Nanopore Sequencing Technology as an Emerging Tool for Diversity Studies of Plant Organellar Genomes
Abstract
1. Introduction
2. Challenges, Limitations, and Recent Improvements in Nanopore Sequencing Technology
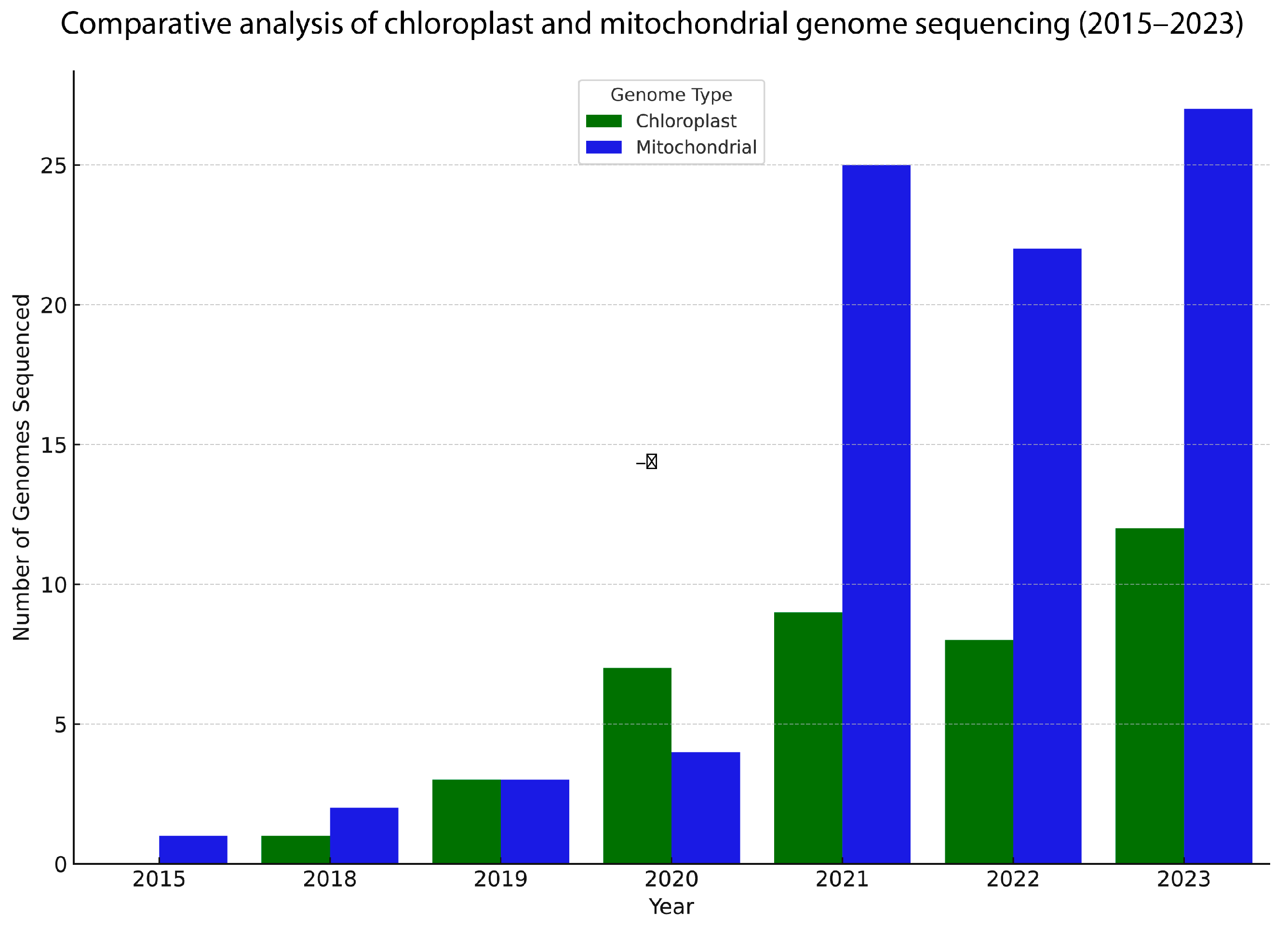
3. Organellar Genomes Sequenced Using Nanopore Technology
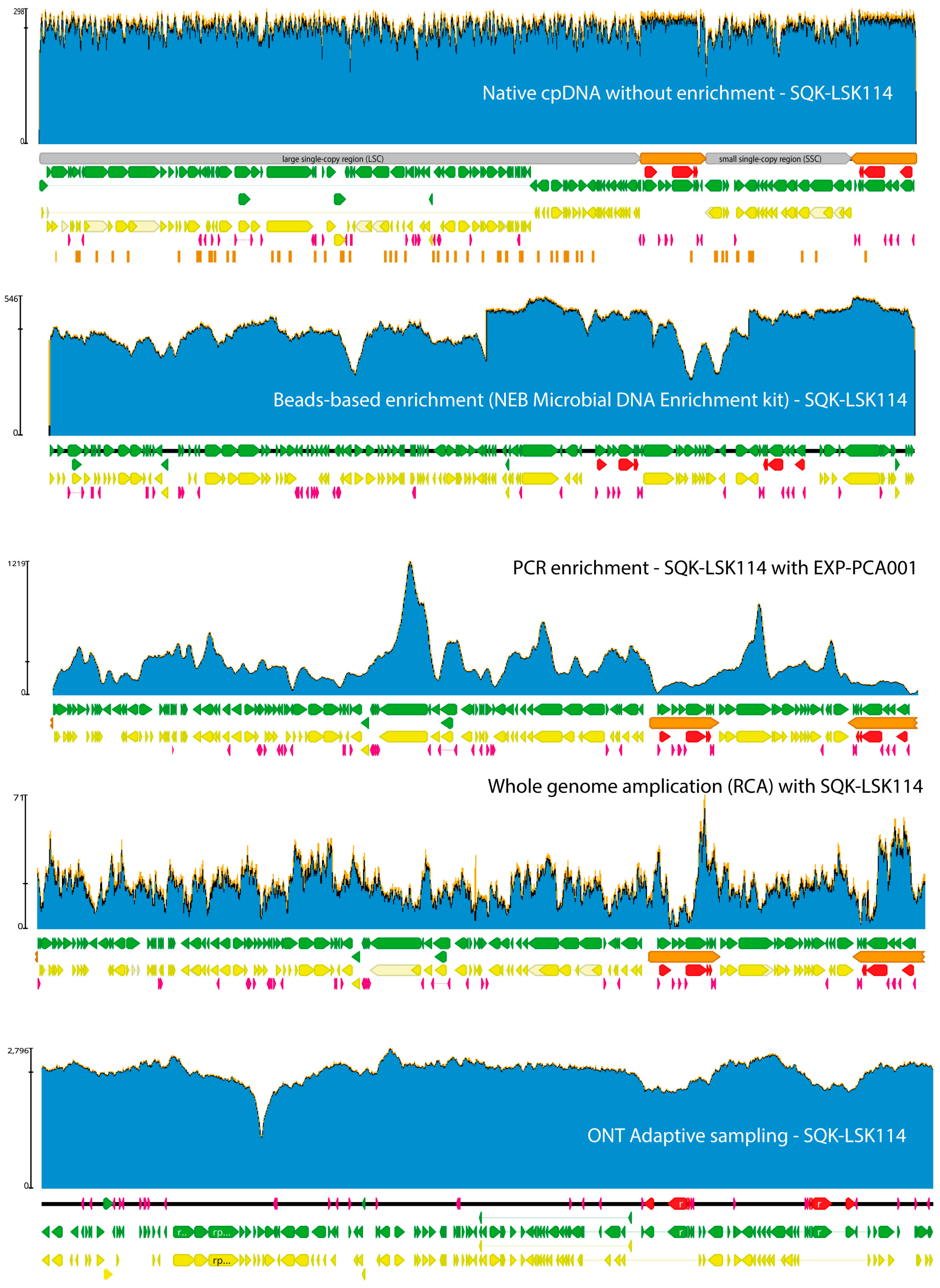
4. Plastid and Mitochondrial DNA Extraction and Enrichment
5. Dedicated Long-Reads Assemblers for Organellar Genomes
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Gray, M.W. The Endosymbiont Hypothesis Revisited. Int. Rev. Cytol. 1992, 141, 233–357. [Google Scholar] [CrossRef] [PubMed]
- Reconstructing Evolution: Gene Transfer from Plastids to the Nucleus—Bock—2008—BioEssays—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/10.1002/bies.20761 (accessed on 1 January 2024).
