Abstract
We report here the design, synthesis, experimental and in silico evaluation of the antibacterial and antifungal activity of some new benzo[f]quinoline derivatives. Two classes of benzo[f]quinolinium derivatives—(benzo[f]quinolinium salts (BQS) and pyrrolobenzo[f]quinolinium cycloadducts (PBQC)—were designed and obtained in two steps via a direct and facile procedure: quaternization followed by a cycloaddition reaction. The synthesized compounds were characterized by elemental and spectral analysis (FT-IR, 1H-NMR, 13C-NMR). The antimicrobial assay reveals that the BQS salts have an excellent quasi-nonselective antifungal activity against the fungus Candida albicans (some of them higher that the control drug nystatin) and very good antibacterial activity against the Gram positive bacterium Staphylococcus aureus. The PBQC compounds are inactive. Analysis of the biological data reveals interesting SAR correlations in the benzo[f]quinolinium series of compounds. The in silico studies furnished important data concerning the pharmacodynamics, pharmacokinetics and ADMET parameters of the BQS salts. Studies of the interaction of each BQS salt 3a–o with ATP synthase in the formed complex, reveal that salts 3j, 3i, and 3n have the best fit in a complex with ATP synthase. Study of the interaction of each BQS salt 3a-o with TOPO II in the formed complex reveals that salts 3j and 3n have the best-fit in complex with TOPO II. The in silico ADMET studies reveal that the BQS salts have excellent drug-like properties, including a low toxicity profile. Overall, the experimental and in silico studies indicate that compounds 3e and 3f (from the aliphatic series), respectively, and 3i, 3j and 3n (from the aromatic series), are promising leading drug candidates.
1. Introduction
According to the WHO, infectious diseases, especially those caused by bacterial (Gram positive and Gram negative) and fungal microorganisms, have become a serious threat to the global health system, being responsible for 22% of all deaths and 27% of disability-adjusted life years worldwide [1]. Antibiotics play a key central role in antimicrobial therapy, and are one of the most effective and successful weapons against different microorganisms. However, the overuse and misuse of antibiotics have led to widespread drug resistance (DR), multi-drug resistance (MDR) and extensive-drug-resistance (EDR), creating an urgent need for new antimicrobial agents [1,2,3,4].
Quinoline and its benzo-fused derivative, benzoquinoline, are crucial scaffolds in medicinal chemistry and they have been reported to possess a large variety of biological activities which include antiplasmodial and antimalarial, antitubercular, antibacterial, antifungal, anti-HIV, anticancer, antinociceptive and anti-inflammatory, antipsychotic, analgesic, anti-Alzheimer’s, antihypertensive properties, etc. [5,6,7,8,9,10,11,12,13,14,15,16,17]. Moreover, the quinoline pharmacophore has proved to be one of the most effective motifs used in antibacterial and antifungal therapy, and many of the existing drugs on the market incorporate a quinoline scaffold e.g., ciprofloxacin (antibacterial), bedaquiline (antitubercular), etc. [18,19], (Scheme 1).
Taking into consideration the above considerations and our expertise in the field of antibacterial and antifungal agents [13,15,20,21,22,23,24,25], we report here the design, synthesis, molecular docking, antibacterial and antifungal evaluation of some new benzo[f]quinoline derivatives.
2. Results and Discussion
2.1. Desingn and Chemistry
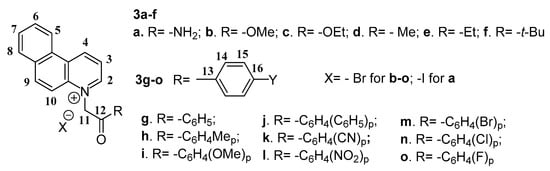
Taking into consideration the abovementioned data, we decided to combine the pharmacophoric antimicrobial capabilities (Scheme 1) of quinoline and its benzo derivatives [26] and also to convert them in salts, in view of the fact that salts usually have better antimicrobial activity and also better pharmacokinetic properties; we also had in mind increasing the number of fused cycles from three in benzo[f]quinoline to four and to see their influence in regards to the antimicrobial activity. In equal measure we were interested in seeing the influence on antimicrobial activity of the substituents on the quaternized nitrogen, taking into account two series an aliphatic and aromatic one. As a result, two series of benzo[f]quinoline derivatives was designed and synthesized: BQS salts and PBQC cycloadducts (Scheme 1).
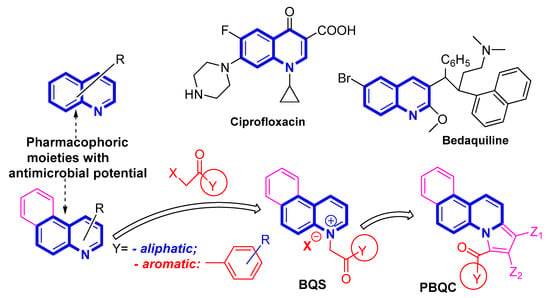
Scheme 1.
Design in the class of benzoquinolines derivatives with antimicrobial activity.
In order to synthesize our fused quinoline derivatives we used a direct and facile two step procedure: quaternization of nitrogen heterocycles followed by a cycloaddition reaction. Using an adaptation of the setup procedure from the literature [27,28,29], in the first step, we performed a quaternization reaction of benzo[f]quinoline (1) with variously activated α-halocarbonyl compounds 2a–o (such as: 1-bromo-alkyl-2-one, 2-iodoacetamide, (un)substituted phenacyl bromides), when the corresponding benzo[f]quinolinium quaternary salts BQS 3a–o were obtained (Scheme 2). The next step consists in a Hüisgen [3 + 2] dipolar cycloaddition of the benzo[f]quinolinium ylides (generated in situ from the corresponding benzo[f]quinolinium quaternary salts BQS 3a–o, in alkaline medium) to alkyne dipolarophiles (non-symmetrically or symmetrically substituted Z-alkynes, methyl propiolate and dimethyl acetylenedicarboxylate, DMAD), when the corresponding fused pyrrolobenzo[f]quinolinium cycloadducts PBQC 4, were obtained (Scheme 2). Initially, the cycloaddition reactions were performed in the case of salts 3e (aliphatic) and 3g (aromatic), and the obtained cycloadducts PBQC (4e1, 4e2, 4g1, and 4g2) was subject to antimicrobial assay. Because the antimicrobial activity of the obtained cycloadducts PBQC 4e and 4g, was negligible, we did not continue our studies in this direction.
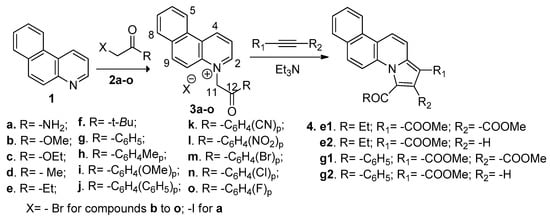
Scheme 2.
Reaction pathways to obtain benzo[f]quinoline salts 3a–o and cycloadducts 4e and 4g.
The structures of compounds were proved by elemental and spectral analysis (FT-IR, 1H-NMR, 13C-NMR, and two-dimensional experiments 2D-COSY, HMQC, HMBC). The main data furnished by FT-IR and NMR spectral analysis are listed in Table 1. In the FT-IR spectra of BQS salts 3a–o, the most important signals are those one of carbonyl groups. In the aromatic BQS salts 3g–o, the signals corresponding to carbonyl ketone group are situated between 1717 cm−1 (R = NO2, 3l) and 1673 cm−1 (R = OCH3, 3i), in accordance with the electronic effects of substituents on the 4-position of a phenyl ring (electron-withdrawing for NO2 and electron-donating for OCH3). In the aliphatic series (BQS salts 3a–f), the signals corresponding to carbonyl group are situated between 1626 cm−1 (amide CO, 3a) and 1743 cm−1 (carboxymethyl CO, 3b), in accordance with the structures of the compounds (amide and aliphatic ester). In the 1H-NMR spectra the most important signals are those of the H-2, H-4, and H-11 (methylene (–CH2, –) hydrogen) atoms. As it can be seen in Table 1 the H-11 methylene hydrogen atoms appear at an unusual very high chemical shift, around 6.50 ppm in the aliphatic series and around 7.00 ppm in the aromatic series. This is in accordance with the powerful electron-withdrawing effect exerted by the positive α–endocyclic nitrogen atom of the benzo[f]quinoline ring. The aromatic/aliphatic difference of about 0.50 ppm is because of the supplementary electron-withdrawing effect exerted by the aromatic phenyl ring. The H-2 and H-4 protons from the pyridine ring appear at high chemical shift, around 10.25 ppm H-4 (positive γ–endocyclic nitrogen) and around 9.50 ppm H-2 (positive α–endocyclic nitrogen).

Table 1.
Main spectral data of BQS salts 3a–o.
The 13C-NMR spectra of BQS salts 3a–o also confirm the structurse and are in accordance with the FT-IR and 1H-NMR data presented above. The carbon from the ketone CO group (C-12) appears around 190 ppm in the aryl-alkyl BQS salts 3g–o; around 166 ppm in the amide and carboxylic BQS salts 3a–c, and around 205 ppm in the in the alkyl-alkyl BQS salts 3d–f. The C-11 methylene carbon atoms appear at high chemical shifts, between 66.0–59.3 ppm, in accordance with the electron-withdrawing effect of the positive α–endocyclic nitrogen atom from benzo[f]quinoline ring and with the structure of the ketone carbonyl group (alkyl or aryl or amide/aliphatic ester). The C-2 and C-4 atoms appear around 148 ppm and 143 ppm, respectively, in accordance with their position in the pyridine ring (α- or γ- endocyclic carbons). The remaining signals in the FT-IR, 1H-NMR and 13C-NMR spectra are also in accordance with the proposed structures.
