The First Complete Chloroplast Genome Sequence of Mortiño (Vaccinium floribundum) and Comparative Analyses with Other Vaccinium Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Genomic DNA Extraction, Library Preparation, and Sequencing
2.3. V. floribundum cp Genome Assembly and Annotation
2.4. SSR Analysis
2.5. Comparison of Vaccinium cp Genomes
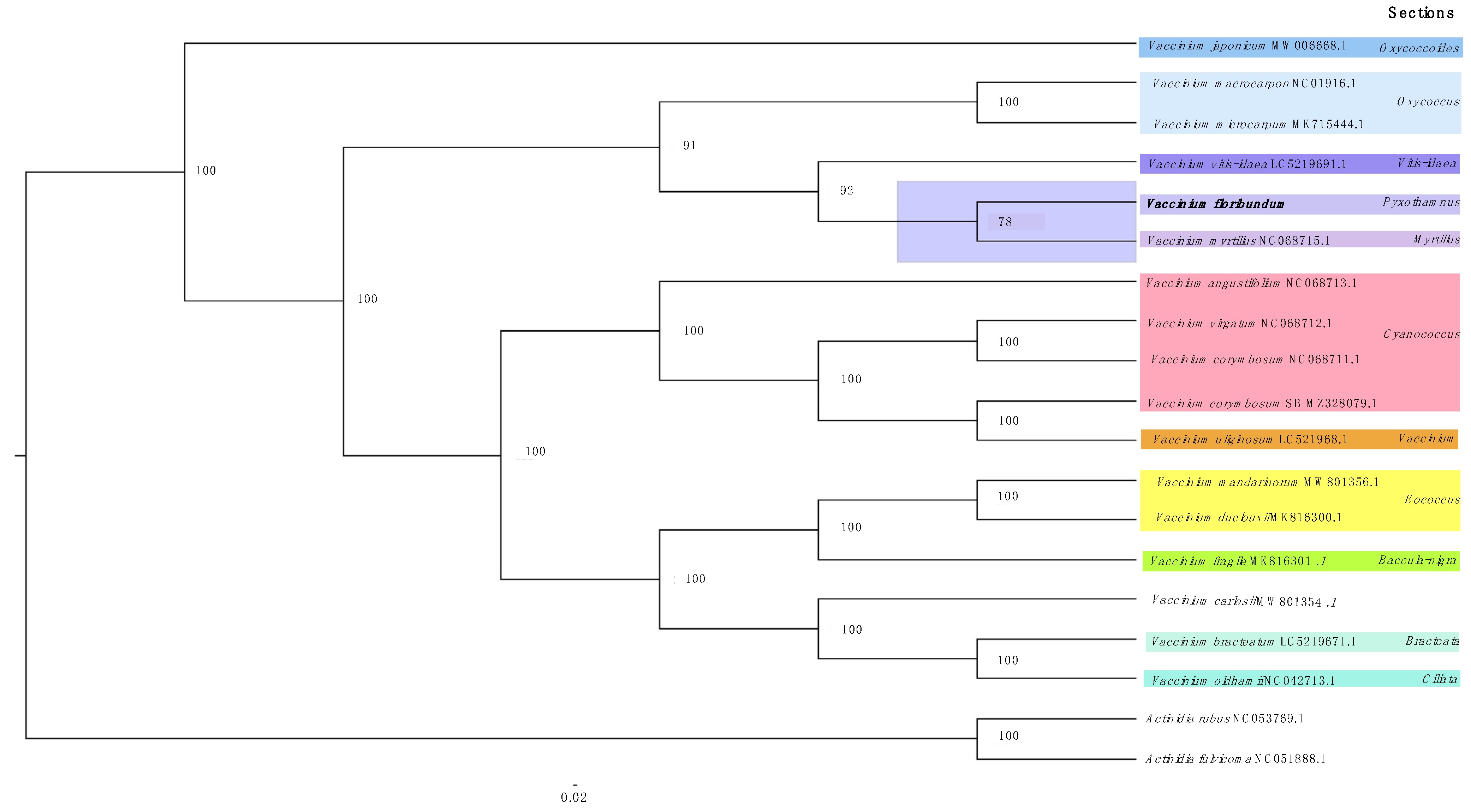
2.6. Phylogenetic Analyses
3. Results
3.1. Vaccinium floribundum cp Genome Sequencing and Assembly