- Palmer, J.D.; Herbon, L.A. Plant Mitochondrial DNA Evolves Rapidly in Structure, but Slowly in Sequence. J. Mol. Evol. 1988, 28, 87–97. [Google Scholar] [CrossRef]
- Mower, J.P.; Stefanović, S.; Hao, W.; Gummow, J.S.; Jain, K.; Ahmed, D.; Palmer, J.D. Horizontal Acquisition of Multiple Mitochondrial Genes from a Parasitic Plant Followed by Gene Conversion with Host Mitochondrial Genes. BMC Biol. 2010, 8, 150. [Google Scholar] [CrossRef] [PubMed]
- Knoop, V. When You Can’t Trust the DNA: RNA Editing Changes Transcript Sequences. Cell. Mol. Life Sci. 2011, 68, 567–586. [Google Scholar] [CrossRef] [PubMed]
- Green, B.R. Chloroplast Genomes of Photosynthetic Eukaryotes. Plant J. 2011, 66, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; dePamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 Genes from 64 Plastid Genomes Resolves Relationships in Angiosperms and Identifies Genome-Scale Evolutionary Patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- The First 50 Plant Genomes—Michael—2013—The Plant Genome—Wiley Online Library. Available online: https://acsess.onlinelibrary.wiley.com/doi/10.3835/plantgenome2013.03.0001in (accessed on 14 January 2024).
- Sugiura, M. The Chloroplast Genome. Plant Mol. Biol. 1992, 19, 149–168. [Google Scholar] [CrossRef]
- Smith, D.R. Plastid Genomes Hit the Big Time. New Phytol. 2018, 219, 491–495. [Google Scholar] [CrossRef]
- Wang, W.; Lanfear, R. Long-Reads Reveal That the Chloroplast Genome Exists in Two Distinct Versions in Most Plants. Genome Biol. Evol. 2019, 11, 3372–3381. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Prudnikow, L.; Pannicke, B.; Wünschiers, R. A Primer on Pollen Assignment by Nanopore-Based DNA Sequencing. Front. Ecol. Evol. 2023, 11, 1112929. [Google Scholar] [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore Sequencing and Assembly of a Human Genome with Ultra-Long Reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of Neural Network Basecalling Tools for Oxford Nanopore Sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and Challenges in Long-Read Sequencing Data Analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Samarakoon, H.; Ferguson, J.M.; Gamaarachchi, H.; Deveson, I.W. Accelerated Nanopore Basecalling with SLOW5 Data Format. Bioinformatics 2023, 39, btad352. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.J. Progress in Plant Genome Sequencing. Appl. Biosci. 2022, 1, 113–128. [Google Scholar] [CrossRef]
- Pucker, B.; Irisarri, I.; de Vries, J.; Xu, B. Plant Genome Sequence Assembly in the Era of Long Reads: Progress, Challenges and Future Directions. Quant. Plant Biol. 2022, 3, e5. [Google Scholar] [CrossRef]
- Sanderson, N.D.; Kapel, N.; Rodger, G.; Webster, H.; Lipworth, S.; Street, T.L.; Peto, T.; Crook, D.; Stoesser, N. Comparison of R9.4.1/Kit10 and R10/Kit12 Oxford Nanopore Flowcells and Chemistries in Bacterial Genome Reconstruction. Microb. Genom. 2023, 9, 000910. [Google Scholar] [CrossRef]
