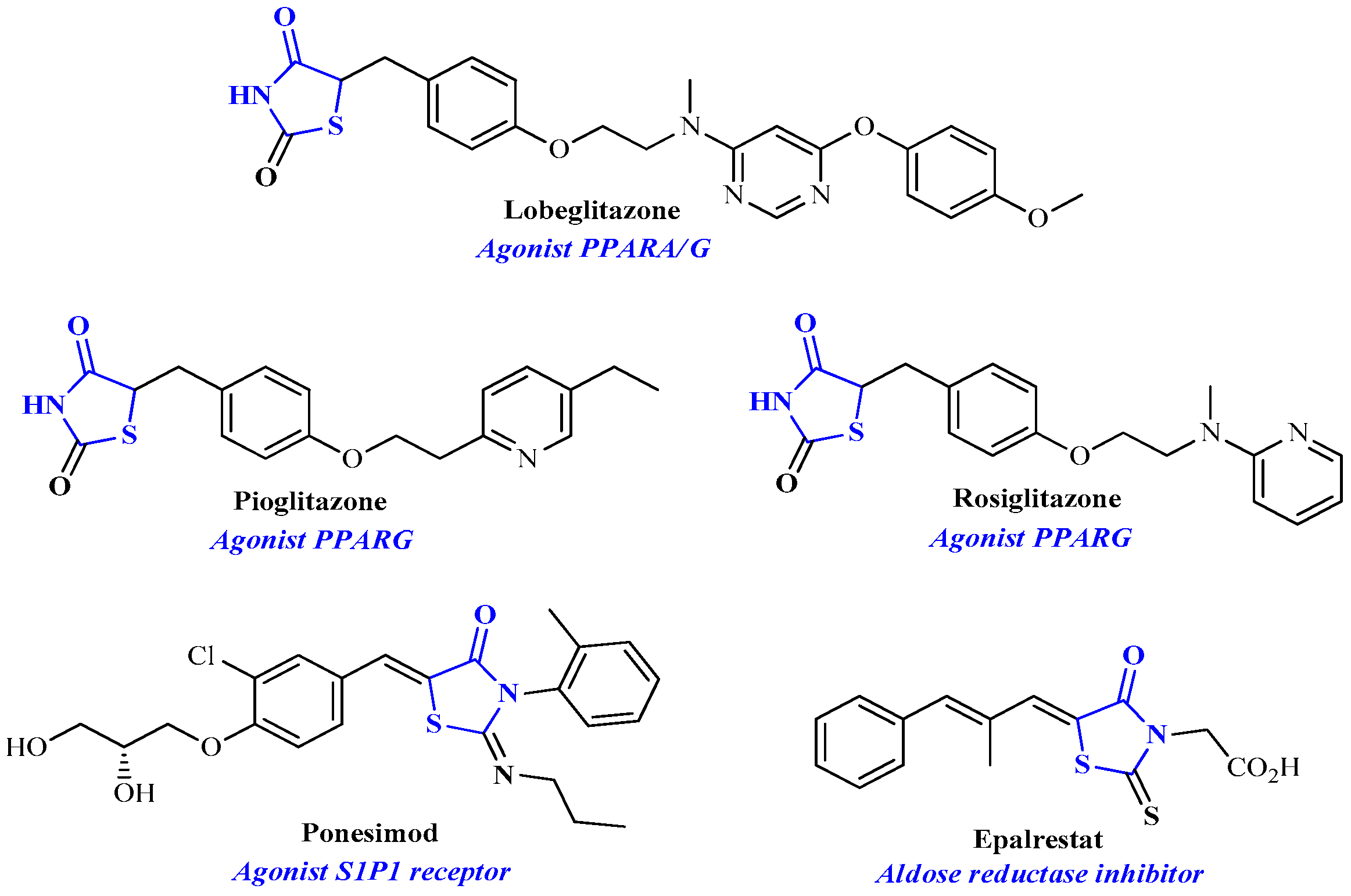
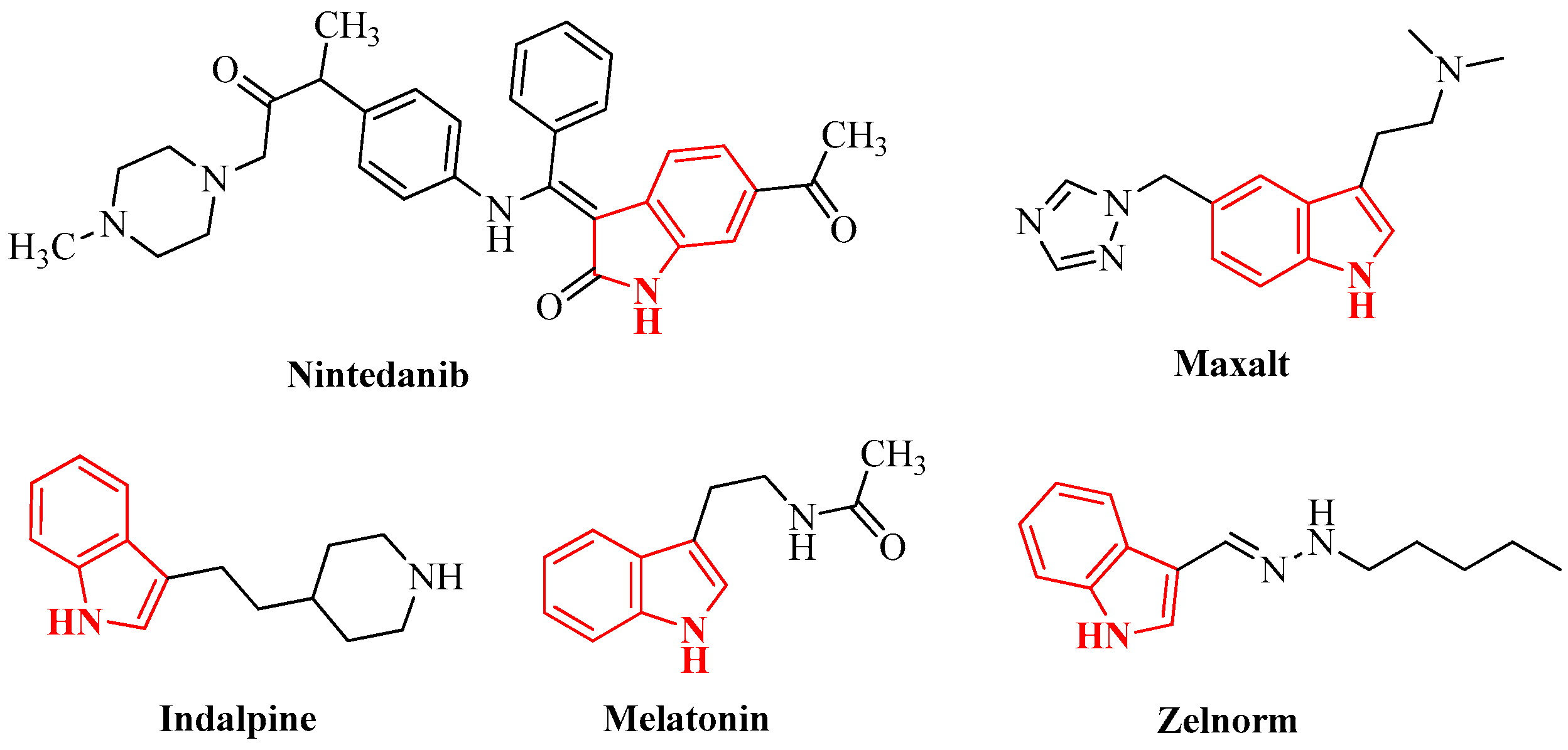
1. Introduction
2. Results and Discussion
2.1. Prediction of Toxicity
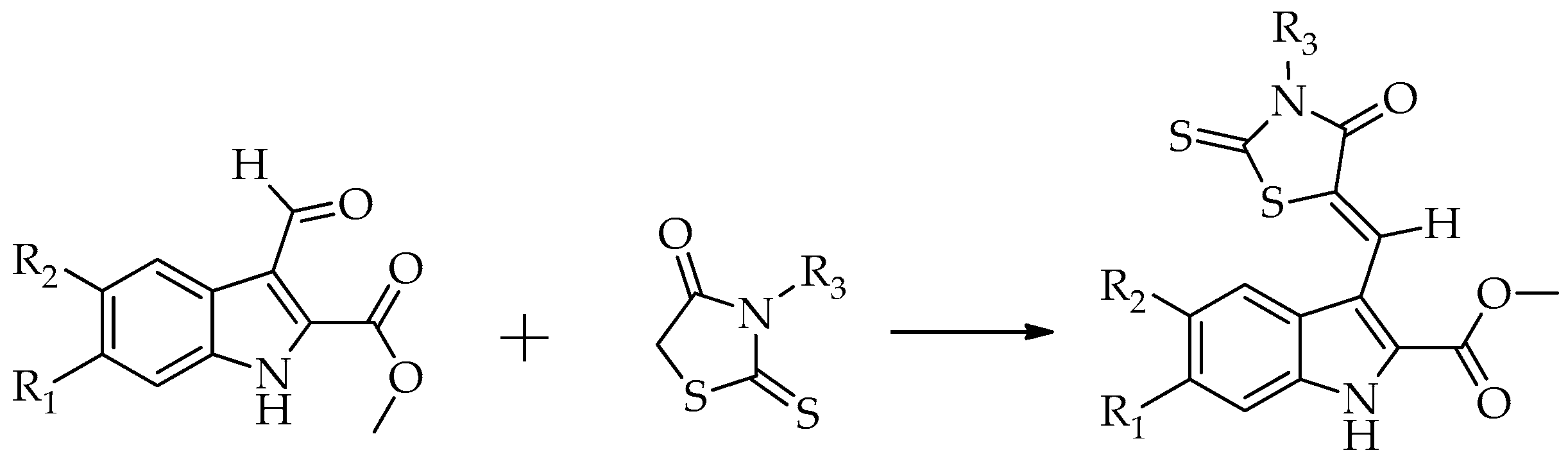
2.2. Chemistry
2.3. Biological Evaluation
2.3.1. Antimicrobial Activity
2.3.2. Antifungal Activity
2.4. Docking Studies
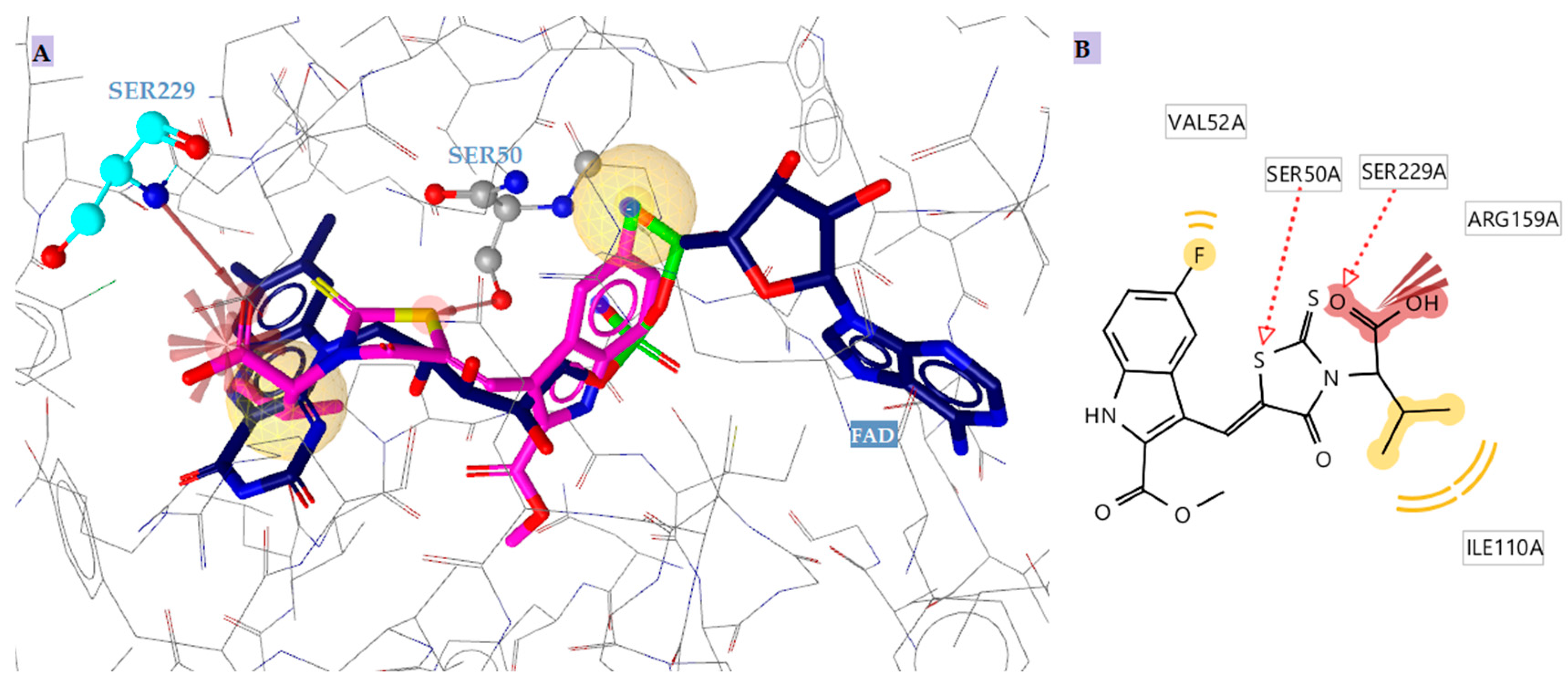
2.4.1. Docking to Antibacterial Targets
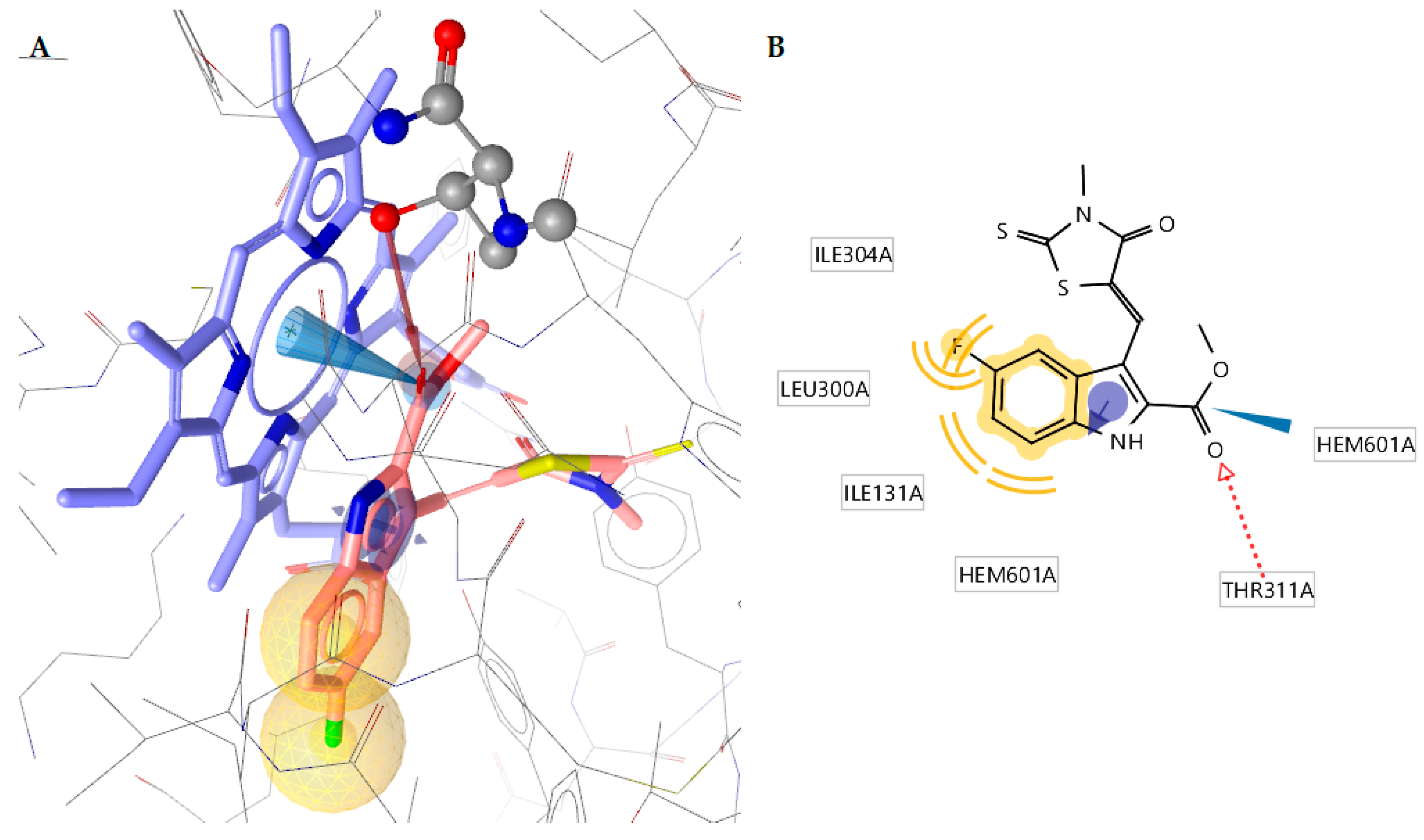
