Synthesis and Anticancer Evaluation of New 1,3,4-Oxadiazole Derivatives
Abstract
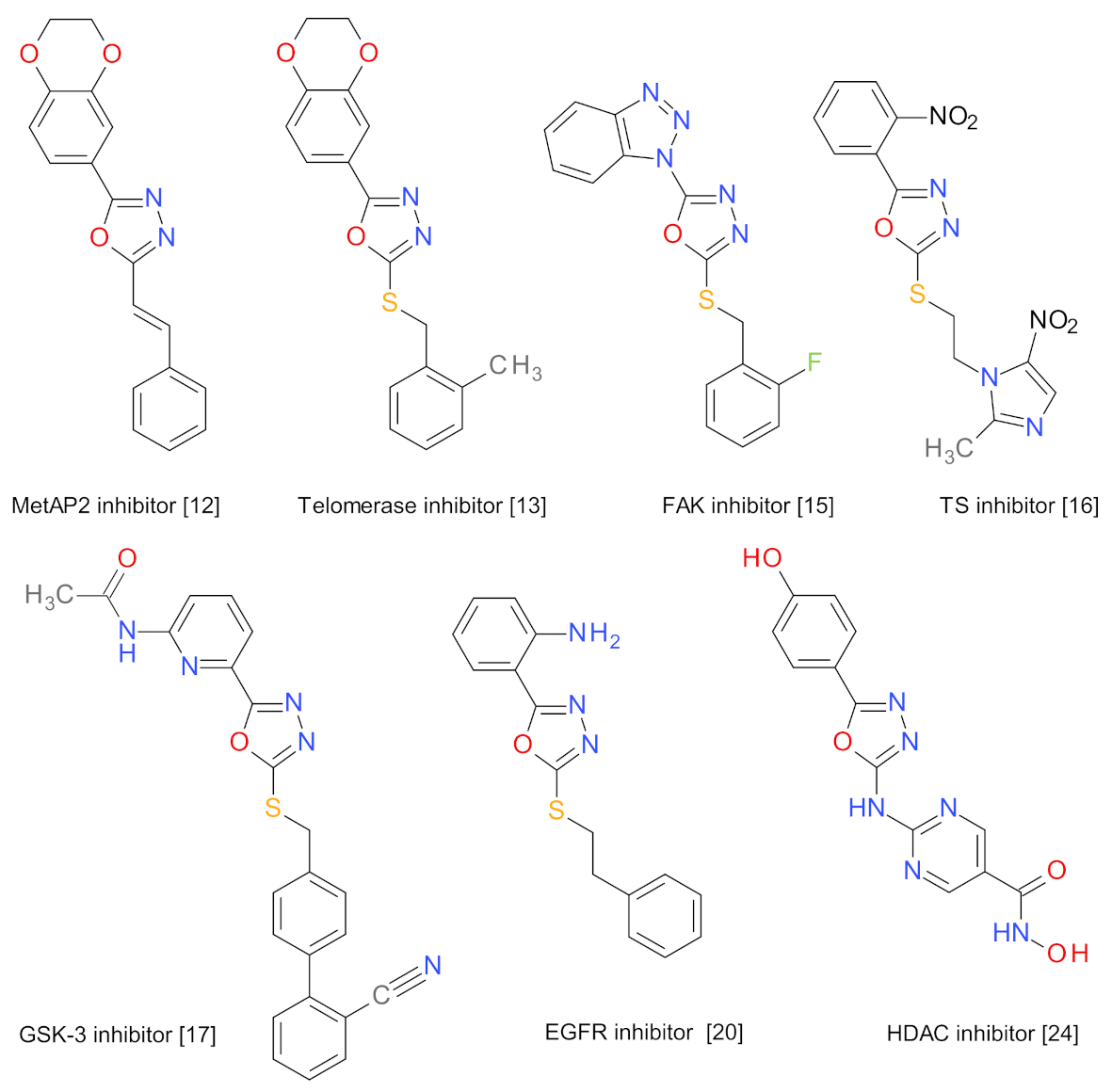
1. Introduction
2. Results
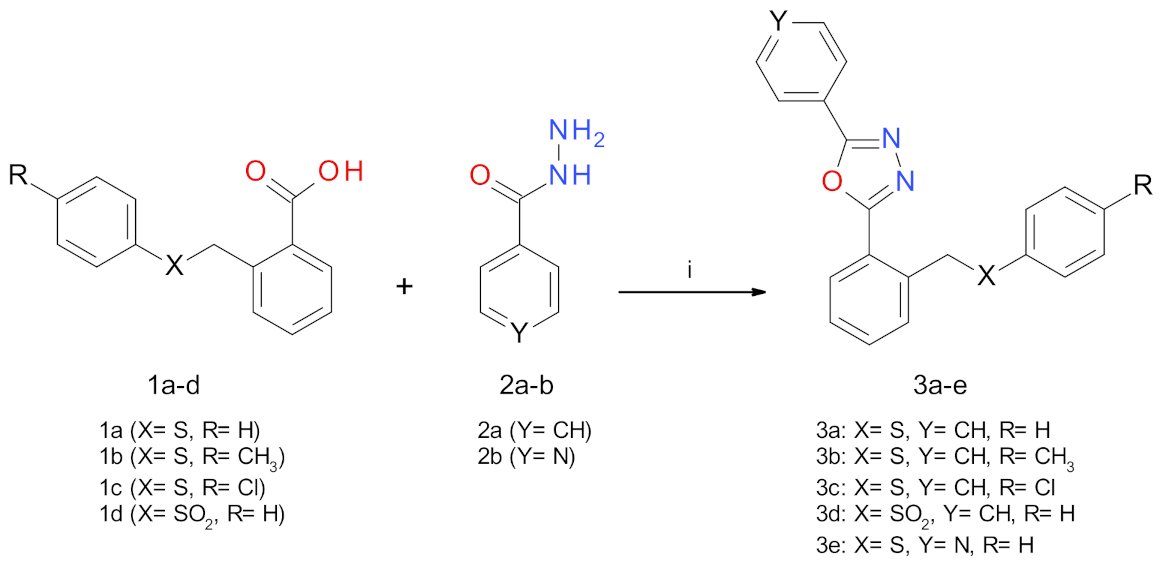
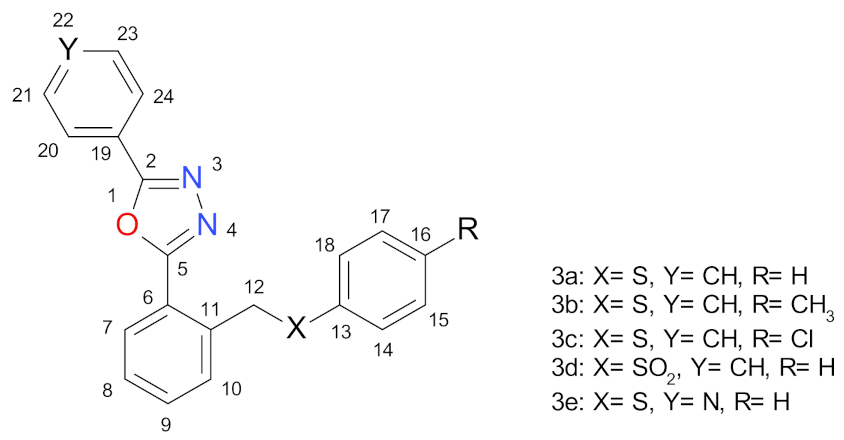
2.1. Synthesis Procedures
2.2. Anticancer Evaluation
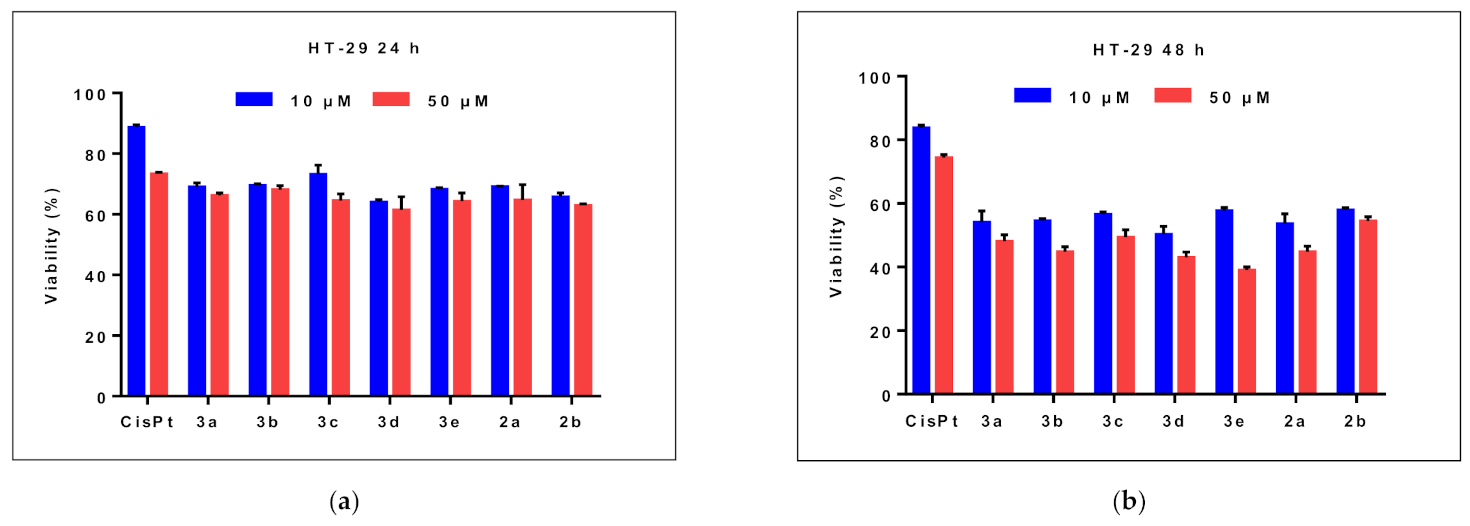
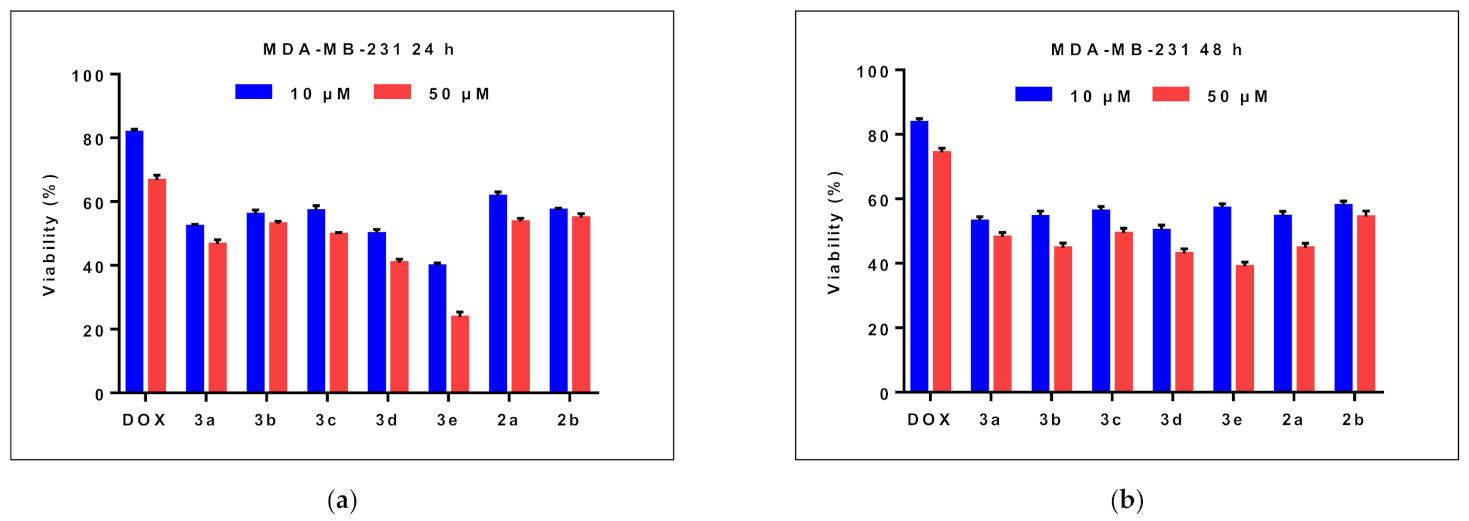
2.2.1. Effects on Cell Viability
2.2.2. Effects on Cell Apoptosis
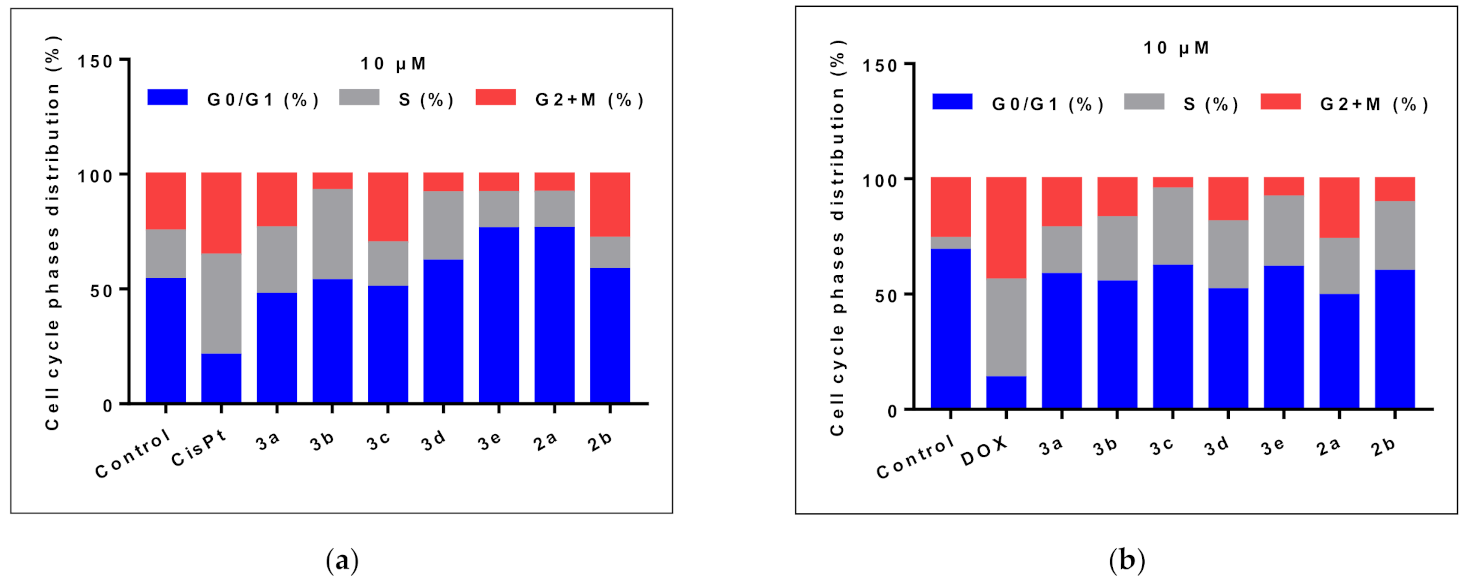
2.2.3. Cell Cycle Analysis
2.3. Daphnia Magna Toxicity Assay
2.4. Prediction of the Molecular Mechanism of Action and Toxicity
2.4.1. PASS Prediction
2.4.2. Structural Similarity Analysis
2.4.3. Predicted Acute Rat Toxicity
3. Discussion
4. Materials and Methods
4.1. Analytical Procedures
4.2. Synthesis Procedures
4.2.1. Synthesis of 2-[(benzenesulfonyl)methyl]benzoic acid (1d)
4.2.2. General Procedure for the Synthesis of the 1,3,4-oxadiazoles Derivatives (3a-e)
4.2.3. 2-Phenyl-5-[2-(phenylsulfanylmethyl)phenyl]-1,3,4-oxadiazole (3a)
4.2.4. 2-Phenyl-5-[2-(p-tolylsulfanylmethyl)phenyl]-1,3,4-oxadiazole (3b)
4.2.5. 2-[2-[(4-Chlorophenyl)sulfanylmethyl]phenyl]-5-phenyl-1,3,4-oxadiazole (3c)
4.2.6. 2-[2-(Benzenesulfonylmethyl)phenyl]-5-phenyl-1,3,4-oxadiazole (3d)
4.2.7. 2-[2-(Phenylsulfanylmethyl)phenyl]-5-(4-pyridyl)-1,3,4-oxadiazole (3e)
4.3. Anticancer Evaluation
4.3.1. Reagents
4.3.2. Cell Culture and Treatments
4.3.3. Cytotoxicity Assay
4.3.4. Apoptosis Analysis
4.3.5. Cell Cycle Analysis
4.4. Daphnia Magna Toxicity Assay
4.5. Prediction of the Molecular Mechanism of Action and Toxicity
4.5.1. PASS Prediction
4.5.2. Structural Similarity Analysis
4.5.3. Prediction of the Compounds’ Toxicity
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2021, 68, 394–424. [Google Scholar]
- Wang, X.; Zhang, H.; Chen, X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019, 2, 141–160. [Google Scholar] [CrossRef]
- Schirrmacher, V. From chemotherapy to biological therapy: A review of novel concepts to reduce the side effects of systemic cancer treatment (Review). Int. J. Oncol. 2019, 54, 407–419. [Google Scholar]
- Ion, G.N.D.; Olaru, O.T.; Nitulescu, G.; Olaru, I.I.; Tsatsakis, A.; Burykina, T.I.; Spandidos, D.A.; Nitulescu, G.M. Improving the odds of success in antitumoral drug development using scoring approaches towards heterocyclic scaffolds. Oncol. Rep. 2020, 44, 589–598. [Google Scholar] [CrossRef]