- Ni, Y.; Liu, X.; Simeneh, Z.M.; Yang, M.; Li, R. Benchmarking of Nanopore R10.4 and R9.4.1 Flow Cells in Single-Cell Whole-Genome Amplification and Whole-Genome Shotgun Sequencing. Comput. Struct. Biotechnol. J. 2023, 21, 2352–2364. [Google Scholar] [CrossRef]
- Sawicki, J.; Krawczyk, K.; Kurzyński, M.; Maździarz, M.; Paukszto, Ł.; Sulima, P.; Szczecińska, M. Nanopore Sequencing of Organellar Genomes Revealed Heteroplasmy in Simple Thalloid and Leafy Liverworts. Acta Soc. Bot. Pol. 2023, 92, 172516. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The Alternative Reality of Plant Mitochondrial DNA: One Ring Does Not Rule Them All. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef] [PubMed]
- Wynn, E.L.; Christensen, A.C. Repeats of Unusual Size in Plant Mitochondrial Genomes: Identification, Incidence and Evolution. G3 GenesGenomesGenetics 2019, 9, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Szandar, K.; Krawczyk, K.; Myszczyński, K.; Ślipiko, M.; Sawicki, J.; Szczecińska, M. Breaking the Limits—Multichromosomal Structure of an Early Eudicot Pulsatilla Patens Mitogenome Reveals Extensive RNA-Editing, Longest Repeats and Chloroplast Derived Regions among Sequenced Land Plant Mitogenomes. BMC Plant Biol. 2022, 22, 109. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Zhu, W.; Sloan, D.B.; Wu, Z. Long-Read Sequencing Characterizes Mitochondrial and Plastid Genome Variants in Arabidopsis Msh1 Mutants. Plant J. Cell Mol. Biol. 2022, 112, 738–755. [Google Scholar] [CrossRef]
- Masutani, B.; Arimura, S.; Morishita, S. Investigating the Mitochondrial Genomic Landscape of Arabidopsis Thaliana by Long-Read Sequencing. PLoS Comput. Biol. 2021, 17, e1008597. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, J.; Bączkiewicz, A.; Buczkowska, K.; Górski, P.; Krawczyk, K.; Mizia, P.; Myszczyński, K.; Ślipiko, M.; Szczecińska, M. The Increase of Simple Sequence Repeats during Diversification of Marchantiidae, An Early Land Plant Lineage, Leads to the First Known Expansion of Inverted Repeats in the Evolutionarily-Stable Structure of Liverwort Plastomes. Genes 2020, 11, 299. [Google Scholar] [CrossRef]
- Liu, J.; Ni, Y.; Liu, C. Polymeric Structure of the Cannabis Sativa L. Mitochondrial Genome Identified with an Assembly Graph Model. Gene 2023, 853, 147081. [Google Scholar] [CrossRef]
- Wang, W.; Schalamun, M.; Morales-Suarez, A.; Kainer, D.; Schwessinger, B.; Lanfear, R. Assembly of Chloroplast Genomes with Long- and Short-Read Data: A Comparison of Approaches Using Eucalyptus Pauciflora as a Test Case. BMC Genom. 2018, 19, 977. [Google Scholar] [CrossRef]
- Scheunert, A.; Dorfner, M.; Lingl, T.; Oberprieler, C. Can We Use It? On the Utility of de Novo and Reference-Based Assembly of Nanopore Data for Plant Plastome Sequencing. PLoS ONE 2020, 15, e0226234. [Google Scholar] [CrossRef]
- The First Genome for the Cape Primrose Streptocarpus Rexii (Gesneriaceae), a Model Plant for Studying Meristem-Driven Shoot Diversity—Nishii—2022—Plant Direct—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/10.1002/pld3.388 (accessed on 1 January 2024).