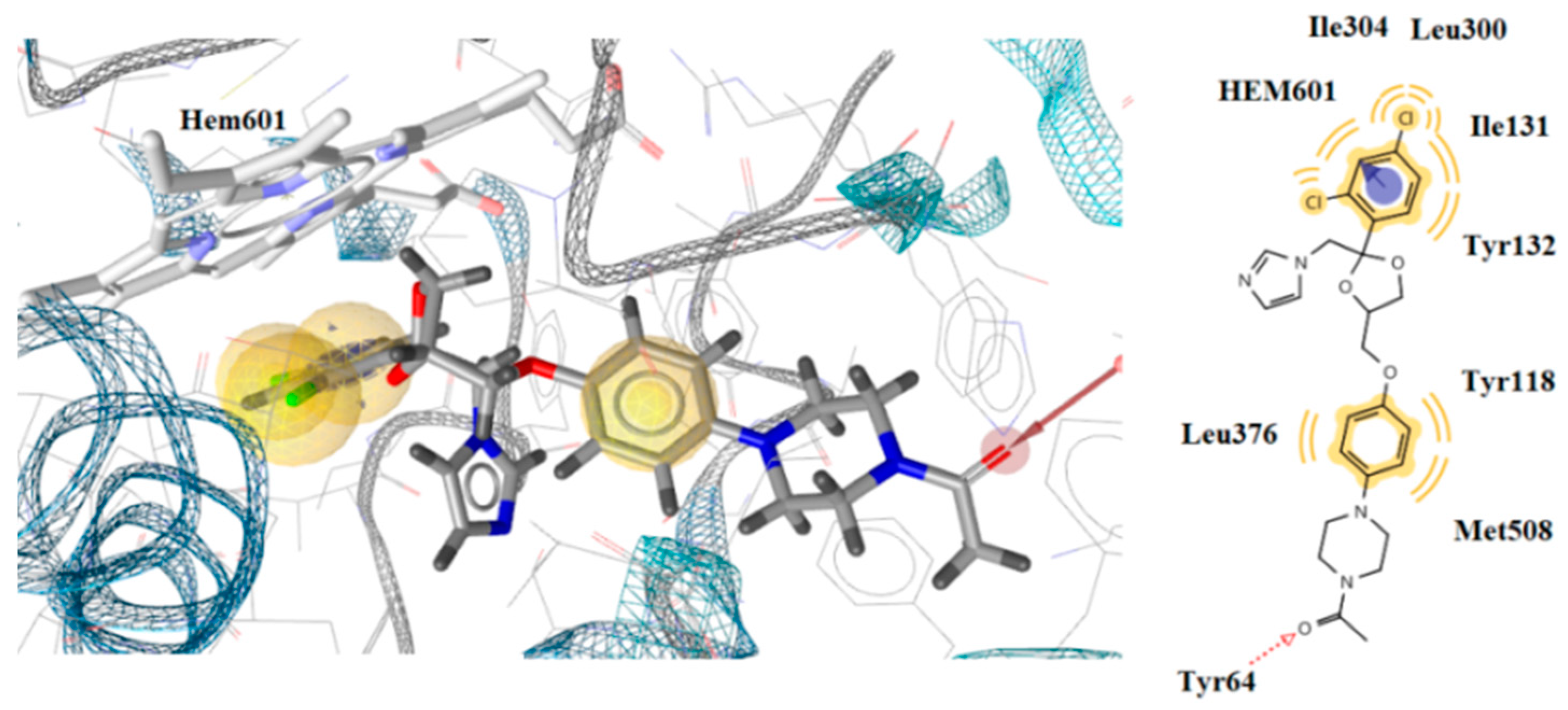
2.4.2. Docking to Antifungal Targets
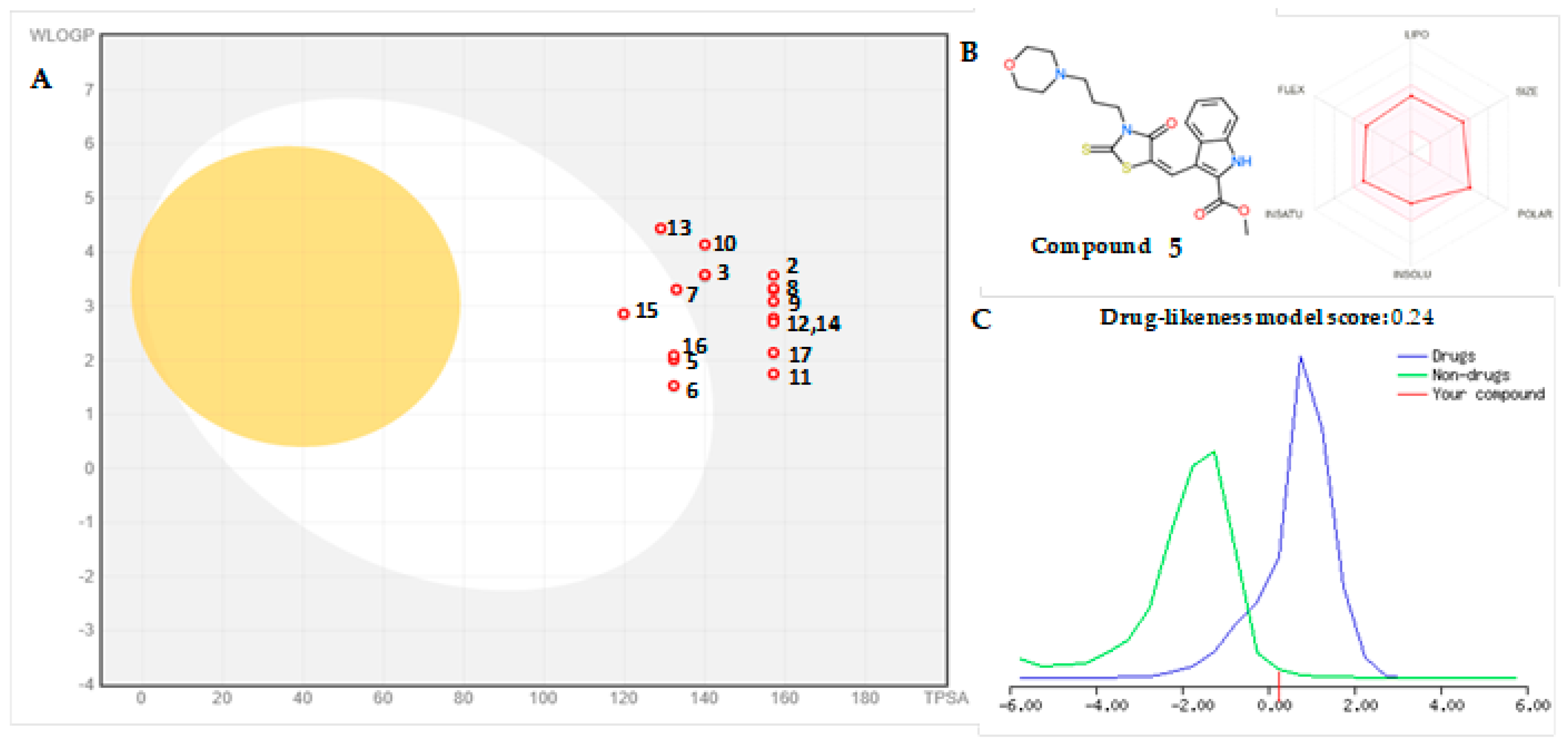
2.5. Drug-Likeness and Bioavailability
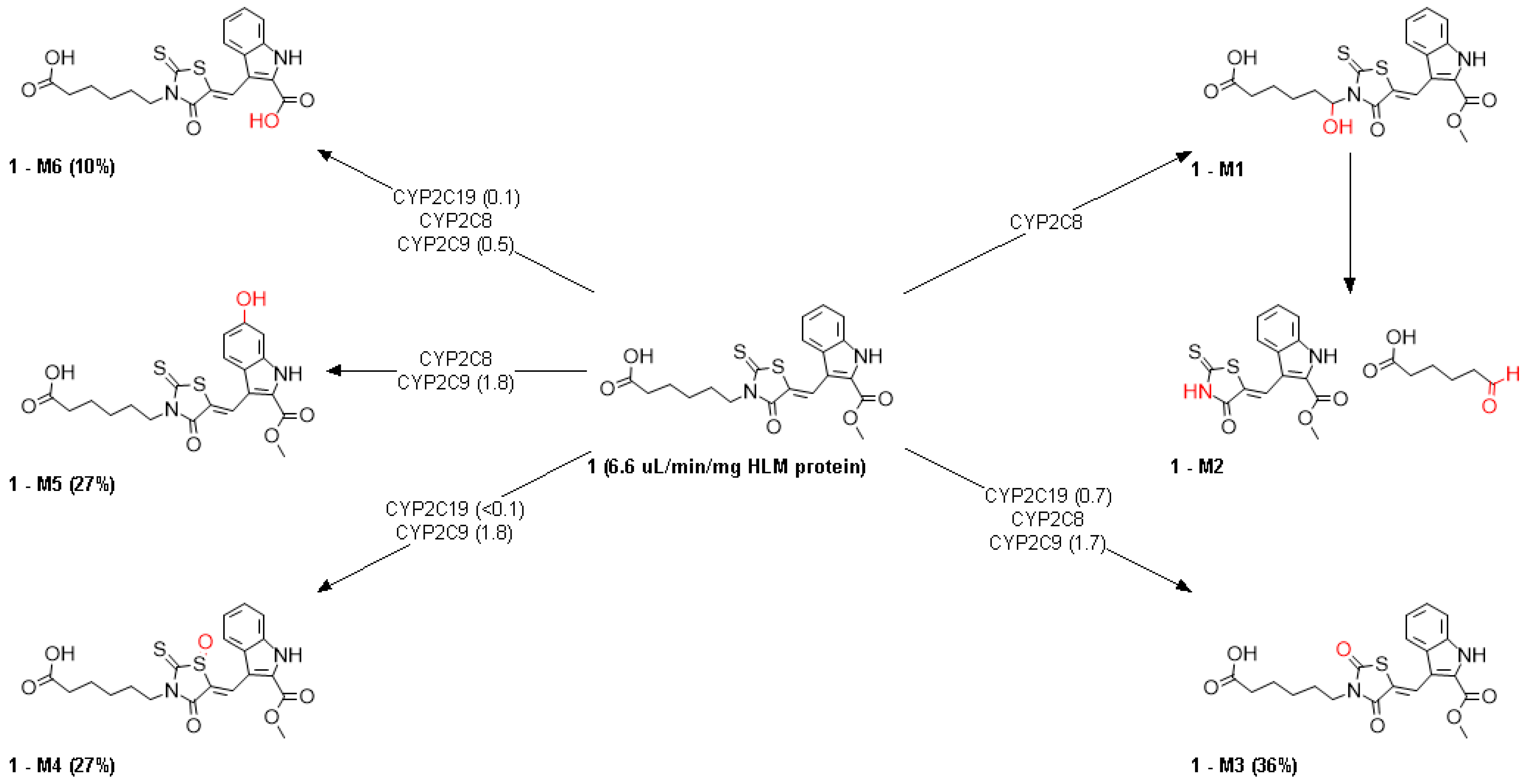
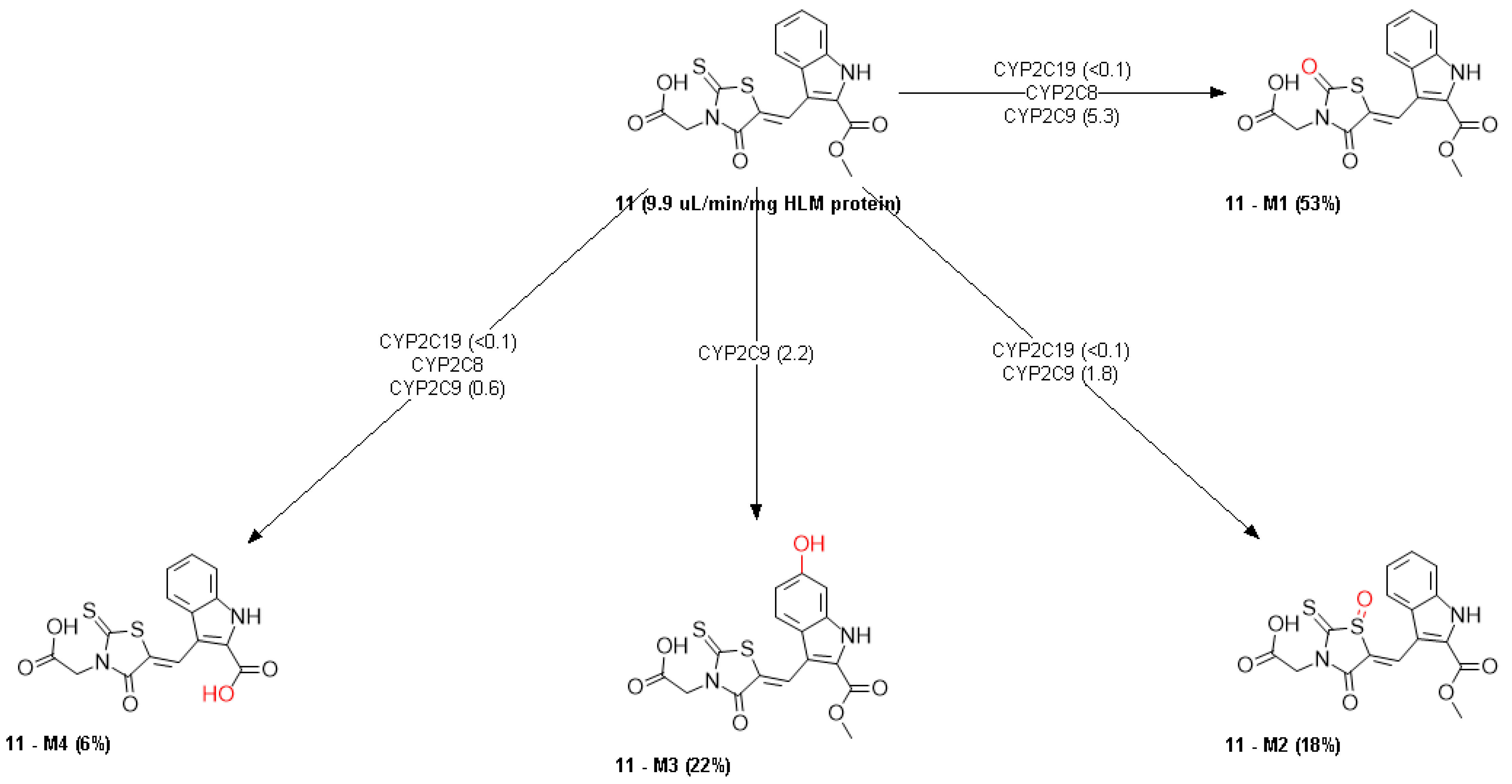
2.6. ADMET Properties
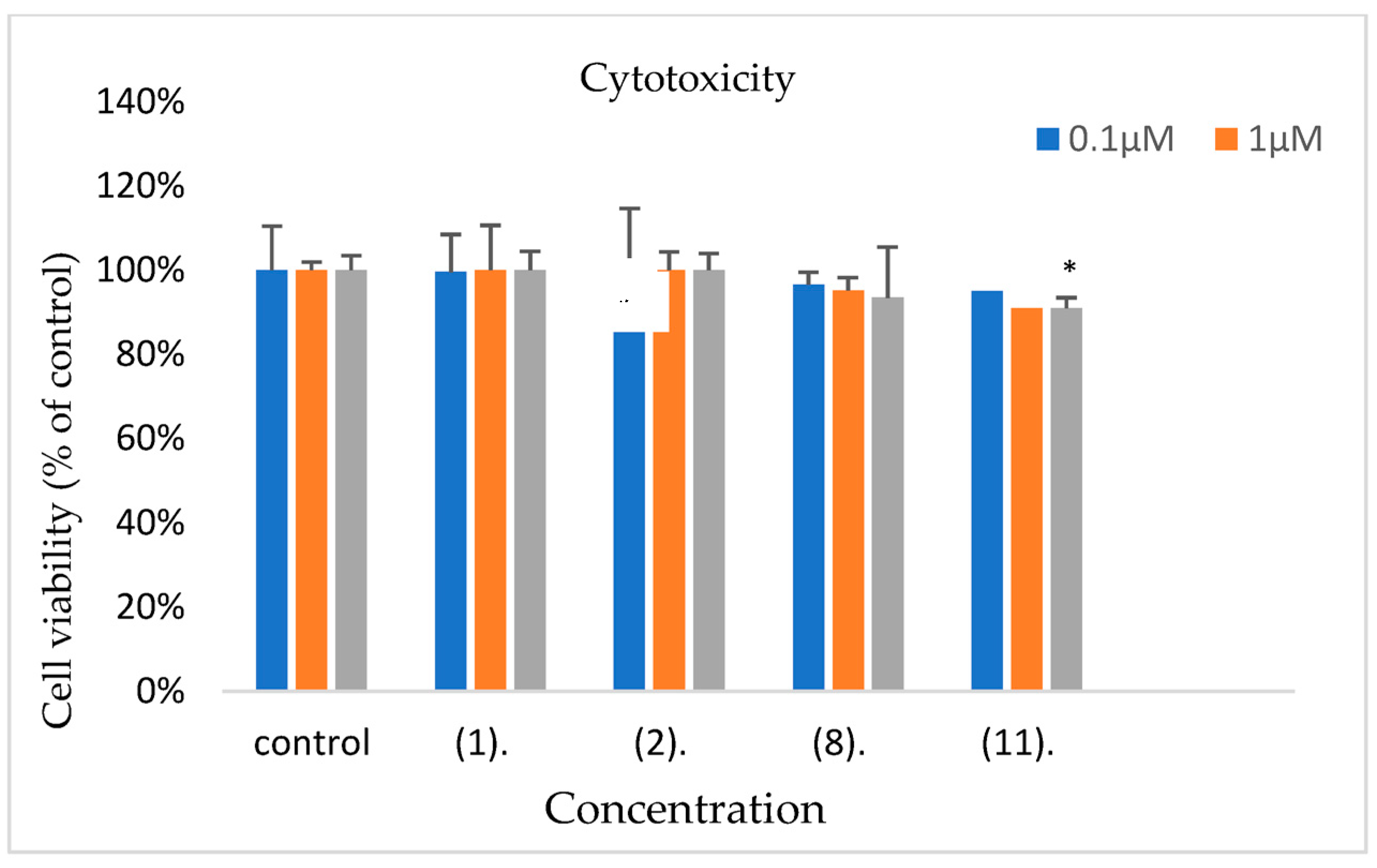
2.7. Cytotoxicity
3. Materials and Methods
3.1. Prediction of Toxicity
3.2. Chemistry
- (Z)-6-(5-((2-(methoxycarbonyl)-1H-indol-3-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)hexanoic acid (1). m.p. 135–138 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.06 (s, 2H,NH, OH), 8.49 (s, 1H), 7.79 (s, 1H), 7.58 (s, 1H), 7.35 (s, 1H), 7.24 (s, 1H), 4.09 (s, 2H), 4.00 (s, 3H, CH3), 2.21 (s, 2H), 1.75 (s, 2H), 1.65 (s, 2H), 1.45 (s, 2H). Anal. Calcd. For C20H20N2O5S2 (%): C, 55.54; H, 4.66; N, 6.48; O, 18.50. Found (%): C, 55.50; H, 4.69; N, 6.52; O, 18.47.