- Lang, D.K.; Kaur, R.; Arora, R.; Saini, B.; Arora, S. Nitrogen-Containing Heterocycles as Anticancer Agents: An Overview. Anti-Cancer Agents Med. Chem. 2020, 20, 2150–2168. [Google Scholar] [CrossRef]
- Boström, J.; Hogner, A.; Llinàs, A.; Wellner, E.; Plowright, A.T. Oxadiazoles in medicinal chemistry. J. Med. Chem. 2012, 55, 1817–1830. [Google Scholar] [CrossRef]
- Benassi, A.; Doria, F.; Pirota, V. Groundbreaking anticancer activity of highly diversified oxadiazole scaffolds. Int. J. Mol. Sci. 2020, 21, 8692. [Google Scholar] [CrossRef]
- Glomb, T.; Szymankiewicz, K.; Świątek, P. Anti-cancer activity of derivatives of 1,3,4-oxadiazole. Molecules 2018, 23, 3361. [Google Scholar] [CrossRef]
- Akhtar, J.; Khan, A.A.; Ali, Z.; Haider, R.; Shahar Yar, M. Structure-activity relationship (SAR) study and design strategies of nitrogen-containing heterocyclic moieties for their anticancer activities. Eur. J. Med. Chem. 2017, 125, 143–189. [Google Scholar] [CrossRef] [PubMed]
- Sankhe, N.M.; Durgashivaprasad, E.; Gopalan Kutty, N.; Venkata Rao, J.; Narayanan, K.; Kumar, N.; Jain, P.; Udupa, N.; Vasanth Raj, P. Novel 2,5-disubstituted-1,3,4-oxadiazole derivatives induce apoptosis in HepG2 cells through p53 mediated intrinsic pathway. Arab. J. Chem. 2019, 12, 2548–2555. [Google Scholar] [CrossRef]
- Hamdy, R.; Elseginy, S.A.; Ziedan, N.I.; El-Sadek, M.; Lashin, E.; Jones, A.T.; Westwell, A.D. Design, synthesis and evaluation of new bioactive oxadiazole derivatives as anticancer agents targeting bcl-2. Int. J. Mol. Sci. 2020, 21, 8980. [Google Scholar] [CrossRef]
- Sun, J.; Li, M.H.; Qian, S.S.; Guo, F.J.; Dang, X.F.; Wang, X.M.; Xue, Y.R.; Zhu, H.L. Synthesis and antitumor activity of 1,3,4-oxadiazole possessing 1,4-benzodioxan moiety as a novel class of potent methionine aminopeptidase type II inhibitors. Bioorg. Med. Chem. Lett. 2013, 23, 2876–2879. [Google Scholar] [CrossRef]
- Zhang, X.M.; Qiu, M.; Sun, J.; Zhang, Y.B.; Yang, Y.S.; Wang, X.L.; Tang, J.F.; Zhu, H.L. Synthesis, biological evaluation, and molecular docking studies of 1,3,4-oxadiazole derivatives possessing 1,4-benzodioxan moiety as potential anticancer agents. Bioorg. Med. Chem. 2011, 19, 6518–6524. [Google Scholar] [CrossRef]