- Plášek, V.; Sawicki, J.; Seppelt, R.D.; Cave, L.H. Orthotrichum Cupulatum Hoffm. Ex Brid. Var. Lithophilum, a New Variety of Epilithic Bristle Moss from Tasmania. Acta Soc. Bot. Pol. 2023, 92, 1–8. [Google Scholar] [CrossRef]
- Ciborowski, K.; Skierkowski, B.; Żukowska, K.; Krawczyk, K.; Sawicki, J. Nanopore Sequencing of Chloroplast Genome of Scapania Undulata (L.) Dumort., 1835 (Scapaniaceae, Jungermanniales). Mitochondrial DNA Part B Resour. 2022, 7, 1424–1426. [Google Scholar] [CrossRef]
- López, K.E.R.; Armijos, C.E.; Parra, M.; Torres, M.d.L. The First Complete Chloroplast Genome Sequence of Mortiño (Vaccinium floribundum) and Comparative Analyses with Other Vaccinium Species. Horticulturae 2023, 9, 302. Available online: https://www.mdpi.com/2311-7524/9/3/302 (accessed on 15 January 2024). [CrossRef]
- Fulton, T.M.; Chunwongse, J.; Tanksley, S.D. Microprep Protocol for Extraction of DNA from Tomato and Other Herbaceous Plants. Plant Mol. Biol. Rep. 1995, 13, 207–209. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Russo, A.; Mayjonade, B.; Frei, D.; Potente, G.; Kellenberger, R.T.; Frachon, L.; Copetti, D.; Studer, B.; Frey, J.E.; Grossniklaus, U.; et al. Low-Input High-Molecular-Weight DNA Extraction for Long-Read Sequencing From Plants of Diverse Families. Front. Plant Sci. 2022, 13, 1494. [Google Scholar] [CrossRef]
- Kang, M.; Chanderbali, A.; Lee, S.; Soltis, D.E.; Soltis, P.S.; Kim, S. High-Molecular-Weight DNA Extraction for Long-Read Sequencing of Plant Genomes: An Optimization of Standard Methods. Appl. Plant Sci. 2023, 11, e11528. [Google Scholar] [CrossRef]
- Bock, R. The Give-and-Take of DNA: Horizontal Gene Transfer in Plants. Trends Plant Sci. 2010, 15, 11–22. [Google Scholar] [CrossRef]
- Smith, D.R.; Keeling, P.J. Mitochondrial and Plastid Genome Architecture: Reoccurring Themes, but Significant Differences at the Extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef] [PubMed]
- Bensasson, D.; Zhang, D.-X.; Hartl, D.L.; Hewitt, G.M. Mitochondrial Pseudogenes: Evolution’s Misplaced Witnesses. Trends Ecol. Evol. 2001, 16, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid Evolution of Enormous, Multichromosomal Genomes in Flowering Plant Mitochondria with Exceptionally High Mutation Rates. PLoS Biol. 2012, 10, e1001241. [Google Scholar] [CrossRef]
- Mariac, C.; Scarcelli, N.; Pouzadou, J.; Barnaud, A.; Billot, C.; Faye, A.; Kougbeadjo, A.; Maillol, V.; Martin, G.; Sabot, F.; et al. Cost-effective Enrichment Hybridization Capture of Chloroplast Genomes at Deep Multiplexing Levels for Population Genetics and Phylogeography Studies. Mol. Ecol. Resour. 2014, 14, 1103–1113. [Google Scholar] [CrossRef]
- Wang, J.; Mu, W.; Yang, T.; Song, Y.; Hou, Y.G.; Wang, Y.; Gao, Z.; Liu, X.; Liu, H.; Zhao, H. Targeted Enrichment of Novel Chloroplast-Based Probes Reveals a Large-Scale Phylogeny of 412 Bamboos. BMC Plant Biol. 2021, 21, 76. [Google Scholar] [CrossRef]
- Uribe-Convers, S.; Duke, J.R.; Moore, M.J.; Tank, D.C. A Long PCR–Based Approach for DNA Enrichment Prior to next-Generation Sequencing for Systematic Studies. Appl. Plant Sci. 2014, 2, 1300063. [Google Scholar] [CrossRef]