- (Z)-4-(5-((2-(methoxycarbonyl)-1H-indol-3-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)benzoic acid (2). m.p. 294–295 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.69 (s, 1H, OH), 8.51 (s, 1H), 8.16 (d, J = 8.1 Hz, 2H), 7.87 (d, J = 8.2 Hz, 1H), 7.60 (d, J = 8.1 Hz, 1H), 7.47 (d, J = 8.2 Hz, 2H), 7.37 (t, J = 7.6 Hz, 1H), 7.28 (t, J = 7.5 Hz, 1H), 4.00 (s, 3H, CH3). Anal. Calcd. For C21H14N2O5S2 (%): C, 57.52; H, 3.22; N, 6.39; O, 18.24. Found (%): C, 57.54; H, 3.20; N, 6.42; O, 18.25.
- (Z)-methyl 3-((3-(3-hydroxyphenyl)-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylate (3). m.p. 245–247 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.66 (s, 1H, OH), 9.61 (s, 1H, NH), 8.48 (s, 1H), 7.87 (d, J = 8.2 Hz, 1H), 7.59 (d, J = 8.3 Hz, 1H), 7.31 (dq, J = 7.2, 22.3 Hz, 3H), 6.90 (d, J = 8.4 Hz, 1H), 6.71 (d, J = 5.2 Hz, 2H), 4.00 (s, 3H,CH3). Anal. Calcd. For C20H14N2O4S2 (%): C, 58.52; H, 3.44; N, 6.82; O, 15.59. Found (%): C, 58.50; H, 3.43; N, 6.87; O, 15.53.
- (Z)-methyl 3-((3-(4-hydroxyphenyl)-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylate (4). m.p. 282–284 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.64 (s, 1H, OH), 9.62 (s, 1H, NH), 8.47 (s, 1H), 7.86 (d, J = 8.2 Hz, 1H), 7.59 (d, J = 8.3 Hz, 1H), 7.36 (t, J = 7.7 Hz, 1H), 7.26 (t, J = 7.6 Hz, 1H), 7.08 (d, J = 8.3 Hz, 2H), 6.90 (d, J = 8.3 Hz, 2H), 4.00 (s, 3H, CH3). Anal. Calcd. For C20H14N2O4S2 (%): C, 58.52; H, 3.44; N, 6.82; O, 15.59. Found (%): C, 58.50; H, 3.47; N, 6.86; O, 15.55.
- (Z)-methyl 3-((3-(3-morpholinopropyl)-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylate (5). m.p. 213–214 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.64 (s, 1H, NH), 8.47 (s, 1H), 7.78 (d, J = 8.2 Hz, 1H), 7.58 (d, J = 8.3 Hz, 1H), 7.35 (t, J = 7.6 Hz, 1H), 7.24 (t, J = 7.6 Hz, 1H), 4.18 (t, J = 7.0 Hz, 2H), 4.00 (s, 2H), 3.55 (t, J = 4.6 Hz, 4H, CH3), 2.58–2.29 (m, 8H), 1.89 (p, J = 6.8 Hz, 2H). Anal. Calcd. For C21H23N3O4S2 (%): C, 56.61; H, 5.20; N, 9.43; O, 14.36. Found (%): C, 56.64; H, 5.18; N, 9.45; O, 14.30.
- (Z)-methyl 3-((3-morpholino-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylate (6). m.p. 278–280 °C. 1H NMR (300 MHz) δ 12.65 (s, 1H, NH), 8.42 (s, 1H), 7.78 (d, J = 8.1 Hz, 1H), 7.57 (d, J = 8.3 Hz, 1H), 7.34 (t, J = 7.7 Hz, 1H), 7.23 (t, J = 7.6 Hz, 1H), 4.00 (s, 3H, CH3), 3.05 (d, J = 22.9 Hz, 4H), 2.50 (s, 3H). Anal. Calcd. For C18H17N3O4S2 (%): C, 53.58; H, 4.25; N, 10.41; O, 15.86. Found (%): C, 53.60; H, 4.21; N, 10.46; O, 15.83.
- (Z)-methyl 3-((3-(furan-2-ylmethyl)-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylate (7). 1H NMR (300 MHz, DMSO-d6) δ 12.65 (s, 1H, NH), 8.52 (s, 1H), 7.79 (d, J = 8.2 Hz, 1H), 7.57 (d, J = 8.2 Hz, 1H), 7.47 (s, 1H), 7.34 (t, J = 7.7 Hz, 1H), 7.23 (t, J = 7.6 Hz, 1H), 6.41 (s, 1H), 6.36 (s, 1H), 5.26 (s, 2H), 4.00 (s, 3H, CH3). m.p. 212–213 °C. Anal. Calcd. For C19H14N2O4S2 (%): C, 57.27; H, 3.54; N, 7.03; O, 16.06. Found (%): C, 57.31; H, 3.50; N, 7.08; O, 16.02.
- (Z)-2-(5-((5-fluoro-2-(methoxycarbonyl)-1H-indol-3-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)-3-methylbutanoic acid (8). 1H NMR (300 MHz, DMSO-d6) δ 8.44 (s, 1H, NH), 7.58 (dd, J = 4.7, 9.0 Hz, 1H), 7.48 (d, J = 9.7 Hz, 1H), 7.15 (t, J = 9.2 Hz, 1H), 5.12 (d, J = 9.6 Hz, 1H), 4.00 (s, 3H, O-CH3), 2.80 (s, 1H, CH-(CH3)2), 1.28 (d, J = 6.2 Hz, 3H, CH-CH3), 0.84 (d, J = 6.7 Hz, 3H, CH-CH3). m.p. 234–235 °C. Anal. Calcd. For C19H17FN2O5S2 (%): C, 52.28; H, 3.93; F, 4.35; N, 6.42; O, 18.33. Found (%): C, 52.23; H, 3.90; F, 4.28; N, 6.54; O, 18.29.
- (Z)-4-(5-((5-fluoro-2-(methoxycarbonyl)-1H-indol-3-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)butanoic acid (9). m.p. 192–193 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.72 (s, 1H, NH), 11.90 (s, 1H, OH), 8.38 (s, 1H), 7.57 (dd, J = 4.7, 9.0 Hz, 1H), 7.46 (dd, J = 2.3, 9.9 Hz, 1H), 7.14 (td, J = 2.5, 9.1 Hz, 1H), 4.14 (t, J = 7.0 Hz, 2H), 3.99 (s, 3H, CH3), 2.32 (t, J = 7.4 Hz, 2H), 1.98 (p, J = 7.4 Hz, 2H). Anal. Calcd. For C18H15FN2O5S2 (%): C, 51.18; H, 3.58; F, 4.50; N, 6.63; O, 18.94. Found (%): C, 51.23; H, 3.55; F, 4.48; N, 6.69; O, 18.90.
- (Z)-methyl 5-fluoro-3-((3-(4-hydroxyphenyl)-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylate (10). m.p. 267–268 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.74 (s, 1H, OH), 9.61 (s, 1H, NH), 8.37 (s, 1H), 7.64 – 7.47 (m, 2H), 7.15 (t, J = 8.7 Hz, 1H), 7.07 (d, J = 8.4 Hz, 2H), 6.90 (d, J = 8.4 Hz, 2H), 3.99 (s, 3H, O-CH3). Anal. Calcd. For C20H13FN2O4S2 (%): C, 56.06; H, 3.06; F, 4.43; N, 6.54; O, 14.94. Found (%): C, 56.12; H, 3.02; F, 4.39; N, 6.58; O, 14.87.
- (Z)-2-(5-((2-(methoxycarbonyl)-1H-indol-3-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)acetic acid (11). m.p. 292–294 °C. 1H NMR (300 MHz, DMSO-d6) δ 8.51 (s, 1H), 7.80 (d, J = 8.3 Hz, 1H), 7.57 (d, J = 8.3 Hz, 1H), 7.34 (t, J = 7.6 Hz, 1H), 7.24 (t, J = 7.6 Hz, 1H), 4.65 (s, 2H), 3.99 (s, 3H,O-CH3). Anal. Calcd. For C16H12N2O5S2 (%): C, 51.05; H, 3.21; N, 7.44; O, 21.25. Found (%): C, 51.11; H, 3.27; N, 7.33; O, 21.20.
- (Z)-2-(5-((2-(methoxycarbonyl)-1H-indol-3-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)-3-methylbutanoic acid (12). m.p. 256–258 °C. 1H NMR (300 MHz, DMSO-d6) δ 8.51 (s, 1H), 7.82 (d, J = 8.2 Hz, 1H), 7.58 (d, J = 8.2 Hz, 1H), 7.34 (t, J = 7.7 Hz, 1H), 7.24 (t, J = 7.6 Hz, 1H), 5.13 (d, J = 9.2 Hz, 1H), 4.00 (s, 3H), 2.79 (s, 1H, CH-(CH3)2), 1.28 (d, J = 6.3 Hz, 3H, CH-CH3), 0.84 (d, J = 6.8 Hz, 3H, CH-CH3). Anal. Calcd. For C19H18N2O5S2 (%): C, 54.53; H, 4.34; N, 6.69; O, 19.12. Found (%): C, 54.49; H, 4.37; N, 6.71; O, 19.10.
- (Z)-methyl 3-((3-(3-fluorophenyl)-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)-6-methoxy-1H-indole-2-carboxylate (13). m.p. 238–240 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.65 (s, 1H, NH), 8.52 (s, 1H), 7.79 (d, J = 8.2 Hz, 1H), 7.57 (d, J = 8.2 Hz, 1H), 7.47 (s, 1H), 7.34 (t, J = 7.7 Hz, 1H), 7.23 (t, J = 7.6 Hz, 1H), 6.41 (s, 1H), 6.36 (s, 1H), 5.26(s, 3H, O-CH3), 4.00 (s, 3H, O-CH3). Anal. Calcd. For C21H15FN2O4S2 (%): C, 57.00; H, 3.42; F, 4.29; N, 6.33; O, 14.46. Found (%): C, 57.12; H, 3.39; F, 4.32; N, 6.38; O, 14.51.
- (Z)-3-(5-((5-fluoro-2-(methoxycarbonyl)-1H-indol-3-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)propanoic acid (14). m.p. 275–276 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.76 (s, 1H, OH), 12.30 (s, 1H, NH), 8.39 (s, 1H), 7.57 (dd, J = 4.6, 9.0 Hz, 1H), 7.44 (d, J = 9.8 Hz, 1H), 7.15 (t, J = 8.9 Hz, 1H), 4.29 (t, J = 7.9 Hz, 2H), 3.99 (s, 3H, CH3), 2.64 (t, J = 8.1 Hz, 2H). Anal. Calcd. For C17H13FN2O5S2 (%): C, 49.99; H, 3.21; F, 4.65; N, 6.86; O, 19.59. Found (%): C, 49.92; H, 3.25; F, 4.60; N, 6.90; O, 19.62.
- (Z)-methyl 5-fluoro-3-((3-methyl-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylate (15). m.p. 234–244 °C. 1H NMR (300 MHz) δ 12.75 (s, 1H, NH), 8.40 (s, 1H), 7.57 (dd, J = 4.7, 9.0 Hz, 1H), 7.43 (d, J = 9.7 Hz, 1H), 7.15 (t, J = 9.0 Hz, 1H), 3.99 (s, 3H, O-CH3), 3.48 (s, 3H, N-CH3). Anal. Calcd. For C15H11FN2O3S2 (%): C, 51.42; H, 3.16; F, 5.42; N, 7.99; O, 13.70. Found (%): C, 51.40; H, 3.21; F, 5.45; N, 7.93; O, 13.75.
- (Z)-methyl 5-fluoro-3-((3-morpholino-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylate (16). m.p. 273–274 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.76 (s, 1H, NH), 8.32 (s, 1H), 7.57 (dd, J = 4.6, 9.1 Hz, 1H), 7.46 (d, J = 9.6 Hz, 1H), 7.15 (t, J = 9.1 Hz, 1H), 4.00 (s, 3H, CH3), 3.77 (dd, J = 16.8, 29.8 Hz, 6H), 3.05 (d, J = 26.1 Hz, 3H). Anal. Calcd. For C18H16FN3O4S2 (%): C, 51.30; H, 3.83; F, 4.51; N, 9.97; O, 15.18. Found (%): C, 51.36; H, 3.79; F, 4.54; N, 9.93; O, 15.21.