- Bajaj, S.; Roy, P.P.; Singh, J. 1,3,4-Oxadiazoles as Telomerase Inhibitor: Potential Anticancer Agents. Anti-Cancer Agents Med. Chem. 2017, 17, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Luo, Y.; He, L.Q.; Liu, Z.J.; Jiang, A.Q.; Yang, Y.H.; Zhu, H.L. Synthesis, biological evaluation, and molecular docking studies of novel 1,3,4-oxadiazole derivatives possessing benzotriazole moiety as FAK inhibitors with anticancer activity. Bioorg. Med. Chem. 2013, 21, 3723–3729. [Google Scholar] [CrossRef]
- Du, Q.R.; Li, D.D.; Pi, Y.Z.; Li, J.R.; Sun, J.; Fang, F.; Zhong, W.Q.; Gong, B.-H.; Zhu, H.L. Novel 1,3,4-oxadiazole thioether derivatives targeting thymidylate synthase as dual anticancer/antimicrobial agents. Bioorg. Med. Chem. 2013, 21, 2286–2297. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, S.; Asati, V.; Singh, J.; Roy, P.P. 1,3,4-Oxadiazoles: An emerging scaffold to target growth factors, enzymes and kinases as anticancer agents. Eur. J. Med. Chem. 2015, 97, 124–141. [Google Scholar] [CrossRef]
- Khan, K.M.; Rani, M.; Ambreen, N.; Ali, M.; Hussain, S.; Perveen, S.; Choudhary, M.I. 2,5-Disubstituted-1,3,4-oxadiazoles: Thymidine phosphorylase inhibitors. Med. Chem. Res. 2013, 22, 6022–6028. [Google Scholar] [CrossRef]
- Alam, M.M.; Almalki, A.S.; Neamatallah, T.; Ali, N.M.; Malebari, A.M.; Nazreen, S. Synthesis of New 1, 3, 4-Oxadiazole-Incorporated 1, 2, 3-Triazole Moieties as Potential Anticancer Agents Targeting Thymidylate Synthase and Their Docking Studies. Pharmaceuticals 2020, 13, 390. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Lu, X.; Zhang, H.J.; Sun, J.; Zhu, H.L. Synthesis, molecular modeling and biological evaluation of 2-(benzylthio)-5-aryloxadiazole derivatives as anti-tumor agents. Eur. J. Med. Chem. 2012, 47, 473–478. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Siddiqui, A.A.; Khan, A.A.; Ali, Z.; Dewangan, R.P.; Pasha, S.; Yar, M.S. Design, synthesis, docking and QSAR study of substituted benzimidazole linked oxadiazole as cytotoxic agents, EGFR and erbB2 receptor inhibitors. Eur. J. Med. Chem. 2017, 126, 853–869. [Google Scholar] [CrossRef] [PubMed]