- Goremykin, V.V.; Viola, R.; Hellwig, F.H. Removal of Noisy Characters from Chloroplast Genome-Scale Data Suggests Revision of Phylogenetic Placements of Amborella and Ceratophyllum. J. Mol. Evol. 2009, 68, 197–204. [Google Scholar] [CrossRef]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative Chloroplast Genomics: Analyses Including New Sequences from the Angiosperms Nuphar Advena and Ranunculus Macranthus. BMC Genomics 2007, 8, 174. [Google Scholar] [CrossRef]
- Potapov, V.; Ong, L.J. Examining Sources of Error in PCR by Single-Molecule Sequencing. PLoS ONE 2017, 12, e0169774. [Google Scholar] [CrossRef] [PubMed]
- Acinas, G.A.; Sarma-Rupavtarm, R.; Klepac-Ceraj, V.; Polz, M.F. PCR-Induced Sequence Artifacts and Bias: Insights from Comparison of Two 16S rRNA Clone Libraries Constructed from the Same Sample. App. Environ. Microb. 2005, 71, 8966–8969. [Google Scholar] [CrossRef]
- Yigit, E.; Hernandez, D.I.; Trujillo, J.T.; Dimalanta, E.; Bailey, C.D. Genome and Metagenome Sequencing: Using the Human Methyl-Binding Domain to Partition Genomic DNA Derived from Plant Tissues. Appl. Plant Sci. 2014, 2, 1400064. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.; Holmes, N.; Clarke, T.; Munro, R.; Debebe, B.J.; Loose, M. Readfish Enables Targeted Nanopore Sequencing of Gigabase-Sized Genomes. Nat. Biotechnol. 2021, 39, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Loose, M.; Malla, S.; Stout, M. Real-Time Selective Sequencing Using Nanopore Technology. Nat. Methods 2016, 13, 751–754. [Google Scholar] [CrossRef]
- Wanner, N.; Larsen, P.A.; McLain, A.; Faulk, C. The Mitochondrial Genome and Epigenome of the Golden Lion Tamarin from Fecal DNA Using Nanopore Adaptive Sequencing. BMC Genom. 2021, 22, 726. [Google Scholar] [CrossRef] [PubMed]
- Gan, M.; Wu, B.; Yan, G.; Li, G.; Sun, L.; Lu, G.; Zhou, W. Combined Nanopore Adaptive Sequencing and Enzyme-Based Host Depletion Efficiently Enriched Microbial Sequences and Identified Missing Respiratory Pathogens. BMC Genom. 2021, 22, 732. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Heavens, D.; Lan, Y.; Horsfield, S.; Clark, M.D.; Leggett, R.M. Nanopore Adaptive Sampling: A Tool for Enrichment of Low Abundance Species in Metagenomic Samples. Genome Biol. 2022, 23, 11. [Google Scholar] [CrossRef] [PubMed]
- Kipp, E.J.; Lindsey, L.L.; Milstein, M.S.; Blanco, C.M.; Baker, J.P.; Faulk, C.; Oliver, J.D.; Larsen, P.A. Nanopore Adaptive Sampling for Targeted Mitochondrial Genome Sequencing and Bloodmeal Identification in Hematophagous Insects. Parasit. Vectors 2023, 16, 68. [Google Scholar] [CrossRef] [PubMed]
- Weilguny, L.; De Maio, N.; Munro, R.; Manser, C.; Birney, E.; Loose, M.; Goldman, N. Dynamic, Adaptive Sampling during Nanopore Sequencing Using Bayesian Experimental Design. Nat. Biotechnol. 2023, 41, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- Soorni, A.; Haak, D.; Zaitlin, D.; Bombarely, A. Organelle_PBA, a Pipeline for Assembling Chloroplast and Mitochondrial Genomes from PacBio DNA Sequencing Data. BMC Genom. 2017, 18, 49. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Miyamoto, M.; Motooka, D.; Gotoh, K.; Imai, T.; Yoshitake, K.; Goto, N.; Iida, T.; Yasunaga, T.; Horii, T.; Arakawa, K.; et al. Performance Comparison of Second- and Third-Generation Sequencers Using a Bacterial Genome with Two Chromosomes. BMC Genom. 2014, 15, 699. [Google Scholar] [CrossRef]