2.2. Antimicrobial Assay
The sensitivity of the microorganisms to the substances under investigation, salts BQS and cycloadducts PBQC, was assessed based on the diameter of the inhibition zone, using the Kirby-Bauer agar disk diffusion method, adopted by the Clinical & Laboratory Standards Institute (CLSI M07-A11, 2018) [30]. The method uses Mueller Hinton nutrient agar medium for antibacterial tests and Sabouraud nutrient agar medium for antifungal tests. The disk diffusion test provides a number of advantages, such as simplicity of the method, low cost, large number of organisms and antimicrobial agents that can be tested, and ease of interpreting the resulting data [30,31]. The in vitro antibacterial activity was evaluated against the Gram-positive bacterium Staphylococcus aureus ATCC 25923 and the Gram-negative Escherichia coli ATCC 25922. The in vitro antifungal activity was evaluated against the fungus Candida albicans ATCC 10231. Penicillin (10 IU), carbenicillin (100 µg/mL) and nystatin (500,000 IU) were used as positive control (C+) for Staphylococcus aureus, Escherichia coli and Candida albicans, respectively. As negative control (C−) sterile filter paper disks (with no antimicrobial compounds) were used. The obtained results were expressed as diameters of inhibition zones (mm). The larger the diameter of the inhibition zones is, the more active the compounds are as antimicrobials and antifungals. The obtained results are listed in Table 2 (and Figures S1–S3, see the Supporting Material for details).

Table 2.
The antibacterial and antifungal activity for BQS salts 3a–o and PBQC cycloadducts 4e and 4g.
The data presented in Table 2 and Figures S1–S3 reveal interesting data concerning the biological properties of salts BQS and cycloadducts PBQC. A first observation is the fact that the tested cycloadducts PBQC did not present any antibacterial and antifungal activity and for this reason we did not continue the assays for this category of benzo[f]quinoline derivatives. On the other hand, the tested BQS salts demonstrated a certain antibacterial and antifungal activity against the tested strains. Practically all the tested the BQS salts manifest a powerful activity against the fungus Candida albicans, with eight compounds being very active with a diameter of inhibition zone up to 20 mm (with two compounds, 3e and 3f, being remarkably active with inhibition zone diameters in the range of 30 mm) and higher that the control drug nystatin. The remaining other BQS salts are also active against C. albicans, having a diameter of inhibition zone between 15 mm (for 3k) and 19 mm (for 3m). Against the Gram positive Staphylococcus aureus bacterial strain, five BQS salts are very active, having an inhibition zone diameter of up to 20 mm (3c, 3f, 3h, 3i, 3n), while the remaining other BQS salts are active (with an inhibition zone diameter between 15 mm (for 3j, 3k, 3l) and 19.5 mm (for 3b)]. Against Gram negative Escherichia coli bacteria, the BQS salts have a moderate activity, six BQS salts being active, with a diameter of inhibition zone between 15.5 mm (for 3f) and 18.5 mm (for 3c).
In the next step of the antimicrobial assays, the minimum inhibitory concentration (MIC) of the ten most active BQS salts (namely 3b–e, 3g–k, 3n) were determined, using the standardized broth microdilution assay procedure [32,33,34,35]. The resulting MIC value is defined as the lowest concentration of the antimicrobial BQS salts under investigation, which prevents visible growth of the tested microorganism. The obtained results are listed in Table 3.

Table 3.
The minimum inhibitory concentration (MIC) for BQS salts 3b–e, 3g–k, n (µg/mL).
Compared with the control drug, the data from Table 3 reveal that BQS salt 3i is active to a low concentration, having a MIC of 30.4 × 10−4 µg/mL in the case of Staphylococcus aureus, 15.2 × 10−4 µg/mL in the case of Escherichia coli and 575 × 10−4 µg/mL in the case of Candida albicans. Significant results were also obtained for the BQS salts 3n (with a MIC of 975 × 10−4 µg/mL for S. aureus and 0.195 µg/mL for E. coli and C. albicans), 3h and 3g (with a MIC in the range of 0.195 µg/mL for all germs). The diameter of inhibition zone data presented in Table 2, MIC data in Table 3 and Figure 1, Figure 2 and Figure 3 reveal some interesting observations and correlations between the compound structures and their antimicrobial activity. The BQS salts 3a–o have an excellent quasi-nonselective antifungal activity against the fungus C. albicans, a very good antibacterial activity against the Gram positive germ S. aureus and are less active against the Gram negative germ E. coli. The antifungal activity is significantly more pronounced in the aliphatic series 3a–f compared with the aromatic one of compounds 3g–o, which demonstrates a certain influence on activity of the aliphatic substituent of carbonyl group. In the aromatic series 3g–o, compounds 3n [Y = -C6H4(Cl)p] and 3i [Y = -C6H4(OMe)p] have the highest antifungal activity, which also indicates an influence of the substituent (chlorine or methoxy) on the para position of the phenyl ring. The same SAR considerations are revealed by the results obtained in the antibacterial assay against the bacteria S. aureus and E. coli. Finally, if we compare the two series of tested compounds, salts BQS 3 and cycloadducts PBQC 4, we may notice that only salts BQS 3 have antimicrobial activity while the cycloadducts PBQC 4 are inactive, which means that in order to have antimicrobial activity it is better to have a mobile substituent linked on the nitrogen atom instead of a new fused cycle.
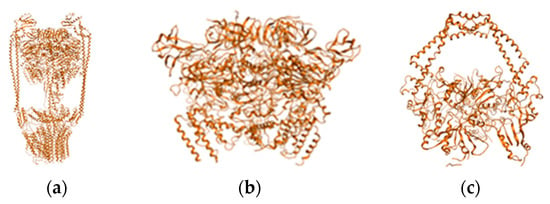
Figure 1.
PDB structures of targeted molecules represented as ribbons in gold orange (atoms are not displayed): (a) ATP synthase structure; (b) ATP synthase outer membrane region; (c) Topoisomerase II.
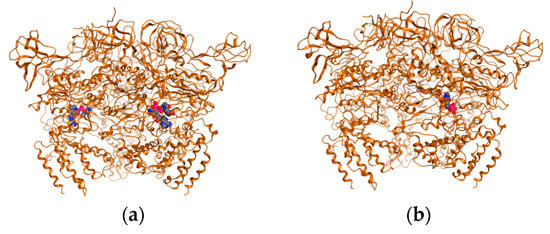
Figure 2.
ATP synthase with ligands: (a) “native”, crystallographic determined structure; (b) redocked ligand (DNA string).
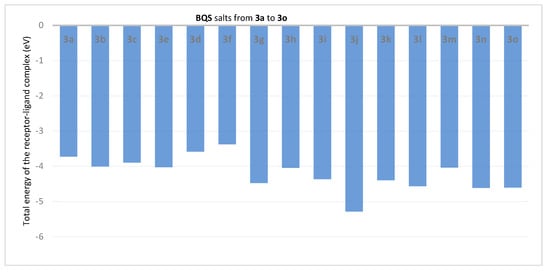
Figure 3.
Total energy (in eV) of the receptor-ligand in BQS 3a–o salts derivative complexes with ATP synthase.
2.3. Molecular Docking
During the last decades molecular docking became an important tool in medicinal chemistry, furnishing relevant information concerning binding sites, binding energy and Absorption, Distribution, Metabolism, Excretion, Toxicity (ADMET) properties. As a result, we decided to perform a computational (in silico) study concerning these parameters for the salts BQS 3. The chemical space was also screened for similar compounds with remarkable bioactivity and ADMET and drug-like properties. Bioactivity is studied with the aid of a computational build = receptor-ligand system. The complex is characterized by its energy, hydrogen bonds, and steric constraints. The targets (binding sites) used for this class of compounds were retrieved from the literature (mainly the work of Hunter et al. [36] and Xavier et al. [37]) and using an online service that suggests ligand predilection for a specific target [38] and are ATP synthase and topoisomerase II (TOPO II). Target structures were retrieved from the PDB database: 6WLZ for ATP synthase [39] and 3KSB for TOPO II [40]. The PDB structures were energetically minimized, charges corrected, names corrected, potential energy recomputed. Cofactors and Aa chains were kept, and ligands and water molecules were removed. BQS 3 salts were introduced computationally as SDF. Files that were energetically minimized and charges corrected. Binding site coordinates (Å) were retrieved from the literature and from an algorithm based on expanded Van der Walls charges [41]. The maximum number of cavities which was set to be detected was 5, corresponding to the binding site molecular theory [42]. The cavity with the most significant volume (Å3) was chosen for each target (see Supporting Materials for details).
First, the two sets of compounds were docked against the two molecular targets. MOE 2009 software and its methodology were used in the docking procedure [43,44,45]. Cartesian coordinates for the two binding sites are as follows: ATP synthase: x45.94 Å, y46.91 Å, z198.20 Å; TOPO II: x-16.33 Å, y43.11 Å, z-34.50 Å. For computational and action mechanism reasons, only protein corona of ATP synthase was used in building the in-silico system (Figure 1).
Docking was validated by redocking the ligands present in PDB target structures. For this step ligands were cut from the crystallographic obtained structures converted to SDF files, minimized and charges corrected. Ligands were then docked on to the free ligand PDB structures. Results are depicted in Figure 2.
Figure 3 and Figure 4, describe the interaction of each BQS salt 3a–o with ATP synthase in the formed complex, in terms of total energies of the complexes (ET) and hydrogen bond energy (EH).
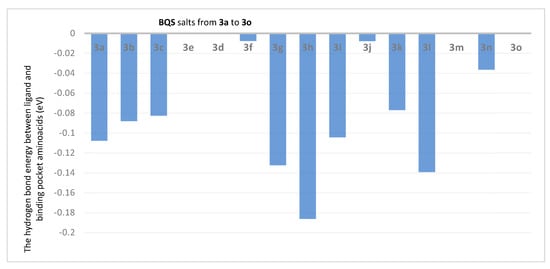
Figure 4.
The hydrogen bond energy between ligand and binding pocket aminoacids (in eV) in BQS 3a–o salt derivative complexes with ATP synthase.