- (Z)-3-(5-((2-(methoxycarbonyl)-1H-indol-3-yl)methylene)-4-oxo-2-thioxothiazolidin-3-yl)propanoic acid (17). m.p. 265–266 °C. 1H NMR (300 MHz, DMSO-d6) δ 12.65 (s, 1H, NH), 12.30 (s, 1H, OH), 8.49 (s, 1H), 7.78 (d, J = 8.2 Hz, 1H), 7.57 (d, J = 8.3 Hz, 1H), 7.35 (t, J = 7.7 Hz, 1H), 7.24 (t, J = 7.4 Hz, 1H), 4.29 (t, J = 7.9 Hz, 2H), 4.00 (s, 3H, O-CH3), 2.64 (t, J = 8.1 Hz, 2H). Anal. Calcd. For C17H14N2O5S2 (%): C, 52.30; H, 3.61; N, 7.17; O, 20.49. Found (%): C, 52.28; H, 3.65; N, 7.21; O, 20.52.
3.3. Biological Evaluation
3.3.1. Antibacterial Activity
3.3.2. Antifungal Activity
3.4. Docking Studies
3.5. Drug Likeness
3.6. ADMET
3.7. Assessment of Cytotoxicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chopra, I.; Schofield, C.; Everett, M.; O’Neill, A.; Miller, K.; Wilcox, M.; Frere, J.-M.; Dawson, M.; Czaplewsky, L.; Urleb, U.; et al. Treatment of Health-Care-Associated Infections Caused by Gram-Negative Bacteria: A Consensus Statement. Lancet Infect. Dis. 2008, 8, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Overbye, K.M.; Barrett, J.F. Antibiotics: Where Did We Go Wrong? Drug Discov. Today 2005, 10, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.W.; Maziarz, E.K.; Arnold, C.J.; Johnson, M.D.; Workman, A.D.; Reynolds, J.M.; Perfect, J.R.; Alexander, B.D. Invasive fungal infection after lung transplantation: Epidemiology in the setting of antifungal prophylaxis. Clin. Infect. Dis. 2020, 70, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Sayed, M.; Kamal El-Dean, A.M.K.; Ahmed, M.; Hassanien, R. Synthesis of some heterocyclic compounds derived from indole as antimicrobial agents. Synth. Commun. 2018, 48, 413–421. [Google Scholar] [CrossRef]
- Roszczenko, P.; Holota, S.; Szewczyk, O.K.; Dudchak, R.; Bielawski, K.; Bielawska, A.; Lesyk, R. 4-Thiazolidinone-Bearing Hybrid Molecules in Anticancer Drug Design. Int. J. Mol. Sci. 2022, 23, 13135. [Google Scholar] [CrossRef]
- Horishny, V.; Kartsev, V.; Matiychuk, V.; Geronikaki, A.; Petrou, A.; Pogodin, P.; Poroikov, V.; Ivanov, M.; Kostic, M.; Soković, M.D.; et al. 3-Amino-5-(indol-3-yl)methylene-4-oxo-2-thioxothiazolidine Derivatives as Antimicrobial Agents: Synthesis, Computational and Biological Evaluation. Pharmaceuticals 2020, 13, 229. [Google Scholar] [CrossRef]
- Barakat, A.; Al-Najjar, H.J.; Al-Majid, A.M.; Soliman, S.M.; Mabkhot, Y.N.; Al-Agamy, M.H.M.; Ghabbour, H.A.; Fun, H.K. Synthesis, molecular structure investigations and antimicrobial activity of 2- thioxothiazolidin-4-one derivatives. J. Mol. Struct. 2015, 1081, 519–529. [Google Scholar] [CrossRef]
- Cebeci, Y.U.; Karaoğlu, Ş.A. Quinolone-Rhodanine Hybrid Compounds: Synthesis and Biological Evaluation as Anti-Bacterial Agents. ChemistrySelect 2022, 7, e202201007. [Google Scholar] [CrossRef]
- Abusetta, A.; Alumairi, J.; Alkaabi, M.Y.; Al Ajeil, R.; Shkaidim, A.A.; Akram, D.; Pajak, J. Design, Synthesis, in Vitro Antibacterial Activity, and Docking Studies of New Rhodanine Derivatives. Open J. Med. Chem. 2020, 10, 15–34. [Google Scholar]
- Kumar, A.S.; Kumar, R.A.; Reddy, E.P.; Satyanarayanaa, V.; Kashannab, J.; Reddy, B.J.M.; Reddy, B.V.S.; Yadav, J.S. Synthesis of Novel 2-Thioxothiazolidin-4-one and Thiazolidine-2, 4-dione.Derivatives as Potential Anticancer Agents. Nat. Prod. Commun. 2018, 13, 589–591. [Google Scholar]
- Manikala, V.; Rao, M.V. Synthesis, Molecular Docking and Anticancer Activity of Novel (E)-5-((1-phenyl-1H-1,2,3-triazol-4-yl)methylene)-2-thioxothiazolidin-4-one Analogues. Iran. J. Chem. Chem. Eng. 2021, 40, 1793–1799. [Google Scholar]
- Battula, H.; Bommi, S.; Bobde, Y.; Tarun Patel, T.; Balaram Ghosh, B.; Jayanty, S. Distinct rhodamine B derivatives exhibiting dual effect of anticancer activity and fluorescence property. J. Photochem. Photobiol. 2021, 6, 100026. [Google Scholar] [CrossRef]
- Tintori, C.; Iovenitt, G.; Ceresola, E.R.; Ferrarese, R.; Zamperini, C.; Brai, A.; Giulio Poli, G.; Dreassi, E.; Cagno, V.; Lembo, D.; et al. Rhodanine derivatives as potent anti-HIV and anti-HSV microbicides. PLoS ONE 2018, 13, e0198478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, L.; Yuan, Y.; Li, Q.; Han, L.; Yang, G.; Hu, H. Rhodanine derivative LJ001 inhibits TGEV and PDCoV replication in vitro. Virus Res. 2020, 289, 198167. [Google Scholar] [CrossRef]
- Khairul, A.; Kamar, D.A.; Yin, L.J.; Liang, C.T.; Fung, G.T.; Avupati, V.R. Rhodanine scaffold: A review of antidiabetic potential and structure–activity relationships (SAR). Med. Drug Dis. 2022, 15, 100131. [Google Scholar]
- Toumi, A.; Boudriga, S.; Hamden, K.; Sobeh, M.; Cheurfa, M.; Askri, M.; Knorr, M.; Strohmann, C.; Brieger, L. Synthesis, antidiabetic activity and molecular docking study of rhodanine-substitued spirooxindole pyrrolidine derivatives as novel α-amylase inhibitors. Bioorg. Chem. 2021, 106, 104507. [Google Scholar] [CrossRef]
- Xu, J.; Wang, T.T.; Yuan, Q.; Duan, Y.T.; Xu, Y.J.; Lv, P.C.; Wang, X.M.; Yang, Y.S.; Zhu, H.L. Discovery and development of novel rhodanine derivatives targeting enoyl-acyl carrier protein reductase. Bioorg. Med. Chem. 2019, 27, 1509–1516. [Google Scholar] [CrossRef] [PubMed]
- Previti, S.; Grasso, S.; Zappalà, M.; Ottanà, R. Identification of 2-thioxoimidazolidin-4-one derivatives as novel noncovalent proteasome and immunoproteasome inhibitors. Bioorg. Med. Chem. Lett. 2018, 28, 278–283. [Google Scholar]
- Vitaku, D.T.; Smith, J.T.N. Analysis of the structural diversity, substitution patterns, and frequency of nitrogen heterocycles among U.S. FDA approved pharmaceuticals. J. Med. Chem. 2014, 57, 10257–10274. [Google Scholar] [CrossRef]
- Dhiman, A.; Sharma, R.; Singh, R.K. Target-based anticancer indole derivatives and insight into structure—Activity relationship: A mechanistic review update (2018–2021). Acta Pharm. Sin. B 2022, 12, 3006–3027. [Google Scholar] [CrossRef]
- Devi, N.; Kaur, K.; Biharee, A.; Vikas Jaitak, V. Recent Development in Indole Derivatives as Anticancer Agent: A Mechanistic Approach. Anti-Cancer Agents Med. Chem. 2021, 21, 1802–1824. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Wen, X.; Gong, Y.; Wang, X. Current scenario of indole derivatives with potential anti-drug-resistant cancer activity. Eur. J. Med. Chem. 2020, 200, 112359. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.A.; Al-Omar, M.A.; Raish, M.; Ansari, M.A.; Abuelizz, H.A.; Bakheit, A.H.; Naglah, A.M. Indole Derivatives as Cyclooxygenase Inhibitors: Synthesis, Biological Evaluation and Docking Studies. Molecules 2018, 23, 1250. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Singh, R.K. Synthesis, Molecular Docking and Biological Evaluation of N-Substituted, Indole Derivatives as Potential Anti-Inflammatory and Antioxidant Agents. Chem. Biodivers. 2022, 19, e202200290. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; He, H.; Zhang, X.; Wu, R.; Gan, L.; Li, D.; Lu, Y.; Wu, P.; Wong, K. The in vitro and in vivo study of oleanolic acid indole derivatives as novel anti-inflammatory agents: Synthesis, biological evaluation, and mechanistic analysis. Bioorg. Chem. 2021, 113, 104981. [Google Scholar]
- Zhu, Y.; Zhao, J.; Luo, L.; Gao, Y.; Bao, H.; Li, P.; Zhang, H. Research progress of indole compound with potential antidiabetic activity. Eur. J. Med. Chem. 2021, 223, 113665. [Google Scholar] [CrossRef]
- Taha, M.; Alrashedy, A.S.; Almandil, N.B.; Iqbal, N.; Anouar, E.H.; Nawaz, M.; Uddin, N.; Chigurupati, S.; Wadood, A.; Rahim, F.; et al. Synthesis of indole derivatives as diabetics II inhibitors and enzymatic kinetics study of α-glucosidase and α-amylase along with their in-silico study. Int. J. Biol. Macromol. 2021, 190, 301–318. [Google Scholar] [CrossRef]
- Dorababu, A. Indole—A promising pharmacophore in recent antiviral drug discovery. RSC Med. Chem. 2020, 11, 1335–1353. [Google Scholar] [CrossRef]
- Sahin, K. Investigation of novel indole-based HIV-1 protease inhibitors using virtual screening and text mining. Investigation of novel indole-based HIV-1 protease inhibitors using virtual screening and text mining. J. Biomol. Struct. Dyn. 2021, 39, 3638–3648. [Google Scholar] [CrossRef]
- Cihan-Üstündağ, G.; Naesens, L.; Şatana, D.; Gonca Erköse-Genç, G.; Mataracı-Kara, E.; Çapan, G. Design, synthesis, antitubercular and antiviral properties of new spirocyclic indole derivatives. Mon. Chem. 2019, 150, 1533–1544. [Google Scholar] [CrossRef]
- Bajad, N.G.; Singh, S.K.; Singh, S.K.; Singh, T.D.; Singh, M. Indole: A promising scaffold for the discovery and development of potential anti-tubercular agents. Curr. Res. Pharmacol. Drug Discov. 2022, 3, 100119. [Google Scholar] [CrossRef] [PubMed]
- Porwal, S.; Gupta, S.; Chauhan, P.M.S. gem-Dithioacetylated indole derivatives as novel antileishmanial agents. Bioorg. Med. Chem. Lett. 2017, 27, 4643–4646. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Kirar, S.; Banerjee, U.C.; Neerupudi, K.B.; Singh, S.; Abdullah, A.; Prasad, V.; Bharatam, V.; Singh, I.P. Synthesis of N-substituted indole derivatives as potential antimicrobial and antileishmanial agents. Bioorg. Chem. 2020, 99, 103787. [Google Scholar] [CrossRef] [PubMed]