- Ruel, R.; Thibeault, C.; L’Heureux, A.; Martel, A.; Cai, Z.W.; Wei, D.; Qian, L.; Barrish, J.C.; Mathur, A.; D’Arienzo, C.; et al. Discovery and preclinical studies of 5-isopropyl-6-(5-methyl-1,3,4-oxadiazol-2-yl)-N-(2-methyl-1H-pyrrolo[2,3-b]pyridin-5-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine (BMS-645737), an in vivo active potent VEGFR-2 inhibitor. Bioorg. Med. Chem. Lett. 2008, 18, 2985–2989. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.; Srikanth, P.S.; Vishnuvardhan, M.V.P.S.; Kumar, G.B.; Suresh Babu, K.; Hussaini, S.M.A.; Kapure, J.S.; Alarifi, A. Combretastatin linked 1,3,4-oxadiazole conjugates as a Potent Tubulin Polymerization inhibitors. Bioorg. Chem. 2016, 65, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Rajak, H.; Agarawal, A.; Parmar, P.; Thakur, B.S.; Veerasamy, R.; Sharma, P.C.; Kharya, M.D. 2,5-Disubstituted-1,3,4-oxadiazoles/thiadiazole as surface recognition moiety: Design and synthesis of novel hydroxamic acid based histone deacetylase inhibitors. Bioorg. Med. Chem. Lett. 2011, 21, 5735–5738. [Google Scholar] [CrossRef]
- Valente, S.; Trisciuoglio, D.; De Luca, T.; Nebbioso, A.; Labella, D.; Lenoci, A.; Bigogno, C.; Dondio, G.; Miceli, M.; Brosch, G.; et al. 1,3,4-Oxadiazole-containing histone deacetylase inhibitors: Anticancer activities in cancer cells. J. Med. Chem. 2014, 57, 6259–6265. [Google Scholar] [CrossRef] [PubMed]
- Terenzi, A.; Fanelli, M.; Ambrosi, G.; Amatori, S.; Fusi, V.; Giorgi, L.; Turco Liveri, V.; Barone, G. DNA binding and antiproliferative activity toward human carcinoma cells of copper(ii) and zinc(ii) complexes of a 2,5-diphenyl[1,3,4]oxadiazole derivative. Dalt. Trans. 2012, 41, 4389–4395. [Google Scholar] [CrossRef]
- Stecoza, C.; Majekova, M.; Majek, P.; Caproiu, M.; Marutescu, L. Novel Dibenzothiepins with Antibiofilm Activity Demonstrated by Microbiological Assays and Molecular Modeling. Curr. Org. Chem. 2013, 17, 113–124. [Google Scholar] [CrossRef]
- Stecoza, C.E.; Ilie, C.; Caproiu, M.T.; Draghici, C. New 2-metyl-O-acyloximino-dibenzo [b,e] thiepins Synthesis and structural characterization. Rev. Chim. 2011, 62, 610–613. [Google Scholar]
- Patra, A.; Ghorai, S.K.; De, S.R.; Mal, D. Regiospecific Synthesis of Benzo[b]fluorenones via Ring Contraction by Benzil-Benzilic Acid Rearrangement of Benz[a]anthracene-5,6-diones. Synthesis 2006, 2006, 2556–2562. [Google Scholar] [CrossRef]