Analysis of the data from Figure 3 reveals that the compounds 3j (ET = −5.39 eV), 3g (ET = −4.48 eV), 3i (ET = −4.37 eV), 3l (ET = −4.57 eV) and 3n (ET = −4.62 eV) show the lowest energies in complex with ATP synthase. The data from Figure 4 reveals that the compounds 3h (EH = −0.1863 eV), 3g (EH = −0.1325 eV), 3i (EH = −0.1044 eV) and 3l (EH = −0.1393 eV) show the lowest hydrogen bond energy of the ATP synthase complex; compounds 3e, 3d, 3m, and 3o don’t form hydrogen bonds.
In Figure 5 is presented the complex with ATP synthase of BQS salt 3f. In the binding site pocket we may notice two powerful H-π interactions between the two benzene rings from the benzo[f]quinoline moiety and the aminoacid Glu 62. Another H-π interaction is formed between Ala 356 and the benzene from the center of the aromatic core. Also, Ala 356 has interresidue contacts with Tyr 353 and Arg 357. A benzene carbon donates electrons to Phe 60, which has an inter-residue contact with Pro 227. All these interactions are stabilizing the BQS salt 3f—ATP synthase complex.
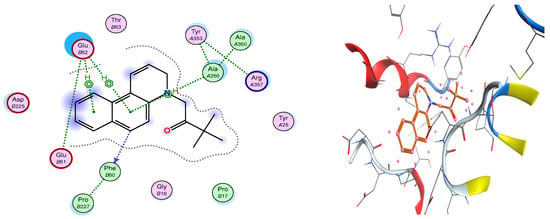
Figure 5.
Compound 3f is represented in complex with ATP synthase as sticks. Surrounding aminoacids at the binding site are represented as ribbons colored by chain type. Also, a schematic view of the binding site pocket is represented.
In the next step, a similar protocol was followed to study the interactions of BQS salts with TOPO II. Figure 6 and Figure 7, describe the interaction of each BQS 3a–o salt with TOPO II in the formed complex, in terms of total energies of the complexes (ET) and hydrogen bond energy (EH).
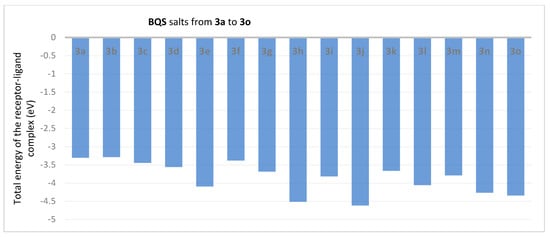
Figure 6.
BQS 3a–o salts in complex with TOPO II: energies of complexes.
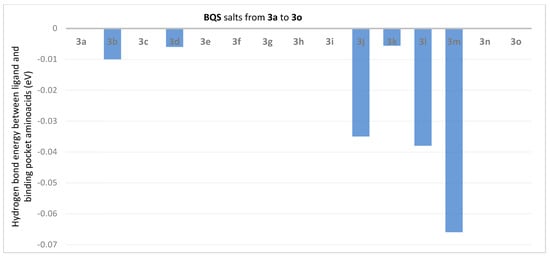
Figure 7.
The hydrogen bond energy (in eV) between ligand and binding pocket aminoacids in BQS 3a–o salts derivates complexes with TOPO II.
Analysis of the data from Figure 6 reveal that the compounds 3j (ET = −4.617 eV), 3h (ET = −4.517 eV), 3e (ET = −4.097 eV) and 3l (ET = −4.059 eV) show the lowest energies in complex with TOPO II. The data from Figure 7 reveal that the compounds 3m (EH = −0.066 eV), 3l (EH = −0.038 eV) and 3j (EH = −0.035 eV) show the lowest hydrogen bond energy of the TOPO II complex; compounds 3a, 3c, 3e, 3f, 3g, 3h, 3i, 3n and 3o don’t form hydrogen bonds.
From the obtained data we may notice that salts 3j and 3n have the best-fit in complex with TOPO II (in Figure 8 the complex of salt 3j is presented). In the binding pocket of TOPO II, a hydrogen bond is formed between the nitrogen atom of the benzo[f]quinoline ring and aminoacid ASP-510, stabilizing the salt BQS 3j—TOPO II complex. We also noticed that TYR 118 and His 76 interact with the aromatic rings. Also, a π bond is observed between His 76 (aromatic) and the positive nitrogen atom.
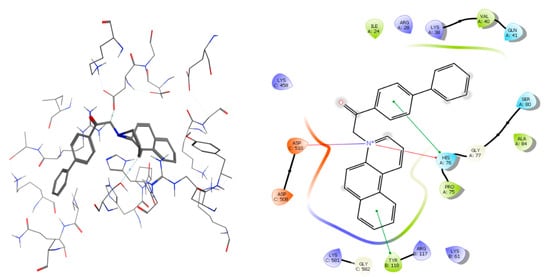
Figure 8.
Compound 3j is represented in complex with TOPO II as sticks. Surrounding aminoacids at the binding site are represented as ribbons colored by chain type. Also, a schematic view of the binding pocket is represented.
Ciprofloxacin, the control molecule, when docked with ATP synthase, shows a total energy of the complex of −2.59 eV and a hydrogen bond energy of −8.31 eV. In a complex with TOPO II, ciprofloxacin shows a total energy of −2.56 eV and a hydrogen bonding energy of −2.037 eV (Figure 9).
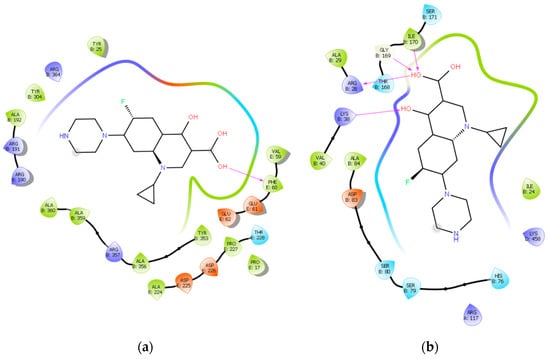
Figure 9.
Ciprofloxacin in complex with: (a) ATP synthase where hydrogen bond is formed between an OH group and Phe 60; (b) TOPO II, where hydrogen bonds are formed between OH groups and Lys 38, Arg 28, Gly 169, and Ile 170, respectively.
The most active antimicrobial compounds 3i and 3n show a similar behaviour in the ATP synthase and TOPO II binding pockets as compound 3j (Figure 10).
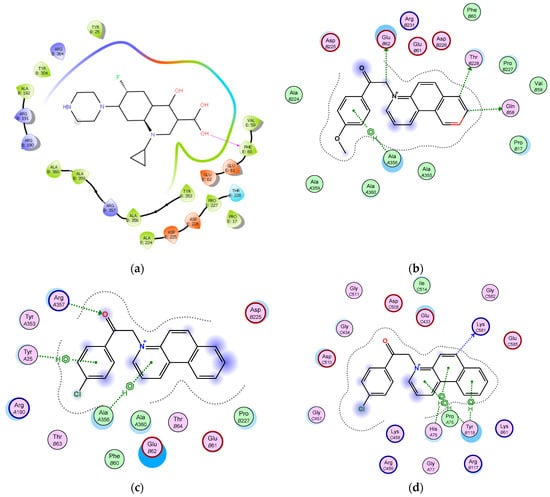
Figure 10.
3i and 3n in complex with ATP synthase and TOPO II in their binding pockets: (a) 3i in complex with ATP synthase arene hydrogen bonds are formed between aromatic core and Ala 366, Glu 62, Thr 228, and Gln 58 accept electrons from aromatic and aliphatic carbons; (b) 3i in complex with TOPO II, nitrogen atom interacts with Asp 508 and Glu 433, heterocyclic aromatic carbon share electrons with Asp 508 and Glu 433; (c) 3n in complex with ATP synthase, arene hydrogen bonds are formed between an aromatic core and Tyr 25 and between heterocycle aromatic core and Ala 356; also keto group shares electrons with Arg 357; (d) 3n in complex with TOPO II, arene hydrogen bonds are formed between each heterocyclic aromatic core and His 76, Pro 75, and Tyr 118, respectively.
Lastly, regarding our computational docking study, compound 3j displayed the best interaction energies. However, experimentally compounds 3i and 3n have the best bioactivities, while 3j has only moderate antimicrobial activity. In comparison with cyprofloxacin where the aromatic centres seem to play no role, in the case of BQS those aromatic centres are crucial in the interaction with the active sites. In the next step, the ADME properties of BQS salts 3a–o were studied (Table 4).

Table 4.
ADME properties computed for BQS 3a–o.
Analysis of the data from Table 4 reveals interesting data concerning the BQS salts’ ADME properties. All salt derivatives show good aqueous solubility. This is indicated by two descriptors: QP logS (which describes the general aqueous solubility) and ClQP logS (which describes the conformation-independent aqueous solubility). The optimal values of these two descriptors are in the range between −6.5 and 0.5. The data from Table 4 indicate for our BQS salt 3a has good solubility. Three compounds (3j, 3m, and 3n) are exceptions being slightly out of limit.
The QP log HERG descriptor is a computational equivalent of zebrafish model toxicity in respect to the mode of action (MOA). This descriptor is linked with the potential K channel-blocking effect due to the electrophilic nature of drugs. The concerned values are below −5. As observed from Table 4 the majority of the synthesized compounds have predictable, convenient values, which means that our BQS quaternary salts 3a-o have low toxicity profiles. The reduced toxicity it is also confirmed by the Lipinski’s rule of five and Jorgensen’s rule of three computational results. From Table 4 we can notice that only compounds 3j, 3m, and 3n present a violation of Lipinski’s rule of 5 and Jorgensen’s rule of 3, respectively. According with Lipinski’s rule of five, the maximum number of violations for a computed compound is five (these five rules are: mol_MW < 500; QP log Po/w < 5; donor HB < 5; accpt. HB < 10) while for Jorgensen’s rule of three, the maximum number of violations for a computed compound is three (QP log s > −5.7; QP PCaco > 22 nm/s; primary metabolites < 7). As a result, all our BQS salts 3a-o have excellent drug-like properties. However, in vitro testing should be performed in order to confirm these theoretical results.