- Elkamhawy, A.; Woo, J.; Nada, H.; Angeli, A.; Bedair, T.M.; Supuran, C.T.; Lee, K. Identification of Novel and Potent Indole-Based Benzenesulfonamides as Selective Human Carbonic Anhydrase II Inhibitors: Design, Synthesis, In Vitro, and In Silico Studies. Int. J. Mol. Sci. 2022, 23, 2540. [Google Scholar] [CrossRef] [PubMed]
- Asati, V.; Bhupal, R.; Bhattacharya, S.; Kaur, K.; Gupta, G.D.; Pathak, A.; Mahapatra, D.K. Recent updates of Indole derivatives as kinase inhibitors in the treatment of cancer. Anti-Cancer Agents Med. Chem. 2022, 23, 404–416. [Google Scholar] [CrossRef]
- Yuan, W.; Yu, Z.; Song, W.; Li, Y.; Fang, Z.; Zhu, B.; Li, X.; Wang, H.; Hong, W.; Sun, N. Indole-core-based novel antibacterial agent targeting FtsZ. Infect. Drug Resist. 2019, 12, 2283–2296. [Google Scholar] [CrossRef]
- Al-Wabli, R.I.; Alsulami, M.A.; Bukhari, S.I.; Moubaye, N.M.S.; Al-Mutairi, M.S.; Attia, M.I. Design, Synthesis, and Antimicrobial Activity of Certain New Indole-1,2,4 Triazole Conjugates. Molecules 2021, 26, 2292. [Google Scholar] [CrossRef]
- Tha, S.; Shakya, S.; Malla, M.; Aryal, P. Prospects of Indole derivatives as methyl transfer inhibitors: Antimicrobial resistance managers. BMC Pharm. Toxicol. 2020, 21, 33–44. [Google Scholar] [CrossRef]
- Kaur, H.; Singh, J.; Narasimhan, B. Indole hybridized diazenyl derivatives: Synthesis, antimicrobial activity, cytotoxicity evaluation and docking studies. BMC Chem. 2019, 13, 65–83. [Google Scholar] [CrossRef]
- Qin, H.-L.; Jing, L.; Fang, W.-Y.; Rakesh, K.P. Indole-based derivatives as potential antibacterial activity against methicillin-resistance Staphylococcus aureus (MRSA). Eur. J. Med. Chem. 2020, 194, 112245. [Google Scholar] [CrossRef]
- Horishny, V.; Geronikaki, A.; Kartsev, V.; Matiychuk, V.; Petrou, A.; Pogodin, P.; Poroikov, V.; Theodoroula, N.F.; Vizirianakis, I.S.; Kostic, M.; et al. Synthesis, biological evaluation and molecular docking studies of 5-indolylmethylen-4-oxo-2-thioxothiazolidine derivatives. Molecules 2020, 25, 1964. [Google Scholar] [CrossRef] [PubMed]
- Michael, G.P.; Chennaiah, A.; Klara, H.; Sven, N.H.; Andrea, V.; Erik, C.B.; David, C. Importance of Co-operative Hydrogen Bonding in the Apramycin-Ribosomal Decoding A-Site Interaction. ChemMedChem 2022, e202200486. [Google Scholar]
- Verma, A.K.; Chennaiah, A.; Dubbu, S.; Vankar, Y.D. Palladium catalyzed synthesis of sugar-fused indolines via C(sp2)-H/N-H activation. Carbohydr. Res. 2019, 473, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Konidala, S.K.; Kotra, V.; Danduga, R.C.S.R.; Kola, P.K.; Bhandare, R.R.; Shaik, A.B. Design, multistep synthesis and in-vitro antimicrobial and antioxidant screening of coumarin clubbed chalcone hybrids through molecular hybridization approach. Arab. J. Chem. 2021, 14, 103154. [Google Scholar] [CrossRef]
- Liu, H.; Sun, D.; Du, H.; Zheng, C.; Li, J.; Piao, H.; Li, J.; Sun, S. Synthesis and biological evaluation of tryptophan-derived rhodanine derivatives as PTP1B inhibitors and anti-bacterial agents. Eur. J. Med. Chem. 2019, 172, 163–173. [Google Scholar] [CrossRef]
- Konechnyi, Y.T.; Lozynskyi1, A.V.; Horishny, V.Y.; Konechna, R.T.; Vynnytska, R.B.; Korniychuk, O.P.; Lesyk, R.B. Synthesis of indoline-thiazolidinone hybrids with antibacterial and antifungal activities. Biopolym. Cell 2020, 36, 381–391. [Google Scholar] [CrossRef]
- Akunuri, R.; Unnissa, T.; Kaul, G.; Akhir, A.; Saxena, D.; Wajidali, M.; Veerareddy, V.; Yaddanapudi, V.M.; Sidharth Chopra, S.; Nanduri, S. Synthesis and Antibacterial Evaluation of Rhodanine and Its Related Heterocyclic Compounds against S. aureus and A. baumannii. Chem. Biodivers. 2022, 19, e202200213. [Google Scholar] [CrossRef]
- Pardasavio, P.T.; Pardasavi, P.; Sherry, D.; Chatarverdi, V. Synthetic and antibacterial studies of rhodanine derivatives with indol-2,3-diones. Ind. J. Chem. 2001, 40B, 1275–1278. [Google Scholar]
- Song, M.X.; Li, S.H.; Peng, J.-Y.; Guo, T.-T.; Xu, W.-H.; Xiong, S.-F.; Deng, X.-Q. Synthesis and Bioactivity Evaluation of N-Arylsulfonylindole Analogs Bearing a Rhodanine Moiety as Antibacterial Agents. Molecules 2017, 22, 970. [Google Scholar] [CrossRef]
- Benson, T.E.; Walsh, C.T.; Massey, V. Kinetic characterization of wild-type and S229A mutant MurB: Evidence for the role of Ser 229 as a general acid. Biochemistry 1997, 36, 796–805. [Google Scholar] [CrossRef]
- Sohlenius-Sternbeck, A.K.; Terelius, Y. Evaluation of ADMET Predictor in Early Discovery Drug Metabolism and Pharmacokinetics Project Work. Drug Metab. Dispos. 2022, 50, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Naga, D.; Parrott, N.; Ecker, G.F.; Morale, A.O. Evaluation of the Success of High-Throughput Physiologically Based Pharmacokinetic (HT-PBPK) Modeling Predictions to Inform Early Drug Discovery. Mol. Pharm. 2022, 19, 2203–2216. [Google Scholar] [CrossRef] [PubMed]
- Kuriwaki, I.; Kameda, M.; Iikubo, K.; Hisamichi, H.; Kawamoto, Y.; Kikuchi, S.; Moritomo, H.; Terasaka, T.; Iwai, Y.; Noda, A.; et al. Discovery of ASP5878: Synthesis and structure–activity relationships of pyrimidine derivatives as pan-FGFRs inhibitors with improved metabolic stability and suppressed hERG channel inhibitory activity. Bioorg. Med. Chem. 2022, 59, 116657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; He, K.; Cai, L.; Chen, Y.C.; Yang, Y.; Shi, Q.; Woolf, T.F.; Ge, W.; Guo, L.; Borlak, J.; et al. Inhibition of bile salt transport by drugs associated with liver injury in primary hepatocytes from human, monkey, dog, rat, and mouse. Chem. Biol. Interact. 2016, 255, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef]
- Kritsi, E.; Matsoukas, M.T.; Potamitis, C.; Detsi, A.; Ivanov, M.; Sokovic, M.; Zoumpoulakis, P. Novel Hit Compounds as Putative Antifungals: The Case of Aspergillus fumigatus. Molecules 2019, 24, 3853. [Google Scholar] [CrossRef]
- Aleksić, M.; Stanisavljević, D.; Smiljković, M.; Vasiljević, P.; Stevanović, M.; Soković, M.; Stojković, D. Pyrimethanil: Between efficient fungicide against Aspergillus rot on cherry tomato and cytotoxic agent on human cell lines. Ann. Appl. Biol. 2019, 175, 228–235. [Google Scholar] [CrossRef]
- Fesatidou, M.; Zagaliotis, P.; Camoutsis, C.; Perou, A.; Eleftheriou, P.; Tratrtat, C.; Haroun, M.; Geronikaki, A.; Ciric, A.; Sokovic, M. 5-Adamantan thiadiazole-based thiazolidinones as antimicrobial agents. Design, synthesis, molecular docking and evaluation. Bioorg. Med. Chem. 2018, 26, 4664–4676. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Tseligka, E.D.; Rova, A.; Amanatiadou, E.P.; Calabrese, G.; Tsibouklis, J.; Fatouros, D.G.; Vizirianakis, I.S. Pharmacological Development of Target-Specific Delocalized Lipophilic Cation-Functionalized Carboranes for Cancer Therapy. Pharm. Res. 2016, 33, 1945–1958. [Google Scholar] [CrossRef]
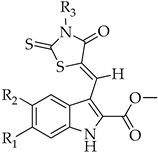 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | R1 | R2 | R3 | N | R1 | R2 | R3 | N | R1 | R2 | R3 |
| 1 | H | H | -(CH2)5COOH | 7 | H | H | 2-methylfuran | 13 | -OCH3 | H | 3-FC6H4 |
| 2 | H | H | 4-COOHC6H4 | 8 | H | F | -C(CH3)2COOH | 14 | H | F | -(CH2)2COOH |
| 3 | H | H | 3-OHC6H4 | 9 | H | F | -(CH2)3COOH | 15 | H | F | -CH3 |
| 4 | H | H | 4-OHC6H4 | 10 | H | F | 4-OHC6H4 | 16 | H | F | morpholine |
| 5 | H | H | 4-propylmorpholine | 11 | H | H | -CH2COOH | 17 | H | H | -(CH2)2COOH |
| 6 | H | H | morpholine | 12 | H | H | -C(CH3)2COOH | ||||
| Compounds | S.a. | B.c. | M.f. | L.m. | P.a. | E. coli | En.cl | S.t. | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MIC | 0.022 ± 0.005 | 0.015 ± 0.009 | 0.022 ± 0.005 | 0.022 ± 0.004 | 0.022 ± 0.005 | 0.015 ± 0.009 | 0.022 ± 0.005 | 0.015 ± 0.009 |
| MBC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 2 | MIC | 0.015 ± 0.009 | 0.015 ± 0.009 | 0.022 ± 0.005 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.022 ± 0.005 | 0.015 ± 0.009 | 0.022 ± 0.005 |
| MBC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 3 | MIC | 0.015 ± 0.009 | 0.015 ± 0.009 | 0.045 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.022 ± 0.005 | 0.015 ± 0.009 | 0.015 ± 0.009 |
| MBC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 4 | MIC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.045 ± 0.001 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.015 ± 0.009 | 0.045 ± 0.003 |
| MBC | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.06 ± 0.006 | |
| 5 | MIC | 0.015 ± 0.009 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.015 ± 0.009 | 0.022 ± 0.005 |
| MBC | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 6 | MIC | 0.015 ± 0.009 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.015 ± 0.009 | 0.03 ± 0.01 |
| MBC | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.12 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | |
| 7 | MIC | 0.015 ± 0.009 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.015 ± 0.009 | 0.015 ± 0.009 |
| MBC | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 8 | MIC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.015 ± 0.001 | 0.004 ± 0.005 | 0.008 ± 0.001 | 0.004 ± 0.005 | 0.008 ± 0.009 |
| MBC | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.008 ± 0.0006 | 0.015 ± 0.009 | 0.008 ± 0.0006 | 0.015 ± 0.009 | |