- Kumar, H.; Javed, S.A.; Khan, S.A.; Amir, M. 1,3,4-Oxadiazole/thiadiazole and 1,2,4-triazole derivatives of biphenyl-4-yloxy acetic acid: Synthesis and preliminary evaluation of biological properties. Eur. J. Med. Chem. 2008, 43, 2688–2698. [Google Scholar] [CrossRef]
- Gamal El-Din, M.M.; El-Gamal, M.I.; Abdel-Maksoud, M.S.; Yoo, K.H.; Oh, C.H. Synthesis and in vitro antiproliferative activity of new 1,3,4-oxadiazole derivatives possessing sulfonamide moiety. Eur. J. Med. Chem. 2015, 90, 45–52. [Google Scholar] [CrossRef]
- Hustedt, N.; Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2017, 19, 1–9. [Google Scholar] [CrossRef]
- Lezaja, A.; Altmeyer, M. Inherited DNA lesions determine G1 duration in the next cell cycle. Cell Cycle 2018, 17, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Bostan, M.; Petrică-Matei, G.G.; Ion, G.; Radu, N.; Mihăilă, M.; Hainăroşie, R.; Braşoveanu, L.I.; Roman, V.; Constantin, C.; Neagu, M.T. Cisplatin effect on head and neck squamous cell carcinoma cells is modulated by ERK1/2 protein kinases. Exp. Ther. Med. 2019, 18, 5041–5051. [Google Scholar] [CrossRef]
- Mihaila, M.; Bostan, M.; Hotnog, D.; Ferdes, M.; Brasoveanu, L.I. Real-time analysis of quercetin, resveratrol and/or doxorubicin effects in mcf-7 cells. Rom. Biotechnol. Lett. 2013, 18, 8106–8114. [Google Scholar]
- Maciuca, A.-M.; Munteanu, A.-C.; Mihaila, M.; Badea, M.; Olar, R.; Nitulescu, G.M.; Munteanu, C.V.A.; Bostan, M.; Uivarosi, V. Rare-Earth Metal Complexes of the Antibacterial Drug Oxolinic Acid: Synthesis, Characterization, DNA/Protein Binding and Cytotoxicity Studies. Molecules 2020, 25, 5418. [Google Scholar] [CrossRef]
- Bostan, M.; Petrică-Matei, G.G.; Radu, N.; Hainarosie, R.; Stefanescu, C.D.; Diaconu, C.C.; Roman, V. The Effect of Resveratrol or Curcumin on Head and Neck Cancer Cells Sensitivity to the Cytotoxic Effects of Cisplatin. Nutrients 2020, 12, 2596. [Google Scholar] [CrossRef]
- Filimonov, D.A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Druzhilovskii, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the biological activity spectra of organic compounds using the pass online web resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar] [CrossRef]
- Choi, S.; Jung, H.J.; Kim, M.W.; Kang, J.H.; Shin, D.; Jang, Y.S.; Yoon, Y.S.; Oh, S.H. A novel STAT3 inhibitor, STX-0119, attenuates liver fibrosis by inactivating hepatic stellate cells in mice. Biochem. Biophys. Res. Commun. 2019, 513, 49–55. [Google Scholar] [CrossRef]