The QP PCaco descriptor predicts permeability at the gut drug barrier. Predictions are for non-active transport. Values below 25 describe a low permeability while values higher than 500 are characteristic of excellent permeability. All described compounds show poor predicted gut barrier permeability.
QP Log BB descriptor predicts brain-blood partition coefficient for oral delivery. An optimal interval is between descriptor values of −3.0 to 1.2. All compounds are off the limit, meaning that potentially they may have poor blood-brain partition coefficients when administered orally.
QP PMDCK is a descriptor designed to simulate MDCK cell permeability, considered to be a good mimic of the blood-brain barrier. Values below 25 describe a poor permeability while values higher than 500 describe an excellent permeability. The data from Table 4 indicate that all BQS salts 3a–o has excellent blood—brain barrier permeability.
QP log KP is a descriptor that predicts skin permeability, with an optimal interval between −8.0 and −1.0. The data from Table 4 indicate that the compounds might not have good skin absorption.
Predicted chemical reactivity (descriptor Metab.) shows very high values for our BQS salts, suggesting a negative behavior as leads (the optimal values for Metab. descriptor are range between 1 to −8).
Interactions of compounds with serum albumin were explored computationally using the QP log Khsa descriptor. The optimal values for QP log Khsa descriptor are in the range between −1.5 to 15. The data from Table 4 indicate that all BQS salts 3a–o have a high potential of interacting with human serum albumin.
%Ab-sorbtion is a descriptor that predicts human oral absorption on a 0 to 100% scale. Values below 25% describe a poor oral absorption while value higher than 80% describe an excellent oral absorption. The data from Table 4 indicate that all BQS salts 3a–o have values higher than 80%, which suggest excellent oral absorption.
3. Materials and Methods
3.1. General Information
Reagents and solvents were purchased from commercial sources and used without further purification. The melting points (uncorrected) of compounds were recorded in open capillary tubes using a MEL-TEMP Electrothermal apparatus (Barnstead International, Dubuque, IA, USA). The nuclear magnetic resonance spectra were recorded on an Avance III 500 MHz spectrometer (Bruker, Vienna, Austria) operating at 500 MHz for 1H and 125 MHz for 13C. Chemical shifts were reported in delta (δ) units (ppm), relative to the residual peak of solvent (ref: DMSO, 1H: 2.50 ppm; 13C: 39.52 ppm), and coupling constants (J) in Hz. In the NMR spectra the multiplicity of signals was indicated using the abbreviations s = singlet, bs = broad singlet, d = doublet, ad = apparent doublet, add = apparent doublet of doublets, t = triplet, at = apparent triplet, td = triplet of doublets, atd = apparent triplet of doublets, q = quartet, m = multiplet. The IR spectra were recorded using a VERTEX 70 FTIR spectrometer (Bruker, Vienna, Austria) equipped with an ATR module. Thin layer chromatography (TLC) was performed on commercial silica gel plates (silica gel 60 F254 plates, Merck, Darmstadt, Germany), the visualization being done using a UV lamp (λmax = 254 or 365 nm). The microanalysis results were in satisfactory agreement (C, ±0.15; H, ±0.10; N, ±0.30) with the calculated values. Compound 3i was already reported in the literature, and its spectral data were in agreement with the reported data [29].
3.2. General Procedure for the Synthesis of Benzo[f]quinoline Salts 3a–o
A solution of halogenated derivatives 2a–f with aliphatic chains and increased reactivity (1.1 mmol) or with aromatic skeletons (compounds 2g–o) (1.5 mmol) in 15 mL of acetone was added dropwise in a solution of benzo[f]quinoline (1, 1 mmol) in 15 mL of acetone. The obtained mixture was heated at reflux during 16 h, and then stirred for 12 h at room temperature. The precipitated quaternary salts 3a–o (Scheme 3) were filtered off, washed two times with 5 mL of acetone, and then dried under vacuum. No further purification was required.
Scheme 3.
Chemical Structures of compounds 3a–o.
3.2.1. 1-(2-Amino-2-oxoethyl)benzo[f]quinolin-1-ium iodide (3a)
Yellowish green powder; yield: 84%; m.p. 238–240 °C; IR, νmax 3335, 3134, 3037, 2966, 1681, 1597, 1412, 1344 cm−1; 1H-NMR (DMSO-d6) δ 10.18 (1H, d, J = 8.5 Hz, H-4), 9.47 (1H, d, J = 6.0 Hz, H-2), 9.10 (1H, d, J = 8.5 Hz, H-5), 8.68 (1H, d, J = 9.5 Hz, H-9), 8.40 (1H, td, J = 8.5 Hz, J = 6.0 Hz, H-3), 8.28 (1H, d, J = 8.0 Hz, H-8), 8.20 (1H, bs, NH), 8.14 (1H, d, J = 9.5 Hz, H-10), 8.01 (1H, t, J = 7.5 Hz, H-6), 7.95 (1H, t, J = 7.5 Hz, H-7), 7.88 (1H, bs, NH), 5.91 (2H, s, CH2); 13C-NMR (DMSO-d6) δ 166.0 (C = O amido), 148.5 (C-2), 142.1 (C-4), 139.6 (C-10a), 138.0 (C-9), 130.7 (C-8a), 130.1 (C-6), 130.0 (C-7), 129.4 (C-8), 127.8 (C-4a, C-4b), 124.2 (C-5), 122.6 (C-3), 115.7 (C-10), 59.3 (CH2); Anal. Calcd. for C15H13IN2O C, 49.47; H, 3.60; N, 7.69. Found C, 49.37; H, 3.70; N, 7.49.
3.2.2. 1-(2-Methoxy-2-oxoethyl)benzo[f]quinolin-1-ium bromide (3b)
Yellowish powder; yield: 70%; m.p. 149–153 °C; IR (KBr), νmax 3029, 2869, 1743, 1600, 1431, 1232, 1068 cm−1; 1H-NMR (400 MHz, DMSO-d6) δ 10.23 (1H, d, J = 8.8 Hz, H-4), 9.62 (1H, d, J = 6.0 Hz, H-2), 9.09 (1H, d, J = 8.0 Hz, H-5), 8.67 (1H, d, J = 9.6 Hz, H-9), 8.44 (1H, atd, J = 8.4 Hz, J = 6.0 Hz, H-3), 8.34 (1H, d, J = 10.0 Hz, H-10), 8.28 (1H, d, J = 7.6 Hz, H-8), 8.01 (2H, m, H-6, H-7), 6.33 (2H, s, CH2), 3.81 (3H, s, CH3); 13C-NMR (100 MHz, DMSO-d6) δ 166.6 (C = O ester), 148.3 (C-2), 142.9 (C-4), 139.5 (C-10a), 138.2 (C-9), 130.7 (C-8a), 130.1 (C-6), 130.0 (C-7), 129.3 (C-8), 127.8 (C-4a), 127.7 (C-4b), 124.1 (C-5), 122.7 (C-3), 115.9 (C-10), 57.9 (CH2), 53.2 (CH3); Anal. Calcd. for C16H14BrNO2 C, 57.85; H, 4.25; N, 4.22. Found C, 57.75; H, 4.20; N, 4.12.
3.2.3. 1-(2-Ethoxy-2-oxoethyl)benzo[f]quinolin-1-ium bromide (3c)
Yellowish powder; yield: 73%; m.p. 125–127 °C; IR (KBr), νmax 3058, 2977, 1741, 1598, 1417, 1209, 1014 cm−1; 1H-NMR (DMSO-d6) δ 10.25 (1H, d, J = 8.4 Hz, H-4), 9.60 (1H, d, J = 5.6 Hz, H-2), 9.12 (1H, d, J = 8.0 Hz, H-5), 8.70 (1H, d, J = 9.6 Hz, H-9), 8.45 (1H, atd, J = 8.4 Hz, J = 6.0 Hz, H-3), 8.32 (2H, m, H-8, H-10), 7.99 (2H, m, H-6, H-7), 6.30 (2H, s, CH2), 4.26 (2H, q, J = 7.2 Hz, CH2-Et), 1.26 (3H, t, J = 7.2 Hz, CH3); 13C-NMR (DMSO-d6) δ 166.1 (C = O ester), 148.4 (C-2), 142.8 (C-4), 139.6 (C-10a), 138.2 (C-9), 130.7 (C-8a), 130.1 (C-6), 130.0 (C-7), 129.4 (C-8), 127.8 (C-4a), 127.7 (C-4b), 124.2 (C-5), 122.7 (C-3), 115.9 (C-10), 62.4 (CH2-Et), 57.9 (CH2), 13.9 (CH3); Anal. Calcd. for C17H16BrNO2 C, 58.97; H, 4.66; N, 4.05. Found C, 58.87; H, 4.60; N, 4.15.
3.2.4. 1-(2-Oxopropyl)benzo[f]quinolin-1-ium bromide (3d)
Beige powder; yield: 67%; m.p. 159–160 °C; IR, νmax 3022, 2855, 1730, 1597, 1511, 1411, 1341, 1173 cm−1; 1H-NMR (DMSO-d6) δ 10.20 (1H, d, J = 8.5 Hz, H-4), 9.39 (1H, d, J = 5.5 Hz, H-2), 9.12 (1H, d, J = 8.0 Hz, H-5), 8.64 (1H, d, J = 9.5 Hz, H-9), 8.44 (1H, dd, J = 5.5 Hz, J = 8.5 Hz, H-3), 8.30 (2H, m, H-8, H-10), 8.03 (1H, t, J = 7.5 Hz, H-6), 7.96 (1H, t, J = 7.5 Hz, H-7), 6.43 (2H, s, CH2), 2.50 (3H, s, CH3); 13C-NMR (DMSO-d6) δ 199.7 (C = O keto), 147.8 (C-2), 144.0 (C-4), 142.3 (C-10a), 139.8 (C-9), 130.8 (C-4a), 130.1 (C-7), 130.0 (C-6), 129.4 (C-8), 127.9 (C-4b), 127.8 (C-9), 124.2 (C-5), 122.7 (C-3), 116.4 (C-10), 66.0 (CH2), 27.6 (CH3); Anal. Calcd. for C16H14BrNO C, 60.78; H, 4.46; N, 4.43. Found C, 60.69; H, 4.42; N, 4.38.