| 9 | MIC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 |
| MBC | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | |
| 10 | MIC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.045 ± 0.003 |
| MBC | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | |
| 11 | MIC | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.011 ± 0.01 | 0.015 ± 0.009 | 0.011 ± 0.01 | 0.015 ± 0.009 |
| MBC | 0.015 ± 0.009 | 0.015 ± 0.009 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 12 | MIC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.015 ± 0.001 | 0.015 ± 0.009 | 0.015 ± 0.009 | 0.03 ± 0.01 | 0.004 ± 0.005 |
| MBC | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.008 ± 0.009 | |
| 13 | MIC | 0.015 ± 0.009 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.015 ± 0.009 | 0.03 ± 0.01 |
| MBC | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.06 ± 0.006 | |
| 14 | MIC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.045 ± 0.003 | 0.015 ± 0.009 | 0.045 ± 0.003 | 0.015 ± 0.009 | 0.045 ± 0.003 |
| MBC | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.06 ± 0.006 | |
| 15 | MIC | 0.022 ± 0.005 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.045 ± 0.003 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.03 ± 0.01 | 0.045 ± 0.003 |
| MBC | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | |
| 16 | MIC | 0.022 ± 0.005 | 0.015 ± 0.009 | 0.045 ± 0.003 | 0.045 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.045 ± 0.003 | 0.045 ± 0.003 |
| MBC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | |
| 17 | MIC | 0.008 ± 0.0006 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.008 ± 0.000 | 0.045 ± 0.003 | 0.008 ± 0.0006 | 0.03 ± 0.01 |
| MBC | 0.015 ± 0.00 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.06 ± 0.006 | 0.015 ± 0.009 | 0.06 ± 0.006 | 0.015 ± 0.009 | 0.06 ± 0.006 | |
| Ampicillin | MIC | 0.10 ± 0.05 | 0.006 ± 0.003 | 0.25 ± 0.09 | 0.15 ± 0.03 | 0.30 ± 0.03 | 0.10 ± 0.05 | 0.25 ± 0.09 | 0.15 ± 0.03 |
| MBC | 0.15 ± 0.05 | 0.025 ± 0.00 | 0.50 ± 0.1 | 0.30 ± 0.03 | 0.50 ± 0.1 | 0.20 ± 0.00 | 0.50 ± 0.1 | 0.20 ± 0.009 | |
| Streptomycin | MIC | 0.10 ± 0.00 | 0.0015 ± 0.0002 | 0.20 ± 0.00 | 0.15 ± 0.03 | 0.10 ± 0.05 | 0.10 ± 0.05 | 0.20 ± 0.005 | 0.10 ± 0.05 |
| MBC | 0.20 ± 0.00 | 0.003 ± 0.0005 | 0.30 ± 0.03 | 0.30 ± 0.03 | 0.20 ± 0.00 | 0.20 ± 0.00 | 0.30 ± 0.03 | 0.20 ± 0.009 |
| Compounds | A.f. | A.v. | A.o. | A.n. | T.v. | P.o. | P.f. | P.v.c. | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MIC | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.008 ± 0.0006 | 0.022 ± 0.005 | 0.015 ± 0.003 | 0.004 ± 0.001 | 0.015 ± 0.003 | 0.03 ± 0.01 |
| MFC | 0.12 ± 0.04 | 0.06 ± 0.006 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.008 ± 0.0006 | 0.03 ± 0.01 | 0.06 ± 0.006 | |
| 2 | MIC | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.11 ± 0.01 | 0.022 ± 0.005 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 |
| MFC | 0.06 ± 0.006 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | |
| 3 | MIC | 0.03 ± 0.01 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.11 ± 0.01 | 0.004 ± 0.001 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.015 ± 0.003 |
| MFC | 0.06 ± 0.006 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 | |
| 4 | MIC | 0.06 ± 0.006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 |
| MFC | 0.12 ± 0.04 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | |
| 5 | MIC | 0.06 ± 0.006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.004 ± 0.001 | 0.11 ± 0.01 | 0.015 ± 0.003 | 0.015 ± 0.003 |
| MFC | 0.12 ± 0.04 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 6 | MIC | 0.06 ± 0.006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.11 ± 0.01 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.015 ± 0.003 |
| MFC | 0.12 ± 0.04 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 7 | MIC | 0.06 ± 0.006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.004 ± 0.001 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.015 ± 0.003 |
| MFC | 0.12 ± 0.04 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 8 | MIC | 0.06 ± 0.006 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.11 ± 0.01 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.06 ± 0.006 |
| MFC | 0.12 ± 0.04 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.12 ± 0.04 | |
| 9 | MIC | 0.09 ± 0.003 | 0.015 ± 0.003 | 0.004 ± 0.001 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.022 ± 0.005 |
| MFC | 0.12 ± 0.04 | 0.03 ± 0.01 | 0.008 ± 0.0006 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.015 ± 0.003 | |
| 10 | MIC | 0.06 ± 0.006 | 0.008 ± 0.0006 | 0.004 ± 0.001 | 0.008 ± 0.0006 | 0.004 ± 0.001 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.015 ± 0.003 |
| MFC | 0.12 ± 0.04 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 | |
| 11 | MIC | 0.06 ± 0.006 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 |
| MFC | 0.12 ± 0.04 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 12 | MIC | 0.06 ± 0.006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.022 ± 0.005 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 |
| MFC | 0.12 ± 0.04 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | |
| 13 | MIC | 0.06 ± 0.006 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.11 ± 0.01 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 |
| MFC | 0.12 ± 0.04 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 14 | MIC | 0.06 ± 0.006 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.11 ± 0.01 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 |
| MFC | 0.12 ± 0.04 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 15 | MIC | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 | 0.008 ± 0.0006 |
| MFC | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | |
| 16 | MIC | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 |
| MFC | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| 17 | MIC | 0.06 ± 0.006 | 0.015 ± 0.003 | 0.008 ± 0.0006 | 0.022 ± 0.005 | 0.11 ± 0.01 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 |
| MFC | 0.12 ± 0.04 | 0.03 ± 0.01 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.06 ± 0.006 | |
| Ketoconazole | MIC | 0.20 ± 0.01 | 0.20 ± 0.01 | 0.25 ± 0.05 | 0.20 ± 0.01 | 1.00 ± 0.1 | 2.50 ± 0.3 | 0.20 ± 0.01 | 0.20 ± 0.01 |
| MFC | 0.50 ± 0.001 | 0.50 ± 0.002 | 0.50 ± 0.006 | 0.50 ± 0.004 | 1.50 ± 0.09 | 3.50 ± 0.03 | 0.50 ± 0.003 | 0.30 ± 0.01 | |
| Bifonazole | MIC | 0.15 ± 0.05 | 0.10 ± 0.002 | 0.15 ± 0.05 | 0.15 ± 0.05 | 0.15 ± 0.05 | 0.20 ± 0.01 | 0.20 ± 0.01 | 0.10 ± 0.009 |
| MFC | 0.20 ± 0.01 | 0.20 ± 0.01 | 0.20 ± 0.01 | 0.20 ± 0.01 | 0.20 ± 0.01 | 0.25 ± 0.05 | 0.25 ± 0.05 | 0.20 ± 0.01 |
| Comp. | Est. Binding Energy (kcal/mol) | Residues Involved in H-Bond Formation in E. coli MurB | ||||
|---|---|---|---|---|---|---|
| E. coli Gyrase 1KZN | Thymidylate Kinase 4QGG | E. coli Primase 1DDE | E. coli MurA JV4T | E. coli MurB 2Q85 | ||
| 1 | −4.15 | −1.68 | - | −5.63 | −9.82 | Arg213, Ser229 |
| 2 | −3.17 | - | - | −5.23 | −10.04 | Arg158, Ser229 |
| 3 | −4.63 | −2.55 | −1.28 | −6.27 | −9.53 | Arg213, Ser229 |
| 4 | −5.10 | - | - | −4.63 | −7.90 | Arg158, Arg213 |
| 5 | −3.51 | - | - | −5.20 | −8.97 | Arg213, Ser229 |
| 6 | −4.15 | - | −1.39 | −4.82 | −7.68 | Arg213 |
| 7 | −4.38 | - | - | −6.12 | −9.14 | Arg158, Ser229 |
| 8 | −5.37 | −2.85 | - | −5.92 | −10.88 | Ser50, Ser229 |
| 9 | −4.12 | - | - | −5.37 | −8.11 | Ser50, Arg158 |
| 10 | −5.47 | - | - | −5.42 | −7.54 | Ser50 |
| 11 | −5.21 | −3.47 | −2.61 | −6.20 | −10.46 | Ser116, Ser229 |
| 12 | −3.50 | −2.51 | − | −5.73 | −9.56 | Ser229 |
| 13 | −4.39 | - | − | −5.14 | −8.55 | Ser229 |
| 14 | −4.26 | - | −1.29 | −5.32 | −8.42 | Ser229 |
| 15 | −3.94 | - | −2.57 | −5.18 | −7.60 | Arg213 |
| 16 | −5.28 | - | - | −4.92 | −8.30 | Ser229 |
| 17 | −4.36 | - | - | −5.16 | −9.46 | Arg158, Ser229 |
| Est. Binding Energy (kcal/mol) | Residues Involved in H-Bond Formation | Residues Involved in Hydrophobic Interactions | Residues Involved in Aromatic Interactions | Interactions with HEM601 | ||
|---|---|---|---|---|---|---|
| Comp. | DNA TopoIV 1S16 | CYP51 of C. albicans 5V5Z | ||||
| 1 | −3.69 | −7.54 | - | Tyr118, Thr311, Phe380, Met508, Hem601 | - | Hydrophobic |
| 2 | −3.18 | −9.70 | Tyr132 | Tyr118, Leu300, Ile304, Thr311, Hem601 | Tyr118 | Hydrophobic |
| 3 | −3.25 | −12.34 | Tyr132 | Tyr118,Ile131, Ile304, Hem601 | Hem601 | Hydrophobic, aromatic, Fe binding |
| 4 | −2.66 | −8.70 | - | Tyr118, Leu300, Thr311, Leu376, Phe380, Met508, Hem601 | Tyr118, Hem601 | Hydrophobic, aromatic |
| 5 | −4.25 | −9.34 | Tyr118 | Tyr118, Leu376, Met508, Hem601 | Tyr118 | Hydrophobic |
| 6 | −3.14 | −9.65 | Tyr64 | Tyr118, Tyr122, Thr311, Leu376, Phe380, Hem601 | Tyr118 | Hydrophobic |
| 7 | −4.17 | −9.82 | Tyr132 | Tyr118, Leu121, Thr311, Phe380, Met508, Hem601 | Hem601 | Hydrophobic, aromatic |
| 8 | −2.59 | −7.11 | - | Tyr118, Leu376, Met508, Hem601 | - | Hydrophobic |
| 9 | −4.39 | −9.54 | Tyr132 | Tyr118, Phe380, Met508, Hem601 | - | Hydrophobic |
| 10 | −2.67 | −10.11 | Tyr64 | Tyr118, Leu300, Thr311, Met508, Hem601 | Tyr118, Hem601 | Hydrophobic, aromatic |