- Malojirao, V.H.; Girimanchanaika, S.S.; Shanmugam, M.K.; Sherapura, A.; Dukanya; Metri, P.K.; Vigneshwaran, V.; Chinnathambi, A.; Alharbi, S.A.; Rangappa, S.; et al. Novel 1,3,4-oxadiazole targets STAT3 signaling to induce antitumor effect in lung cancer. Biomedicines 2020, 8, 368. [Google Scholar] [CrossRef] [PubMed]
- Khanam, R.; Hejazi, I.I.; Shahabuddin, S.; Bhat, A.R.; Athar, F. Pharmacokinetic evaluation, molecular docking and in vitro biological evaluation of 1, 3, 4-oxadiazole derivatives as potent antioxidants and STAT3 inhibitors. J. Pharm. Anal. 2019, 9, 133–141. [Google Scholar] [CrossRef]
- Verhoeven, Y.; Tilborghs, S.; Jacobs, J.; De Waele, J.; Quatannens, D.; Deben, C.; Prenen, H.; Pauwels, P.; Trinh, X.B.; Wouters, A.; et al. The potential and controversy of targeting STAT family members in cancer. Semin. Cancer Biol. 2020, 60, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, K.; Masuda, Y.; Uehara, Y.; Sato, H.; Muroya, A.; Takahashi, O.; Yokotagawa, T.; Furuya, T.; Okawara, T.; Otsuka, M.; et al. Identification of a New Series of STAT3 Inhibitors by Virtual Screening. ACS Med. Chem. Lett. 2010, 1, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, Z.; Ding, C.; Chu, L.; Zhang, Y.; Terry, K.; Liu, H.; Shen, Q.; Zhou, J. Fragment-based drug design and identification of HJC0123, a novel orally bioavailable STAT3 inhibitor for cancer therapy. Eur. J. Med. Chem. 2013, 62, 498–507. [Google Scholar] [CrossRef]
- Corvinus, F.M.; Orth, C.; Moriggl, R.; Tsareva, S.A.; Wagner, S.; Pfitzner, E.B.; Baus, D.; Kaufmann, R.; Huber, L.A.; Zatloukal, K.; et al. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia 2005, 7, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.X.; Wu, Q.N.; Zhang, Y.; Li, Y.Y.; Liao, D.Z.; Hou, J.H.; Fu, J.; Zeng, M.S.; Yun, J.P.; Wu, Q.L.; et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, L.; Chen, Y.; He, T.; Chen, X.; Mao, J.; Li, C.; Lyu, J.; Meng, Q.H. MicroRNA-21 promotes proliferation, migration, and invasion of colorectal cancer, and tumor growth associated with down-regulation of sec23a expression. BMC Cancer 2016, 16, 605. [Google Scholar] [CrossRef]