3.2.5. 1-(2-Oxobutyl)benzo[f]quinolin-1-ium bromide (3e)
Yellow powder; yield: 71%; m.p. 165–166 °C; IR, νmax 3052, 2964, 1723, 1599, 1413, 1340, 1106 cm−1; 1H-NMR (DMSO-d6) δ 10.21 (1H, d, J = 8.5 Hz, H-4), 9.40 (1H, d, J = 6.0 Hz, H-2), 9.12 (1H, d, J = 8.5 Hz, H-5), 8.63 (1H, d, J = 10.0 Hz, H-9), 8.42 (1H, dd, J = 6.0 Hz, J = 8.5 Hz, H-3), 8.30 (2H, m, H-8, H-10), 8.02 (1H, t, J = 7.5 Hz, H-6), 7.96 (1H, t, J = 7.5 Hz, H-7), 6.42 (2H, s, CH2), 2.93 (2H, q, J = 7.5 Hz, CH2-Et), 1.06 (3H, t, J = 7.5 Hz, CH3-Et); 13C-NMR (DMSO-d6) δ 202.3 (C = O keto), 147.8 (C-2), 142.2 (C-4), 139.8 (C-10a), 137.9 (C-9), 130.8 (C-4a), 130.1 (C-7), 130.0 (C-6), 129.4 (C-8), 127.9 (C-4b), 127.8 (C-9), 124.2 (C-5), 122.7 (C-3), 116.3 (C-10), 65.3 (CH2), 32.8 (CH2-Et), 6.9 (CH3-Et); Anal. Calcd. for C17H16BrNO C, 61.83; H, 4.88; N, 4.24. Found C, 61.73; H, 4.81; N, 4.17.
3.2.6. 1-(3,3-Dimethyl-2-oxobutyl)benzo[f]quinolin-1-ium bromide (3f)
Yellow powder; yield: 89%; m.p. 129–130 °C; IR, νmax 3051, 2942, 1708, 1578, 1414, 1342, 1057 cm−1; 1H-NMR (DMSO-d6) δ 10.24 (1H, d, J = 8.5 Hz, H-4), 9.46 (1H, d, J = 6.0 Hz, H-2), 9.15 (1H, d, J = 8.5 Hz, H-5), 8.68 (1H, d, J = 9.5 Hz, H-9), 8.45 (1H, dd, J = 6.0 Hz, J = 8.5 Hz, H-3), 8.30 (1H, d, J = 7.5 Hz, H-8), 8.08 (1H, d, J = 9.5 Hz, H-10), 8.04 (1H, t, J = 7.5 Hz, H-6), 7.98 (1H, t, J = 7.5 Hz, H-7), 6.68 (2H, s, CH2), 1.36 (9H, s, 3 × CH3-tBu); 13C-NMR (DMSO-d6) δ 207.0 (C = O keto), 148.2 (C-2), 142.3 (C-4), 139.6 (C-10a), 138.1 (C-9), 130.8 (C-4a), 130.2 (C-7), 130.1 (C-6), 129.4 (C-8), 128.0 (C-4b), 127.9 (C-9), 124.3 (C-5), 122.8 (C-3), 115.7 (C-10), 63.0 (CH2), 43.4 (C(CH3)3-tBu), 25.7 (3 × CH3-tBu); Anal. Calcd. for C19H20BrNO C, 63.70; H, 5.63; N, 3.91. Found C, 63.59; H, 5.56; N, 3.86.
3.2.7. 1-(Phenacyl)benzo[f]quinolin-1-ium bromide (3g)
Cream-colored powder; yield: 61%; m.p. 234–236 °C; IR, νmax 2989, 2898, 1682, 1577, 1343, 1222, 1065 cm−1; 1H-NMR (500 MHz, DMSO-d6) δ 10.20 (1H, d, J = 8.5 Hz, H-4), 9.45 (1H, d, J = 5.5 Hz, H-2), 9.11 (1H, d, J = 8.5 Hz, H-5), 8.56 (1H, d, J = 10 Hz, H-9), 8.39 (1H, dd, J = 8.5 Hz, J = 6.0 Hz, H-3), 8.23 (2H, d, J = 9.5 Hz, J = 7.5 Hz, H-8, H-10), 8.17 (2H, d, J = 7.0 Hz, 2 × H-14), 8.02 (1H, atd, J = 8.0 Hz, J = 7.0 Hz, H-6), 7.95 (1H, t, J = 8.0 Hz, J = 7.0 Hz, H-7), 7.80 (1H, d, J = 7.5 Hz, H-16), 7.67 (2H, t, J = 8.0 Hz, 2 × H-15), 7.05 (2H, s, CH2); 13C-NMR (125 MHz, DMSO-d6) δ 190.6 (C = O keto), 148.2 (C-2), 142.5 (C-4), 140.3 (C-10a), 138.4 (C-9), 135.0 (C-16), 133.6 (C-13), 131.0 (C-8a), 130.3 (C-7), 130.2 (C-6), 129.5 (C-8), 129.1 (2 × C-15), 128.7 (2 × C-14), 128.3 (C-4a), 128.1 (C-4b), 124.2 (C-5), 122.9 (C-3), 116.1 (C-10), 63.9 (CH2); Anal. Calcd. for C21H16BrNO C, 66.68; H, 4.26; N, 3.70. Found C, 66.78; H, 4.16; N, 3.65.
3.2.8. 1-(4-Methylphenacyl)benzo[f]quinolin-1-ium bromide (3h)
Yellowish powder; yield: 57%; m.p. 220–224 °C; IR, νmax 3027, 2946, 1677, 1600, 1344, 1231, 1185 cm−1; 1H-NMR (DMSO-d6) δ 10.25 (1H, d, J = 8.5 Hz, H-4), 9.55 (1H, d, J = 5.5 Hz, H-2), 9.15 (1H, d, J = 8.0 Hz, H-5), 8.61 (1H, d, J = 9.5 Hz, H-9), 8.46 (1H, atd, J = 8.5 Hz, J = 6.0 Hz, H-3), 8.28 (2H, m, H-8, H-10), 8.08 (2H, d, J = 8.0 Hz, 2 × H-14), 8.02 (1H, t, J = 8.0 Hz, J = 7.0 Hz, H-6), 7.96 (1H, t, J = 7.5 Hz, H-7), 7.50 (2H, d, J = 8.0 Hz, 2 × H-15), 7.12 (2H, s, CH2), 2.46 (3H, s, CH3); 13C-NMR (DMSO-d6) δ 190.2 (C = O keto), 148.2 (C-2), 145.6 (C-16), 142.4 (C-4), 140.0 (C-10a), 138.0 (C-9), 131.1 (C-13), 130.8 (C-8a), 130.1 (C-7), 130.0 (C-6), 129.5 (2 × C-15), 129.4 (C-8), 128.8 (2 × C-14), 127.9 (C-4a), 127.8 (C-4b), 124.2 (C-5), 122.8 (C-3), 116.3 (C-10), 63.8 (CH2), 21.4 (CH3); Anal. Calcd. for C22H18BrNO C, 67.36; H, 4.62; N, 3.57. Found C, 67.47; H, 4.52; N, 3.47.
3.2.9. 1-(4-Methoxyphenacyl)benzo[f]quinolin-1-ium bromide (3i)
Yellowish powder; yield: 70%; m.p. 139–142 °C; IR, νmax 2968, 2900, 1673, 1598, 1418, 1344, 1237, 1179, 1021 cm−1; 1H-NMR (DMSO-d6) δ 10.25 (1H, d, J = 8.5 Hz, H-4), 9.52 (1H, d, J = 5.5 Hz, H-2), 9.15 (1H, d, J = 8.0 Hz, H-5), 8.62 (1H, d, J = 9.5 Hz, H-9), 8.46 (1H, t, J = 8.0 Hz, J = 6.5 Hz, H-3), 8.27 (2H, at, J = 8.0 Hz, H-8, H-10), 8.15 (2H, d, J = 8.5 Hz, 2 × H-14), 8.03 (1H, t, J = 8.0 Hz, J = 7.0 Hz, H-6), 7.96 (1H, t, J = 7.5 Hz, J = 7.0 Hz, H-7), 7.22 (2H, d, J = 8.5 Hz, 2 × H-15), 7.08 (2H, s, CH2), 3.92 (3H, s, CH3); 13C-NMR (DMSO-d6) δ 188.9 (C = O keto), 164.4 (C-16), 148.2 (C-2), 142.4 (C-4), 140.0 (C-10a), 138.0 (C-9), 131.2 (2 × C-14), 130.8 (C-8a), 130.1 (C-7), 130.0 (C-6), 129.4 (C-8), 127.9 (C-4a), 127.8 (C-4b), 126.4 (C-13), 124.2 (C-5), 122.8 (C-3), 116.3 (C-10), 114.3 (2 × C-15), 63.5 (CH2), 55.9 (CH3); Anal. Calcd. for C22H18BrNO2 C, 64.72; H, 4.44; N, 3.43. Found C, 64.62; H, 4.40; N, 3.63.