- Denisov, G.; Walenz, B.; Halpern, A.L.; Miller, J.; Axelrod, N.; Levy, S.; Sutton, G. Consensus Generation and Variant Detection by Celera Assembler. Bioinformatics 2008, 24, 1035–1040. [Google Scholar] [CrossRef]
- Syme, A.E.; McLay, T.G.B.; Udovicic, F.; Cantrill, D.J.; Murphy, D.J.; McLay, T.G.B.; Udovicic, F.; Cantrill, D.J.; Murphy, D.J. Long-Read Assemblies Reveal Structural Diversity in Genomes of Organelles—An Example with Acacia Pycnantha. Gigabyte 2021, 2021, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Shafin, K.; Pesout, T.; Lorig-Roach, R.; Haukness, M.; Olsen, H.E.; Bosworth, C.; Armstrong, J.; Tigyi, K.; Maurer, N.; Koren, S.; et al. Nanopore Sequencing and the Shasta Toolkit Enable Efficient de Novo Assembly of Eleven Human Genomes. Nat. Biotechnol. 2020, 38, 1044–1053. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-Resolved de Novo Assembly Using Phased Assembly Graphs with Hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xie, P.; Guo, Y.; Zhou, W.; Liu, E.; Yu, Y. Easy353: A Tool to Get Angiosperms353 Genes for Phylogenomic Research. Mol. Biol. Evol. 2022, 39, msac261. [Google Scholar] [CrossRef]
- Zhou, W.; Armijos, C.E.; Lee, C.; Lu, R.; Wang, J.; Ruhlman, T.A.; Jansen, R.K.; Jones, A.M.; Jones, C.D. Plastid Genome Assembly Using Long-Read Data. Mol. Ecol. Resour. 2023, 23, 1442–1457. [Google Scholar] [CrossRef]
- Lee, C.; Ruhlman, T.A.; Jansen, R.K. Unprecedented Intraindividual Structural Heteroplasmy in Eleocharis (Cyperaceae, Poales) Plastomes. Genome Biol. Evol. 2020, 12, 641–655. [Google Scholar] [CrossRef]
- Ruhlman, T.A.; Zhang, J.; Blazier, J.C.; Sabir, J.S.M.; Jansen, R.K. Recombination-Dependent Replication and Gene Conversion Homogenize Repeat Sequences and Diversify Plastid Genome Structure. Am. J. Bot. 2017, 104, 559–572. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawicki, J.; Krawczyk, K.; Paukszto, Ł.; Maździarz, M.; Kurzyński, M.; Szablińska-Piernik, J.; Szczecińska, M. Nanopore Sequencing Technology as an Emerging Tool for Diversity Studies of Plant Organellar Genomes. Diversity 2024, 16, 173. https://doi.org/10.3390/d16030173
Sawicki J, Krawczyk K, Paukszto Ł, Maździarz M, Kurzyński M, Szablińska-Piernik J, Szczecińska M. Nanopore Sequencing Technology as an Emerging Tool for Diversity Studies of Plant Organellar Genomes. Diversity. 2024; 16(3):173. https://doi.org/10.3390/d16030173
Chicago/Turabian StyleSawicki, Jakub, Katarzyna Krawczyk, Łukasz Paukszto, Mateusz Maździarz, Mateusz Kurzyński, Joanna Szablińska-Piernik, and Monika Szczecińska. 2024. "Nanopore Sequencing Technology as an Emerging Tool for Diversity Studies of Plant Organellar Genomes" Diversity 16, no. 3: 173. https://doi.org/10.3390/d16030173
APA StyleSawicki, J., Krawczyk, K., Paukszto, Ł., Maździarz, M., Kurzyński, M., Szablińska-Piernik, J., & Szczecińska, M. (2024). Nanopore Sequencing Technology as an Emerging Tool for Diversity Studies of Plant Organellar Genomes. Diversity, 16(3), 173. https://doi.org/10.3390/d16030173