- Botezatu, A.; Iancu, I.V.; Plesa, A.; Manda, D.; Popa, O.; Bostan, M.; Mihaila, M.; Albulescu, A.; Fudulu, A.; Vladoiu, S.V.; et al. Methylation of tumour suppressor genes associated with thyroid cancer. Cancer Biomark. 2019, 25, 53–65. [Google Scholar] [CrossRef]
- Hotnog, D.; Mihaila, M.; Botezatu, A.; Matei, G.G.; Hotnog, C.; Anton, G.; Bostan, M.; Brasoveanu, L.I. Genistein potentiates the apoptotic effect of 5-fluorouracyl in colon cancer cell lines. Rom. Biotechnol. Lett. 2013, 18, 7151–7160. [Google Scholar]
- Nitulescu, G.; Mihai, D.P.; Nicorescu, I.M.; Olaru, O.T.; Ungurianu, A.; Zanfirescu, A.; Nitulescu, G.M.; Margina, D. Discovery of natural naphthoquinones as sortase A inhibitors and potential anti-infective solutions against Staphylococcus aureus. Drug Dev. Res. 2019, 80, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Seremet, O.C.; Olaru, O.T.; Gutu, C.M.; Nitulescu, G.M.; Ilie, M.; Negres, S.; Zbarcea, C.E.; Purdel, C.N.; Spandidos, D.A.; Tsatsakis, A.M.; et al. Toxicity of plant extracts containing pyrrolizidine alkaloids using alternative invertebrate models. Mol. Med. Rep. 2018, 17, 7757–7763. [Google Scholar] [CrossRef] [PubMed]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program for Chemistry Aware Data Visualization and Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Zakharov, A.V.; Peach, M.L.; Sitzmann, M.; Nicklaus, M.C. A new approach to radial basis function approximation and its application to QSAR. J. Chem. Inf. Model. 2014, 54, 713–719. [Google Scholar] [CrossRef] [PubMed]

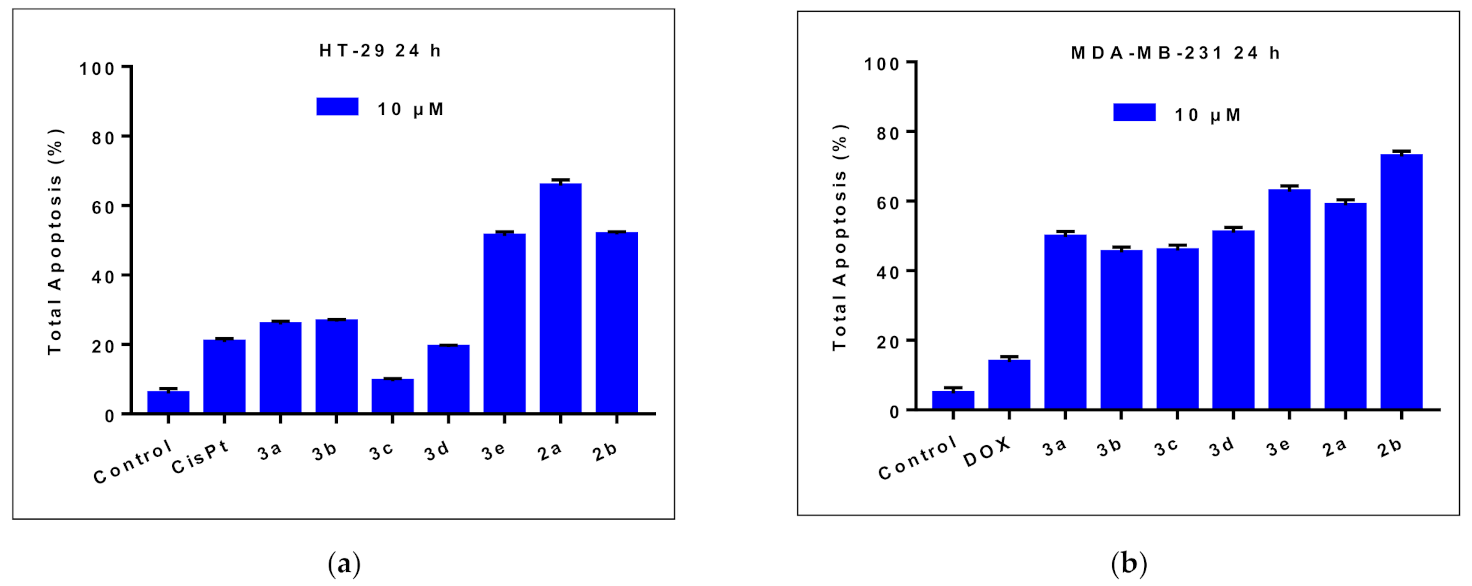

| HT-29 Cells | MDA-MB-231 Cells | |||||
|---|---|---|---|---|---|---|
| Early Apoptosis (%) | Late Apoptosis (%) | Total Apoptosis (%) | Early Apoptosis (%) | Late Apoptosis (%) | Total Apoptosis (%) | |
| Control | 4.8 | 1.1 | 5.9 | 4.2 | 0.5 | 4.7 |
| CisPt 1 | 16.5 | 4.2 | 20.7 | - | - | - |
| DOX 1 | - | - | - | 11.4 | 2.3 | 13.7 |
| 3a | 23.5 | 2.2 | 25.7 | 41.7 | 8.0 | 49.7 |
| 3b | 21.7 | 4.8 | 26.5 | 40.5 | 4.7 | 45.2 |
| 3c | 6.4 | 3 | 9.4 | 40.8 | 5.0 | 45.8 |
| 3d | 16.5 | 2.7 | 19.2 | 44.3 | 6.6 | 50.9 |
| 3e | 43.9 | 7.3 | 51.2 | 51.8 | 10.9 | 62.7 |
| 2a | 53.0 | 12.7 | 65.7 | 45.9 | 12.9 | 58.8 |
| 2b | 44.8 | 6.8 | 51.6 | 55.5 | 17.3 | 72.8 |
| Compound | 24 h | 48 h | ||
|---|---|---|---|---|
| LC50 (µM) | 95%CI of LC50 (µM) | LC50 (µM) | 95%CI of LC50 (µM) | |
| 3a | 115.8 | 43.6 to 307.4 | ND* | ND* |