3.2.10. 1-(4-Phenylphenacyl)benzo[f]quinolin-1-ium bromide (3j)
Yellowish powder; yield: 80%; m.p. 242–245 °C; IR, νmax 3016, 2895, 1689, 1601, 1339, 1225 cm−1; 1H-NMR (DMSO-d6) δ 10.28 (1H, d, J = 8.5 Hz, H-4), 9.56 (1H, add, J = 6.5 Hz, H-2), 9.17 (1H, d, J = 8.5 Hz, H-5), 8.64 (1H, d, J = 10 Hz, H-9), 8.48 (1H, dd, J = 8.5 Hz, J = 6.0 Hz, H-3), 8.36 (1H, d, J = 10.0 Hz, H-10), 8.30 (1H, d, J = 7.5 Hz, H-8), 8.26 (2H, d, J = 8.5 Hz, 2 × H-14), 8.03 (3H, m, 2 × H-15, H-6), 7.97 (1H, atd, J = 8.0 Hz, J = 7.5 Hz, H-7), 7.84 (2H, d, J = 8.5 Hz, 2 × H-18), 7.55 (2H, t, J = 8.0 Hz, J = 7.0 Hz, 2 × H-19), 7.48 (1H, atd, J = 8.5 Hz, J = 7.0 Hz, H-20), 7.18 (2H, s, CH2); 13C-NMR (DMSO-d6) δ 190.3 (C = O keto), 148.2 (C-2), 146.0 (C-16), 142.5 (C-4), 140.1 (C-10a), 138.5 (C-17), 138.9 (C-9), 132.4 (C-13), 130.8 (C-8a), 130.1 (C-6), 130.0 (C-7), 129.4 (2 × C-14, C-8), 129.2 (2 × C-19), 128.8 (C-20), 127.9 (C-4a), 127.8 (C-4b), 127.1 (2 × C-18, 2 × C-15), 124.3 (C-5), 122.8 (C-3), 116.4 (C-10), 63.9 (CH2); Anal. Calcd. for C27H20BrNO C, 71.37; H, 4.44; N, 3.08. Found C, 71.47; H, 4.34; N, 3.18.
3.2.11. 1-(4-Cyanophenacyl)benzo[f]quinolin-1-ium bromide (3k)
Yellow powder; yield: 85%; m.p. 199–202 °C; IR, νmax 3015, 2952, 2232, 1706, 1401, 1340, 1214 cm−1; 1H-NMR (DMSO-d6) δ 10.28 (1H, d, J = 8.5 Hz, H-4), 9.51 (1H, d, J = 6.0 Hz, H-2), 9.17 (1H, d, J = 8.5 Hz, H-5), 8.64 (1H, d, J = 9.5 Hz, H-9), 8.48 (1H, atd, J = 8.5 Hz, J = 6.0 Hz, H-3), 8.41 (1H, d, J = 9.5 Hz, H-10), 8.31 (3H, t, J = 8.5 Hz, H-8, 2 × H-14), 8.20 (2H, d, J = 8.5 Hz, 2 × H-15), 8.04 (1H, t, J = 8.0 Hz, J = 7.5 Hz, H-6), 7.98 (1H, t, J = 8.0 Hz, J = 7.0 Hz, H-7), 7.15 (2H, s, CH2); 13C-NMR (DMSO-d6) δ 190.3 (C = O keto), 148.2 (C-2), 142.6 (C-4), 140.2 (C-10a), 138.1 (C-9), 136.9 (C-13), 132.9 (2 × C-15), 130.8 (C-8a), 130.1 (C-7, C-6), 129.4 (C-8), 129.3 (2 × C-14), 127.9 (C-4a), 127.8 (C-4b), 124.3 (C-5), 122.8 (C-3), 118.0 (CN), 116.4 (C-10), 116.3 (C-16), 64.1 (CH2); Anal. Calcd. for C22H15BrN2O C, 65.52; H, 3.75; N, 6.95. Found C, 65.62; H, 3.70; N, 6.85.
3.2.12. 1-(4-Nitrophenacyl)benzo[f]quinolin-1-ium bromide (3l)
Yellowish powder; yield: 74%; m.p. 156–158 °C; IR, νmax 3023, 2963, 1717, 1599, 1520, 1349, 1217 cm−1; 1H-NMR (DMSO-d6) δ 10.29 (1H, d, J = 8.5 Hz, H-4), 9.52 (1H, d, J = 5.5 Hz, H-2), 9.17 (1H, d, J = 8.5 Hz, H-5), 8.65 (1H, d, J = 9.5 Hz, H-9), 8.52 (2H, d, J = 9.0 Hz, 2 × H-15), 8.48 (1H, at, J = 8.5 Hz, J = 6.0 Hz, H-3), 8.43 (1H, d, J = 10.0 Hz, H-10), 8.40 (2H, d, J = 8.5 Hz, 2 × H-14), 8.13 (1H, d, J = 7.5 Hz, H-8), 8.50 (1H, t, J = 8.0 Hz, J = 7.0 Hz, H-6), 7.98 (1H, t, J = 7.5 Hz, J = 7.0 Hz, H-7), 7.18 (2H, s, CH2); 13C-NMR (DMSO-d6) δ 190.1 (C = O keto), 150.6 (C-16), 148.2 (C-2), 142.7 (C-4), 140.2 (C-10a), 138.3 (C-13), 138.1 (C-9), 130.8 (C-8a), 130.2 (2 × C-14), 130.1 (C-7, C-6), 129.4 (C-8), 128.0 (C-4a), 127.8 (C-4b), 124.2 (C-5), 124.0 (2 × C-15), 122.8 (C-3), 116.4 (C-10), 64.1 (CH2); Anal. Calcd. for C21H15BrN2O3 C, 59.59; H, 3.57; N, 6.62. Found C, 59.49; H, 3.50; N, 6.52.
3.2.13. 1-(4-Bromophenacyl)benzo[f]quinolin-1-ium bromide (3m)
Cream-colored powder; yield: 80%; m.p. 229–232 °C; IR, νmax 3018, 2911, 1696, 1583, 1347, 1223 cm−1; 1H-NMR (DMSO-d6) δ 10.27 (1H, d, J = 9.0 Hz, H-4), 9.52 (1H, d, J = 6.0 Hz, H-2), 9.16 (1H, d, J = 8.5 Hz, H-5), 8.63 (1H, d, J = 10.0 Hz, H-9), 8.47 (1H, atd, J = 8.5 Hz, J = 6.0 Hz, H-3), 8.35 (1H, d, J = 9.5 Hz, H-10), 8.29 (1H, d, J = 7.5 Hz, H-8), 8.10 (2H, d, J = 8.5 Hz, 2 × H-14), 8.36 (1H, t, J = 7.5 Hz, J = 7.0 Hz, H-6), 7.97 (1H, t, J = 7.5 Hz, H-7), 7.94 (2H, d, J = 8.5 Hz, 2 × H-15), 7.11 (2H, s, CH2); 13C-NMR (DMSO-d6) δ 190.1 (C = O keto), 148.2 (C-2), 142.5 (C-4), 140.1 (C-10a), 138.0 (C-9), 132.7 (C-13), 132.1 (2 × C-15), 130.8 (C-8a), 130.6 (2 × C-14), 130.1 (C-7), 130.0 (C-6), 129.4 (C-8), 128.9 (C-16), 127.9 (C-4a), 127.8 (C-4b), 124.2 (C-5), 122.8 (C-3), 116.4 (C-10), 63.8 (CH2); Anal. Calcd. for C21H15Br2NO C, 55.17; H, 3.31; N, 3.06. Found C, 55.07; H, 3.36; N, 3.16.
3.2.14. 1-(4-Chlorophenacyl)benzo[f]quinolin-1-ium bromide (3n)
Yellowish powder; yield: 79%; m.p. 192–196 °C; IR, νmax 3020, 2931, 1688, 1586, 1341, 1088, 759 cm−1; 1H-NMR (DMSO-d6) δ 10.28 (1H, d, J = 8.5 Hz, H-4), 9.47 (1H, d, J = 5.5 Hz, H-2), 9.18 (1H, d, J = 8.5 Hz, H-5), 8.64 (1H, d, J = 9.5 Hz, H-9), 8.47 (1H, atd, J = 8.5 Hz, J = 6.0 Hz, H-3), 8.36 (1H, d, J = 9.5 Hz, H-10), 8.31 (1H, d, J = 8.0 Hz, H-8), 8.18 (2H, d, J = 8.5 Hz, 2 × H-14), 8.05 (1H, t, J = 8.0 Hz, J = 7.0 Hz, H-6), 7.99 (1H, t, J = 8.0 Hz, J = 7.0 Hz, H-7), 7.80 (2H, d, J = 8.5 Hz, 2 × H-15), 7.08 (2H, s, CH2); 13C-NMR (DMSO-d6) δ 189.9 (C = O keto), 148.2 (C-2), 142.6 (C-4), 140.1 (C-10a), 139.6 (C-16), 138.1 (C-9), 132.4 (C-13), 130.8 (C-8a), 130.6 (2 × C-14), 130.1 (C-7, C-6), 129.4 (C-8), 129.2 (2 × C-15), 128.0 (C-4a), 127.9 (C-4b), 124.3 (C-5), 122.8 (C-3), 116.4 (C-10), 63.8 (CH2); Anal. Calcd. for C21H15BrClNO C, 61.11; H, 3.66; N, 3.39. Found C, 61.01; H, 3.60; N, 3.29.