| 11 | −2.73 | −9.31 | Tyr118 | Leu300, Met508, Hem601 | Hem601 | Hydrophobic, aromatic |
| 12 | −4.37 | −8.62 | - | Tyr118, Tyr122, Thr311, Met508, Hem601 | Tyr122, Hem601 | Hydrophobic, aromatic |
| 13 | −3.56 | −8.81 | - | Tyr118, Tyr122, Thr311, Leu376, Phe380, Hem601 | Tyr118 | Hydrophobic |
| 14 | −3.28 | −7.64 | - | Tyr118, Thr311, Leu376, Met508, Hem601 | - | Hydrophobic |
| 15 | −3.10 | −12.95 | Thr311 | Ile131, Leu300, Ile304, Hem601 | Hem601 | Hydrophobic, aromatic, Fe binding |
| 16 | −2.67 | −10.26 | Tyr132 | Tyr118, Tyr122, Leu300, Ile304, Hem601 | Hem601 | Hydrophobic, aromatic |
| 17 | −2.57 | −7.96 | - | Tyr118, Tyr122, Thr311, Leu376, Met508, Hem601 | Tyr118, Hem601 | Hydrophobic, aromatic |
| Ketoconazole | - | −8.23 | Tyr64 | Tyr118, Ile131, Tyr132, Leu300, Ile304, Leu376, Met508, Hem601 | Hem601 | Hydrophobic, aromatic |
| Comp. | MW a (≤500) | HBAs b (≤10) | HBDs c (≤5) | Lipophilicity (ClogP) d (≤5) | RBs e (≤10) | TPSA f (≤140) | Lipinski Violations | Bioavailability Score | Drug-Likeness Score |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 432.51 | 5 | 2 | 3.30 | 9 | 157.09 | 0 | 0.55 | −0.14 |
| 2 | 438.48 | 5 | 2 | 3.45 | 5 | 157.09 | 0 | 0.11 | −0.64 |
| 3 | 410.47 | 4 | 2 | 3.46 | 4 | 140.02 | 0 | 0.55 | −0.78 |
| 4 | 410.47 | 4 | 2 | 3.47 | 4 | 140.02 | 0 | 0.55 | −0.81 |
| 5 | 445.56 | 5 | 1 | 2.86 | 7 | 132.26 | 0 | 0.55 | +0.24 |
| 6 | 403.48 | 5 | 1 | 2.48 | 4 | 132.26 | 0 | 0.55 | −0.38 |
| 7 | 398.46 | 4 | 1 | 3.28 | 5 | 132.93 | 0 | 0.55 | −0.69 |
| 8 | 436.48 | 6 | 2 | 3.31 | 6 | 157.09 | 0 | 0.55 | −0.70 |
| 9 | 422.45 | 6 | 2 | 2.99 | 7 | 157.09 | 0 | 0.11 | −0.37 |
| 10 | 428.46 | 5 | 2 | 3.78 | 4 | 140.02 | 0 | 0.55 | −0.77 |
| 11 | 376.41 | 5 | 2 | 2.11 | 5 | 157.09 | 0 | 0.55 | −0.71 |
| 12 | 418.49 | 5 | 2 | 3.02 | 6 | 157.09 | 0 | 0.11 | −0.24 |
| 13 | 442.48 | 5 | 1 | 4.15 | 5 | 129.02 | 0 | 0.55 | −0.95 |
| 14 | 408.42 | 6 | 2 | 2.65 | 6 | 157.09 | 0 | 0.11 | −0.40 |
| 15 | 350.39 | 4 | 1 | 3.02 | 3 | 119.79 | 0 | 0.55 | −0.15 |
| 16 | 421.47 | 6 | 1 | 2.79 | 4 | 132.26 | 0 | 0.55 | −0.14 |
| 17 | 390.43 | 5 | 2 | 2.35 | 6 | 157.09 | 0 | 0.11 | −0.59 |
| No | Absorption | Distribution | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peff a | MDCK b | Sw c | SpH d | FaSSGF e | FaSSIF f | FeSSIF g | Absorption Risk | BBB h | LogBBB i | fu% j | Vd k | RBP l | |
| 1 | 1.642 | 18.188 | 23 | 10.009 | 0.009 | 0.067 | 0.160 | 0.000 | Low (59%) | −1.231 | 4.588 | 0.328 | 0.661 |
| 2 | 2.232 | 112.633 | 5 | 1.295 | 0.008 | 0.040 | 0.115 | 1.000 | Low (66%) | −0.793 | 4.446 | 0.244 | 0.692 |
| 3 | 2.491 | 160.415 | 2 | 0.002 | 0.026 | 0.010 | 0.155 | 1.000 | High (89%) | −0.071 | 4.548 | 0.691 | 0.738 |
| 4 | 2.439 | 192.624 | 2 | 0.002 | 0.026 | 0.008 | 0.111 | 1.000 | High (88%) | −0.127 | 4.449 | 0.742 | 0.737 |
| 5 | 1.979 | 244.487 | 30 | 0.049 | 0.914 | 0.031 | 0.089 | 0.000 | High (99%) | −0.006 | 9.925 | 1.232 | 0.748 |
| 6 | 2.608 | 289.456 | 8 | 0.008 | 0.124 | 0.071 | 0.197 | 0.329 | High (99%) | −0.130 | 9.631 | 1.097 | 0.767 |
| 7 | 2.846 | 695.382 | 2 | 0.002 | 0.013 | 0.013 | 0.159 | 1.000 | High (94%) | −0.393 | 5.233 | 0.780 | 0.744 |
| 8 | 1.713 | 20.994 | 28 | 6.904 | 0.009 | 0.035 | 0.381 | 0.000 | Low (84%) | −0.964 | 5.525 | 0.325 | 0.680 |
| 9 | 1.824 | 11.524 | 37 | 14.568 | 0.014 | 0.090 | 0.207 | 0.000 | Low (66%) | −1.151 | 5.491 | 0.328 | 0.677 |
| 10 | 2.552 | 238.505 | 2 | 0.002 | 0.024 | 0.005 | 0.104 | 1.000 | High (77%) | −0.046 | 4.722 | 0.746 | 0.741 |
| 11 | 1.557 | 18.324 | 37 | 7.429 | 0.017 | 0.092 | 0.380 | 0.000 | Low (74%) | −1.047 | 5.954 | 0.313 | 0.684 |
| 12 | 1.624 | 16.857 | 29 | 7.179 | 0.010 | 0.051 | 0.308 | 0.000 | Low (84%) | −1.056 | 5.402 | 0.327 | 0.677 |
| 13 | 3.755 | 512.700 | 1 | 0.001 | 0.018 | 0.006 | 0.085 | 1.918 | High (96%) | 0.129 | 4.768 | 0.861 | 0.731 |
| 14 | 1.864 | 39.838 | 38 | 13.699 | 0.014 | 0.178 | 0.322 | 0.000 | Low (90%) | −0.997 | 5.963 | 0.313 | 0.683 |
| 15 | 3.536 | 572.262 | 4 | 0.004 | 0.099 | 0.018 | 0.211 | 1.000 | High (99%) | 0.137 | 9.910 | 0.779 | 0.783 |
| 16 | 3.229 | 352.377 | 8 | 0.008 | 0.122 | 0.061 | 0.169 | 0.370 | High (99%) | −0.037 | 10.236 | 1.141 | 0.769 |
| 17 | 1.652 | 29.583 | 38 | 13.731 | 0.015 | 0.236 | 0.260 | 0.000 | Low (84%) | −1.085 | 5.838 | 0.314 | 0.680 |
| No | Metabolism | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP 1A2 Inh. | CYP1A2 Substr. Km | CYP 2C9 Inh | CYP2C9 Substr. Km | CYP 2C19 Inh. | CYP2C19 Substr. Km | CYP 2D6 Inh. | CYP2D6 Substr. Km | CYP 3A4 Inh. | CYP3A4 Substr. Km | CYP CLint a | HEP CLint b | UGTs Subs. c | CYP Risk | |
| 1 | No | Non Subs. | No | 86.146 | No | 24.703 | No | Non Subs. | No | Non Subs. | 6.568 | 11.362 | 1A3, 1A9 | 0.000 |
| 2 | No | 23.682 | No | 8.127 | No | 39.264 | No | Non Subs. | No | Non Subs. | 100.398 | 16.359 | 1A8, 1A9, 1A10 | 2.000 |
| 3 | No | 30.916 | No | 14.403 | No | 29.495 | No | 127.051 | Yes | Non Subs. | 65.013 | 71.146 | 1A1, 1A8, 1A9, 1A10, 2B15 | 2.372 |
| 4 | No | 29.485 | No | 8.423 | No | 93.004 | No | 204.398 | Yes | Non Subs. | 80.365 | 89.557 | 1A1, 1A8, 1A9, 1A10, 2B15 | 2.985 |
| 5 | No | 18.129 | No | 83.216 | No | 20.465 | No | 13.006 | Yes | 22.767 | 78.710 | 23.739 | 1A4, 1A9 | 1.147 |
| 6 | No | 7.486 | No | 87.569 | No | 44.972 | No | 28.012 | Yes | 31.235 | 101.603 | 18.418 | Non Subs. | 1.000 |
| 7 | No | 26.192 | No | 15.906 | No | 4.882 | No | 38.409 | Yes | 21.943 | 89.261 | 21.857 | 1A9 | 0.981 |
| 8 | No | Non Subs. | No | 20.918 | No | 203.738 | No | Non Subs. | No | Non Subs. | 19.358 | 7.088 | 1A3, 1A8, 1A9, 1A10 | 0.935 |
| 9 | No | Non Subs. | No | 123.319 | No | 16.370 | No | Non Subs. | Yes | Non Subs. | 6.868 | 8.300 | 1A3, 1A9 | 0.000 |
| 10 | No | 34.151 | No | 7.744 | No | 94.454 | No | 255.204 | Yes | Non Subs. | 131.921 | 93.639 | 1A1, 1A8, 1A9, 1A10, 2B15 | 3.000 |
| 11 | No | Non Subs. | No | 36.930 | No | 220.331 | No | Non Subs. | No | Non Subs. | 9.918 | 5.080 | 1A3, 1A8, 1A9 | 0.000 |
| 12 | No | Non Subs. | No | 25.953 | No | 199.456 | No | Non Subs. | No | Non Subs. | 12.348 | 6.689 | 1A3, 1A8, 1A9 | 0.234 |
| 13 | No | 3.514 | No | 15.364 | No | 66.722 | No | 27.176 | Yes | 11.550 | 259.955 | 80.554 | 1A1, 1A9 | 3.228 |
| 14 | No | Non Subs. | No | 96.500 | No | 5.165 | No | Non Subs. | Yes | Non Subs. | 13.286 | 6.370 | 1A3, 1A9 | 0.000 |
| 15 | No | 20.385 | No | 167.240 | No | 60.458 | No | 91.889 | Yes | 130.324 | 57.430 | 21.782 | 1A1, 1A9 | 0.740 |
| 16 | No | 9.212 | No | 64.353 | No | 48.228 | No | 32.873 | Yes | 24.123 | 102.341 | 18.506 | 1A8, 1A9 | 1.055 |
| 17 | No | Non Subs. | No | 100.364 | No | 4.815 | No | Non Subs. | Yes | Non Subs. | 10.276 | 5.966 | 1A3, 1A9 | 0.000 |
| No | Pgp Sub. | Pgp Inh. | BCRP Sub. | BCRP Inh. | BSEP Inh. |
|---|---|---|---|---|---|
| 1 | Yes | No (49%) | Yes | No | No |
| 2 | Yes | Yes (62%) | Yes | No | No |
| 3 | Yes | Yes (97%) | Yes | No | No |
| 4 | Yes | Yes (97%) | Yes | No | No |
| 5 | Yes | Yes (88%) | Yes | No | No |
| 6 | Yes | Yes (60%) | Yes | No | No |
| 7 | Yes | Yes (97%) | Yes | No | No |
| 8 | Yes | No (93%) | Yes | No | No |
| 9 | Yes | No (49%) | Yes | No | No |
| 10 | Yes | Yes (97%) | Yes | No | No |
| 11 | Yes | No (93%) | Yes | No | No |
| 12 | Yes | No (93%) | No | No | No |
| 13 | Yes | Yes (97%) | Yes | No | No |
| 14 | Yes | No (68%) | Yes | No | No |
| 15 | Yes | No (46%) | Yes | No | No |
| 16 | Yes | Yes (57%) | Yes | No | No |
| 17 | Yes | No (78%) | Yes | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrou, A.; Geronikaki, A.; Kartsev, V.; Kousaxidis, A.; Papadimitriou-Tsantarliotou, A.; Kostic, M.; Ivanov, M.; Sokovic, M.; Nicolaou, I.; Vizirianakis, I.S. N-Derivatives of (Z)-Methyl 3-(4-Oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylates as Antimicrobial Agents—In Silico and In Vitro Evaluation. Pharmaceuticals 2023, 16, 131. https://doi.org/10.3390/ph16010131
Petrou A, Geronikaki A, Kartsev V, Kousaxidis A, Papadimitriou-Tsantarliotou A, Kostic M, Ivanov M, Sokovic M, Nicolaou I, Vizirianakis IS. N-Derivatives of (Z)-Methyl 3-(4-Oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylates as Antimicrobial Agents—In Silico and In Vitro Evaluation. Pharmaceuticals. 2023; 16(1):131. https://doi.org/10.3390/ph16010131
Chicago/Turabian StylePetrou, Anthi, Athina Geronikaki, Victor Kartsev, Antonios Kousaxidis, Aliki Papadimitriou-Tsantarliotou, Marina Kostic, Marija Ivanov, Marina Sokovic, Ioannis Nicolaou, and Ioannis S. Vizirianakis. 2023. "N-Derivatives of (Z)-Methyl 3-(4-Oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylates as Antimicrobial Agents—In Silico and In Vitro Evaluation" Pharmaceuticals 16, no. 1: 131. https://doi.org/10.3390/ph16010131
APA StylePetrou, A., Geronikaki, A., Kartsev, V., Kousaxidis, A., Papadimitriou-Tsantarliotou, A., Kostic, M., Ivanov, M., Sokovic, M., Nicolaou, I., & Vizirianakis, I. S. (2023). N-Derivatives of (Z)-Methyl 3-(4-Oxo-2-thioxothiazolidin-5-ylidene)methyl)-1H-indole-2-carboxylates as Antimicrobial Agents—In Silico and In Vitro Evaluation. Pharmaceuticals, 16(1), 131. https://doi.org/10.3390/ph16010131