| 3b | ND* | ND* | ND* | ND* |
| 3c | ND* | ND* | 11.5 | ND* |
| 3d | ND* | ND* | 2.34 | ND* |
| 3e | ND* | ND* | 3.5 | 2.0–7.3 |
| 2a | 332.5 | 201.1–549.8 | 35.9 | 23.0–56.1 |
| 2b | 296.0 | 205.3–426.7 | 21.8 | 11.5–41.1 |
| Target | 3a | 3b | 3c | 3d | 3e |
|---|---|---|---|---|---|
| Transcription factor inhibitor | 0.58 | 0.62 | 0.55 | 0.32 | 0.58 |
| Transcription factor STAT inhibitor | 0.61 | 0.64 | 0.61 | 0.42 | 0.65 |
| Transcription factor STAT3 inhibitor | 0.55 | 0.58 | 0.56 | 0.26 | 0.54 |
| JAK2 expression inhibitor | 0.40 | 0.33 | 0.40 | 0.33 | 0.22 |
| Focal adhesion kinase inhibitor | 0.23 | 0.22 | 0.22 | 0.24 | 0.27 |
| Focal adhesion kinase 2 inhibitor | 0.35 | 0.34 | 0.34 | 0.36 | 0.38 |
| MAP3K5 inhibitor | 0.28 | 0.26 | - | - | 0.29 |
| Vascular endothelial growth factor 1 antagonist | 0.27 | 0.22 | 0.22 | 0.30 | 0.30 |
| Rat Acute Toxicity | 3a | 3b | 3c | 3d | 3e | 2a | 2b |
|---|---|---|---|---|---|---|---|
| Oral LD50 (mg/kg) | 1970 | 1977 | 1317 | 1417 | 1924 | 967 | 900 |
| Oral LD50 Class | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| IV LD50 (mg/kg) | 321.1 | 289.6 | 253.3 | 383.0 | 298.8 | 135.7 | 256.1 |
| IV LD50 Class | 5 | 4 | 4 | 5 | 4 | 4 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stecoza, C.E.; Nitulescu, G.M.; Draghici, C.; Caproiu, M.T.; Olaru, O.T.; Bostan, M.; Mihaila, M. Synthesis and Anticancer Evaluation of New 1,3,4-Oxadiazole Derivatives. Pharmaceuticals 2021, 14, 438. https://doi.org/10.3390/ph14050438
Stecoza CE, Nitulescu GM, Draghici C, Caproiu MT, Olaru OT, Bostan M, Mihaila M. Synthesis and Anticancer Evaluation of New 1,3,4-Oxadiazole Derivatives. Pharmaceuticals. 2021; 14(5):438. https://doi.org/10.3390/ph14050438
Chicago/Turabian StyleStecoza, Camelia Elena, George Mihai Nitulescu, Constantin Draghici, Miron Teodor Caproiu, Octavian Tudorel Olaru, Marinela Bostan, and Mirela Mihaila. 2021. "Synthesis and Anticancer Evaluation of New 1,3,4-Oxadiazole Derivatives" Pharmaceuticals 14, no. 5: 438. https://doi.org/10.3390/ph14050438
APA StyleStecoza, C. E., Nitulescu, G. M., Draghici, C., Caproiu, M. T., Olaru, O. T., Bostan, M., & Mihaila, M. (2021). Synthesis and Anticancer Evaluation of New 1,3,4-Oxadiazole Derivatives. Pharmaceuticals, 14(5), 438. https://doi.org/10.3390/ph14050438