3.2.15. 1-(4-Fluorophenacyl)benzo[f]quinolin-1-ium bromide (3o)
Cream-colored powder; yield: 87%; m.p. 143–145 °C; IR, νmax 3048, 2969, 1694, 1593, 1335, 1221, 1152 cm−1; 1H-NMR (DMSO-d6) δ 10.27 (1H, d, J = 8.5 Hz, H-4), 9.51 (1H, d, J = 6.0 Hz, H-2), 9.16 (1H, d, J = 8.0 Hz, H-5), 8.63 (1H, d, J = 10.0 Hz, H-9), 8.47 (1H, atd, J = 8.5 Hz, J = 6.5 Hz, H-3), 8.34 (1H, d, J = 10.0 Hz, H-10), 8.28 (3H, m, H-8, 2 × H-14), 8.04 (1H, t, J = 7.5 Hz, H-6), 7.97 (1H, t, J = 7.5 Hz, H-7), 7.56 (2H, t, J = 8.5 Hz, J = 9.0 Hz, 2 × H-15), 7.11 (2H, s, CH2); 13C-NMR (DMSO-d6) δ 189.5 (C = O keto), 165.93 (d, J = 101 Hz, C-16), 148.2 (C-2), 142.5 (C-4), 140.1 (C-10a), 138.1 (C-9), 131.9 (d, J = 4.0 Hz, 2 × C-14), 130.8 (C-8a), 130.4 (ad, J = 2.0 Hz, C-13), 130.1 (C-7, C-6), 129.4 (C-8), 128.0 (C-4a), 127.9 (C-4b), 124.3 (C-5), 122.8 (C-3), 116.4 (C-10), 116.26 (d, J = 9.0 Hz, 2 × C-15), 63.8 (CH2); Anal. Calcd. for C21H15BrFNO C, 63.65; H, 3.82; N, 3.53. Found C, 63.75; H, 3.77; N, 3.43.
3.3. Antimicrobial Assay
3.3.1. Disk-Diffusion Method
For inoculum preparation, reference microbial cultures of bacteria (Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922) and fungi (Candida albicans ATCC 10231) were employed. A number of approximately five colonies from each type of culture were used to inoculate 10 mL of Mueller Hinton (MH) agar (for antibacterial tests) and Sabouraud agar (for antifungal tests). Using a DU 730 spectrophotometer (Beckman Coulter, city, state abbrev if USA, country, λ = 600 nm), the turbidity of the inoculum was adjusted to a 0.5 McFarland standard (1–2 × 108 CFU/mL for bacteria and 1–5 × 106 CFU/mL for Candida), and the inoculum was transferred, in a 1 mL volume, onto the surface of the growth media specific for bacteria (MH) and fungi (Sabouraud). Once the inoculum was absorbed, sterile paper disks of approximately 6 mm in diameter and impregnated with 10 µL of antibacterial compound (dissolved in 3% DMSO) were placed on the surface of the culture media; for all the tested compounds, the concentration used was 25 mg/mL. Following incubation at the optimal temperatures for bacteria and fungi, of 37 °C and 28 °C, respectively, for 24 h (bacteria) and 72 h (fungi), the diameters of the inhibition zones were measured using a ruler. The controls were prepared in the same growth conditions (i.e., C+: sterile filter paper disks impregnated with antibiotics inducing sensitivity in the organisms under investigation, namely penicillin 10 IU for Staphylococcus aureus, carbenicillin 100 µg/mL for Escherichia coli and nystatin 500,000 IU for Candida albicans, and C−: sterile filter paper disks with no antimicrobial compounds).
3.3.2. Broth Microdilution Method for Determining the Minimum Inhibitory Concentration (MIC)
The working technique involves the use of a 96-well microtiter plate (microdilution). In each well of the plate, 80 µL of growth medium MH, 10 µL of microbial inoculum (Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, or Candida albicans ATCC 10231) prepared in the same manner as in the diffusion test (i.e., by diluting the standardized microbial suspension adjusted to a 0.5 McFarland standard), and 100 µL of antimicrobial substance to be tested were transferred by pipetting, in different concentrations. For this purpose, double dilutions of the antimicrobial agent were made in the DMSO 3%, starting with the 25 µg/mL dilution (e.g., 12.5 µg/mL, 6.25 µg/mL, 3.12 µg/mL, 1.56 µg/mL, 0.78 µg/mL and so on). For each tested microorganism, a positive control C+ (containing 80 µL of MH growth medium, 10 µL of diluted microbial culture, 100 µL successive double dilutions of antibiotic) and a negative one C− (containing 80 µL of MH growth medium and 10 µL of diluted microbial culture) were prepared. Following the incubation of the microplates at 37 °C for 24 h (for Staphylococcus aureus ATCC 25923 and Escherichia coli ATCC 25922) and at 28 °C for 72 h (for Candida albicans ATCC 10231), 10 µL of resazurin were added in each well. The samples were incubated once again at the optimal temperature for each microorganism for one hour. The colour of the indicator turned from purple to pink. Resazurin is a colorimetric indicator for cell viability widely applied for monitoring cell proliferation. The redox dye, resazurin, enters the cytosol in the oxidized form (purple-blue) and is converted to the reduced form, resorufin (pink).
4. Conclusions
We report herein the design, synthesis, experimental and in silico evaluation of the antibacterial and antifungal activity of some new benzo[f]quinoline derivatives. Two classes of benzo[f]quinolinium derivatives (salts BQS and cycloadducts PBQC) were designed and obtained via a direct and facile two step procedure: quaternization followed by a cycloaddition reaction. The synthesized compounds were characterized by elemental and spectral analysis (FT-IR, 1H-NMR, 13C-NMR). The antifungal assays revealed that the BQS salts have an excellent quasi-nonselective antifungal activity against the fungus Candida albicans, some of them higher that the control drug nystatin. The antibacterial assay revealed that the BQS salts have a very good antibacterial activity against the Gram positive germ Staphylococcus aureus while the activity against the Gram negative germ Escherichia coli is negligible. The cycloadducts PBQC 4 are inactive. Analysis of the biological data reveals some interesting SAR correlations between the structures and their antimicrobial activity. The in silico studies furnished important data concerning the pharmacodynamics, pharmacokinetics and ADMET parameters of the BQS salts. Study of the de interaction of each BQS salt 3a-o with ATP synthase in the formed complex, reveal that salts 3j, 3i, and 3n have the best-fit in complex with ATP synthase. Study of the de interaction of each BQS salt 3a-o with TOPO II in the formed complex, revealed that salts 3j and 3n have the best-fit in complex with TOPO II. The in silico ADMET studies reveal that the BQS salts have excellent drug-like properties, low toxicity profiles, excellent blood—brain barrier permeability, an excellent oral absorption and a good solubility. Overall, the experimental and in silico studies indicated that compounds 3e and 3f (from the aliphatic series) and 3i, 3j and 3n (from the aromatic series), are promising leading drug candidates.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ph14040335/s1, Figures S1–S3: The antibacterial activity for BQS salts 3i (OL22), 3h (OL23), and QBSC cycloadducts 4e1 (VB10-29), 4g1 (VB10-30), Tables S1–S4: Docking data used in ligand evaluation and QSAR model, Table S5: Molecular descriptors used in QSAR models evaluation, Table S6: Screening results for the best fit hypothesis of BQS3 interaction with ATP synthase and Topoisomerase II, Figures S4–S33: 1H-NMR spectra of compounds 3a–o.
Author Contributions
Design, conception and writing were performed by G.Z. and I.I.M. Biological assay was performed by S.D. Molecular docking was performed by C.N.L. Synthesis, structure elucidation, biological data analysis and molecular docking interpretations were performed by all authors, which also reviewed and approved the final version. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant of the Romanian Ministry of Education and Research, CNCS—UEFISCDI, project number PN-III-P4-ID-PCE-2020-0371, within PNCDI III.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Authors are also gratefully to CERNESIM center, for NMR experiments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO Global Strategy for Containment of Antimicrobial Resistance. Available online: https://www.who.int/drugresistance/WHO_Global_Strategy_English.pdf (accessed on 24 February 2021).
- Silverman, R.B.; Holladay, M.W. The Organic Chemistry of DRUG design and Drug Action, 3rd ed.; Academic Press: London, UK, 2014; ISBN 9780123820303. [Google Scholar]
- Brunton, L.; Knollmann, B.; Hilal-Dandan, R. Goodman & Gilman’s the Pharmacological Basis of Therapeutics, 13th ed.; McGraw-Hill: New York, NY, USA, 2013; ISBN 9781259584732. [Google Scholar]
- Eicher, T.; Hauptmann, S.; Speicher, A. The Chemistry of Heterocycles: Structures, Reactions, Synthesis, and Applications, 3rd ed.; Wiley-VCH: Weinheim, Germany, 2013; ISBN 978-3-527-32747-8. [Google Scholar]
- Kumari, L.S.; Mazumder, A.; Kumar, V.; Gupta, S. Synthesis and biological potentials of quinoline analogues: A review of literature. Mini-Rev. Org. Chem. 2019, 16, 653–688. [Google Scholar] [CrossRef]
- Song, Y.; Xu, H.; Chen, W.; Zhan, P.; Liu, X. 8-Hydroxyquinoline: A privileged structure with a broad-ranging pharmacological potential. Med. Chem. Commun. 2015, 6, 61–74. [Google Scholar] [CrossRef]
- Kalaria, P.N.; Karad, S.C.; Raval, D.K. A review on diverse heterocyclic compounds as the privileged scaffolds in antimalarial drug discovery. Eur. J. Med. Chem. 2018, 158, 917–936. [Google Scholar] [CrossRef]
- Hu, Y.Q.; Gao, C.; Zhang, S.; Xu, L.; Xu, Z.; Feng, L.S.; Wu, X.; Zhao, F. Quinoline hybrids and their antiplasmodial and antimalarial activities. Eur. J. Med. Chem. 2017, 139, 22–47. [Google Scholar] [CrossRef]
- Mantu, D.; Antoci, V.; Moldoveanu, C.; Zbancioc, G.; Mangalagiu, I.I. Hybrid imidazole (benzimidazole)/pyridine(quinoline) derivatives and evaluation of their anticancer and antimycobacterial activity. J. Enz. Inhib. Med. Chem. 2016, 31, 96–103. [Google Scholar] [CrossRef]
- Afzal, O.; Kumar, S.; Haider, M.R.; Haider, R.; Ali, R.; Kumar, R.; Jaggi, M.; Bawa, S. A review on anticancer potential of bioactive heterocycle quinoline. Eur. J. Med. Chem. 2015, 97, 871–910. [Google Scholar] [CrossRef] [PubMed]
- Pinz, M.P.; Reis, A.S.; Oliveira, R.L.; Voss, G.T.; Vogt, A.G.; Sacramento, M.D.; Roehrs, J.A.; Alves, D.; Luchese, C.; Wilhelm, E.A. 7-Chloro-4-phenylsulfonylquinoline, a new antinociceptive and anti-inflammatory molecule: Structural improvement of a quinoline derivate with pharmacological activity. Regul. Toxicol. Pharmacol. 2017, 90, 72–77. [Google Scholar] [CrossRef]
- Zhong, F.; Geng, G.; Chen, B.; Pan, T.; Li, Q.; Zhang, H.; Bai, C. Identification of benzenesulfonamide quinoline derivatives as potent HIV-1 replication inhibitors targeting Rev protein. Org. Biomol. Chem. 2015, 13, 1792–1799. [Google Scholar] [CrossRef]
- Diaconu, D.; Mangalagiu, V.; Amariucai-Mantu, D.; Antoci, V.; Giuroiu, C.L.; Mangalagiu, I.I. Hybrid quinoline-sulfonamide complexes (M2+) derivatives with antimicrobial activity. Molecules 2020, 25, 2946. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, S.; Ba, Y.; Xu, Z. 1,2,4-Triazole-quinoline/quinolone hybrids as potential anti-bacterial agents. Eur. J. Med. Chem. 2019, 174, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Al Matarneh, C.; Sardaru, M.; Apostu, M.; Rosca, I.; Ciobanu, C.; Mangalagiu, I.I.; Danac, R. Synthesis and antibacterial evaluation of new pyrrolo[3′,4′:3,4]pyrrolo[1,2-a]quinoline and pyrrolo[3′,4′:3,4] pyrrolo[1,2-a]isoquinoline derivatives. Studia UBB Chemia 2019, LXIV, 67–80. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, S.; Pan, B.; Liu, X.; Feng, L.S. 4-Quinolone derivatives and their activities against Gram positive pathogens. Eur. J. Med. Chem. 2018, 143, 710–723. [Google Scholar] [CrossRef] [PubMed]
- Ajani, O.O.; Iyaye, K.T.; Audu, O.Y.; Kuye, A.O.; Olanrewaju, I.O. Microwave assisted synthesis and antimicrobial potential of quinoline based 4-hydrazide-hydrazone derivatives. J. Heterocycl. Chem. 2018, 55, 302–312. [Google Scholar] [CrossRef]
- Nainwal, L.M.; Tasneem, S.; Akhtar, W.; Verma, G.; Khan, M.F.; Parvez, S.; Shaquiquzzaman, M.; Akhter, M.; Alam, M.M. Green recipes to quinoline: A review. Eur. J. Med. Chem. 2019, 164, 121–170. [Google Scholar] [CrossRef]
- Gattu, R.; Bagdi, P.R.; Sidick Basha, R.; Khan, A.T. Camphorsulfonic acid catalyzed one-pot three-component reaction for the synthesis of fused quinoline and benzoquinoline derivatives. J. Org. Chem. 2017, 82, 12416–12429. [Google Scholar] [CrossRef]
- Lungu, L.; Ciocarlan, A.; Simigon, C.; Ozer, I.; Shova, S.; Gutu, I.; Vornicu, N.; Mangalagiu, I.I.; D’Ambrosio, M.; Aricu, A. Synthesis and evaluation of biological activity of homodrimane sesquiterpenoids bearing 1,3,4-oxadiazole or 1,3,4-thiadiazole units. Chem. Heterocycl. Compd. 2020, 56, 578–585. [Google Scholar] [CrossRef]
- Aricu, A.; Ciocarlan, A.; Lungu, L.; Barba, A.; Shova, S.; Zbancioc, G.; Mangalagiu, I.I.; D’Ambrosio, M.; Vornicu, N. Synthesis of new antibacterial and antifungal drimane sesquiterpenoids with azaheterocyclic units. Med. Chem. Res. 2016, 25, 2316–2323. [Google Scholar] [CrossRef]
- Balan, A.M.; Miron, A.; Tuchilus, C.; Rotinberg, P.; Mihai, C.T.; Mangalagiu, I.I.; Zbancioc, G. Synthesis and in vitro analysis of novel dihydroxyacetophenone derivatives with antimicrobial and antitumor activities. Med. Chem. 2014, 10, 476–483. [Google Scholar]
- Kuchkova, K.; Aricu, A.; Barba, A.; Vlad, P.; Shova, S.; Secara, E.; Ungur, N.; Tuchilus, C.; Zbancioc, G.; Mangalagiu, I.I. Design, syntheses and antimicrobial activity of some novel homodrimanese squiterpenoids with diazine skeleton. Med. Chem. Res. 2014, 23, 1559–1568. [Google Scholar] [CrossRef]
- Tucaliuc, R.; Cotea, V.; Niculaua, M.; Tuchilus, C.; Mantu, D.; Mangalagiu, I.I. New pyridazine–fluorine derivatives: Synthesis, chemistry and biological activity. Part II. Eur. J. Med. Chem. 2013, 67, 367–372. [Google Scholar] [CrossRef]
- Balan, A.M.; Florea, O.; Moldoveanu, C.; Zbancioc, G.; Iurea, D.; Mangalagiu, I.I. Diazinium salts with dihydroxyacetophenone skeleton: Syntheses and antimicrobial activity. Eur. J. Med. Chem. 2009, 44, 2275–2279. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.K.; Grondard, L.; Couprie, J.; Desoize, B.; Comoe, L.; Jardillier, J.C.; Riou, J.F. The antileukemic alkaloid fagaronine is an inhibitor of DNA topoisomerases I and II. Biochem. Pharmacol. 1993, 46, 1403–1412. [Google Scholar] [CrossRef]
- Van, D.N.; Rucinschi, E.; Druta, I.; Zugravescu, I. Reaction de benzo[f]quinoline avec sels. Bul. Inst. Politeh. Iasi Sect. 2 Chim. 1977, 23, 51–57. [Google Scholar]
- Mangalagiu, I.I.; Mangalagiu, G.; Roman, M.; Olariu, I.; Petrovanu, M. Synthesis and spectral characterization of some new diazine salts. An. Stiint. Univ. Al. I. Cuza Iasi 1999, 7, 137–142. [Google Scholar]
- Georgescu, E.; Draghici, C.; Iuhas, P.C.; Georgescu, F. A new approach for the synthesis of benzo[f]pyrrolo [1,2-a]-quinolines. Arkivoc 2005, X, 95–104. [Google Scholar] [CrossRef]
- Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 11th ed.; Approved Standard, CLSI Document M07-A11; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; Available online: https://clsi.org/media/1928/m07ed11_sample.pdf (accessed on 24 February 2021).
- Konaté, K.; Mavoungou, J.F.; Lepengué, A.N.; Aworet-Samseny, R.R.R.; Hilou, A.; Souza, A.; Dicko, M.H.; M’Batchi, B. Antibacterial activity against β- lactamase producing Methicillin and Ampicillin-resistants Staphylococcus aureus: Fractional Inhibitory Concentration Index (FICI) determination. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 1–12. [Google Scholar] [CrossRef]
- Kavanagh, A.; Ramu, S.; Gong, Y.; Copper, M.A.; Blaskovich, M.A.T. Effects of microplate type and broth additives on microdilution MIC susceptibility assays. Antimicrob. Agents Chemother. 2019, 63, 1–17. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, K.S. Methods for in vitro evaluating antimicrobial activity. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Thakur, D.; Sahani, K. In vitro antimicrobial activity and MIC of the extracellular ethyl acetate crude extract of endophytic fungi Fusarium sp. isolated from Tephrosia purpurea root. Int. J. Pharm. Pharm. Sci. 2019, 11, 48–53. [Google Scholar] [CrossRef]
- Osaka, I.; Hefty, P.S. Simple resazurin-based microplate assay for measuring chlamydia infections. Antimicrob. Agents Chemother. 2013, 57, 2838–2840. [Google Scholar] [CrossRef]
- Hunter Lindsey, R.; Bromberg, K.D.; Felix, C.A.; Osheroff, N. 1,4-Benzoquinone is a topoisomerase II poison. Biochemistry 2004, 43, 7563–7574. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, M.; Xavier, A.S. Bedaquiline—The first ATP synthase inhibitor against multi drug resistant tuberculosis. J. Young Pharm. 2013, 5, 112–115. [Google Scholar] [CrossRef]
- SwissTargetPrediction. Available online: http://www.swisstargetprediction.ch/ (accessed on 4 December 2020).
- Wang, L.; Wu, D.; Robinson, C.V.; Wu, H.; Fu, T.M. Structures of a complete human V-ATPase reveal mechanisms of its assembly. Mol. Cell. 2020, 80, 501–5011. [Google Scholar] [CrossRef]
- Laponogov, I.; Pan, X.S.; Veselkov, D.A.; McAuley, K.E.; Fisher, L.M.; Sanderson, M.R. Structural basis of gate-DNA breakage and resealing by Type II Topoisomerases. PLoS ONE 2010, 5, e11338. [Google Scholar] [CrossRef]
- Dos Santos Maia, M.; Soares Rodrigues, G.C.; de Sousa, N.F.; Scotti, M.T.; Scotti, L.; Mendonça-Junior, F.J.B. Identification of new targets and the virtual screening of lignans against Alzheimer’s disease. Oxid. Med. Cell. Longev. 2020, 2020, 3098673. [Google Scholar] [CrossRef] [PubMed]
- Levy, Y.; Wolynes, P.G.; Onuchic, J.N. Protein topology determines binding mechanism. Proc. Natl. Acad. Sci. USA 2004, 101, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef]
- Rollinger, J.M.; Stuppner, H.; Langer, T. Virtual screening for the discovery of bioactive natural products. In Natural Compounds as Drugs Volume I; Progress in Drug Research; Springer: Berlin/Heidelberg, Germany, 2008; Volume 65, pp. 211–249. [Google Scholar]
- Bohacek, R.S.; McMartin, C.; Guida, W.C. The art and practice of structure-based drug design: A molecular modeling perspective. Med. Res. Rev. 1999, 16, 3–50. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).