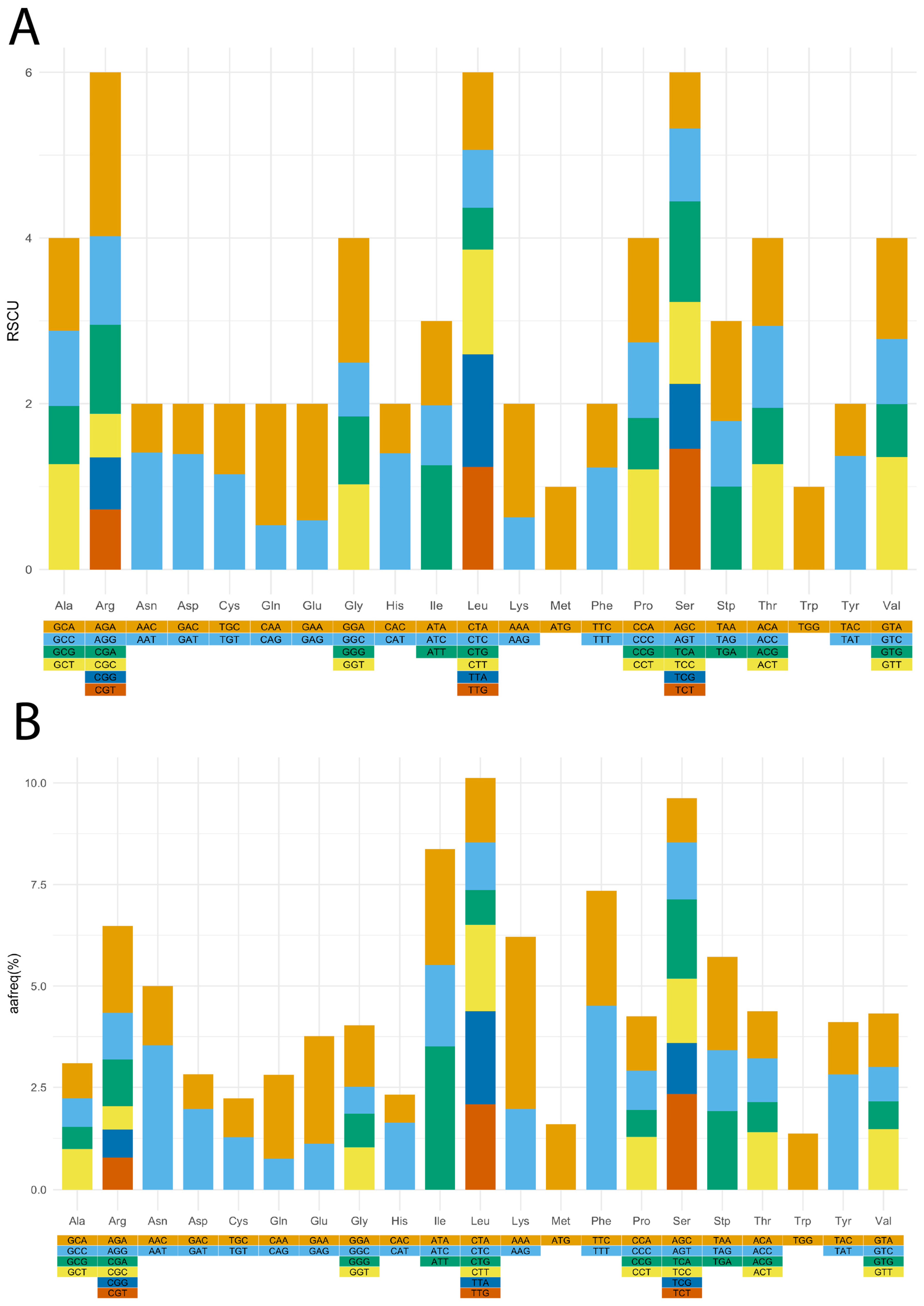
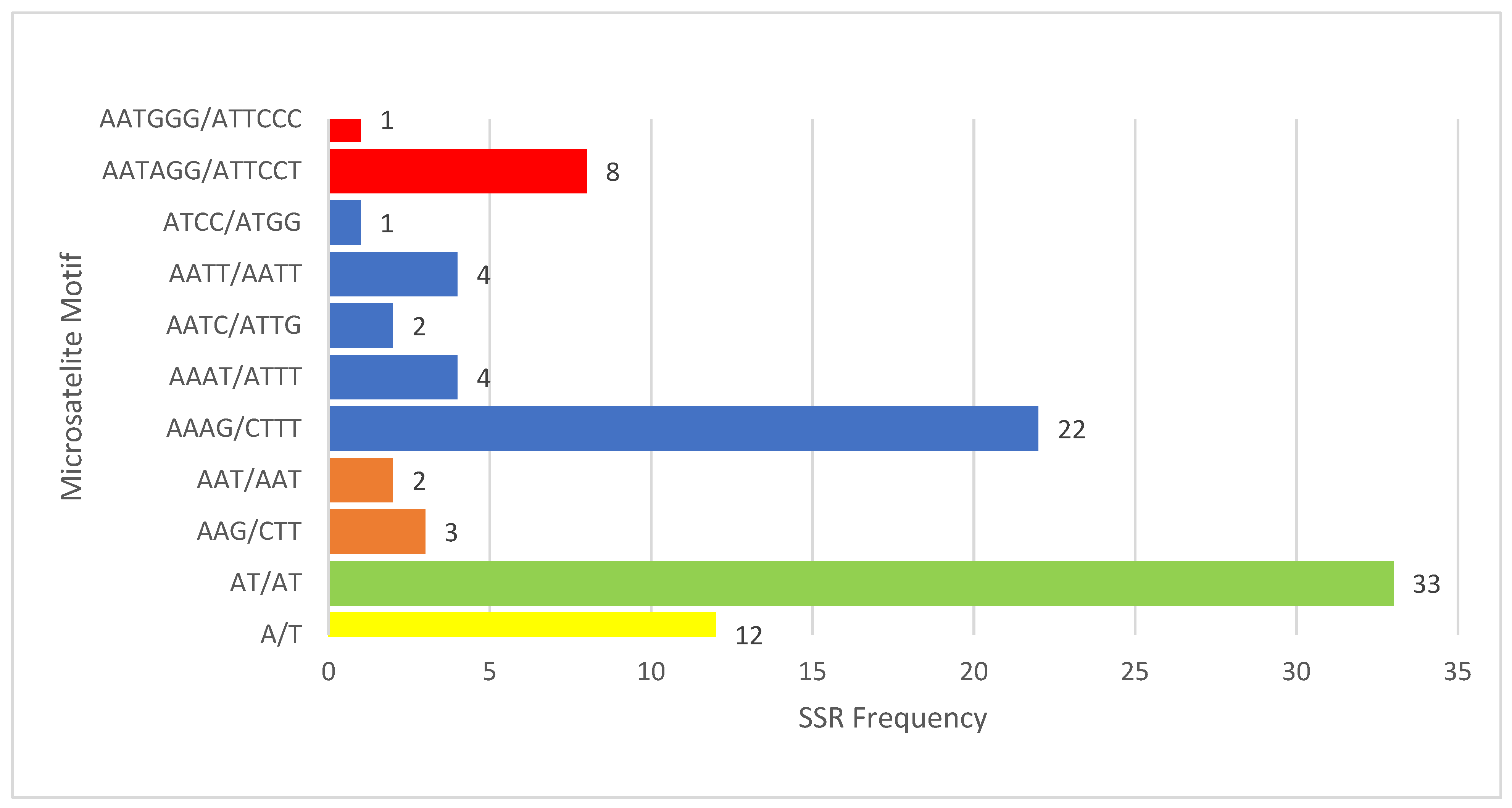
3.2. Relative Synonymous Codon Usage (RSCU), Amino Acid Frequency, and Simple Sequence Repeats (SSR) Identification in V. floribundum cp Genome
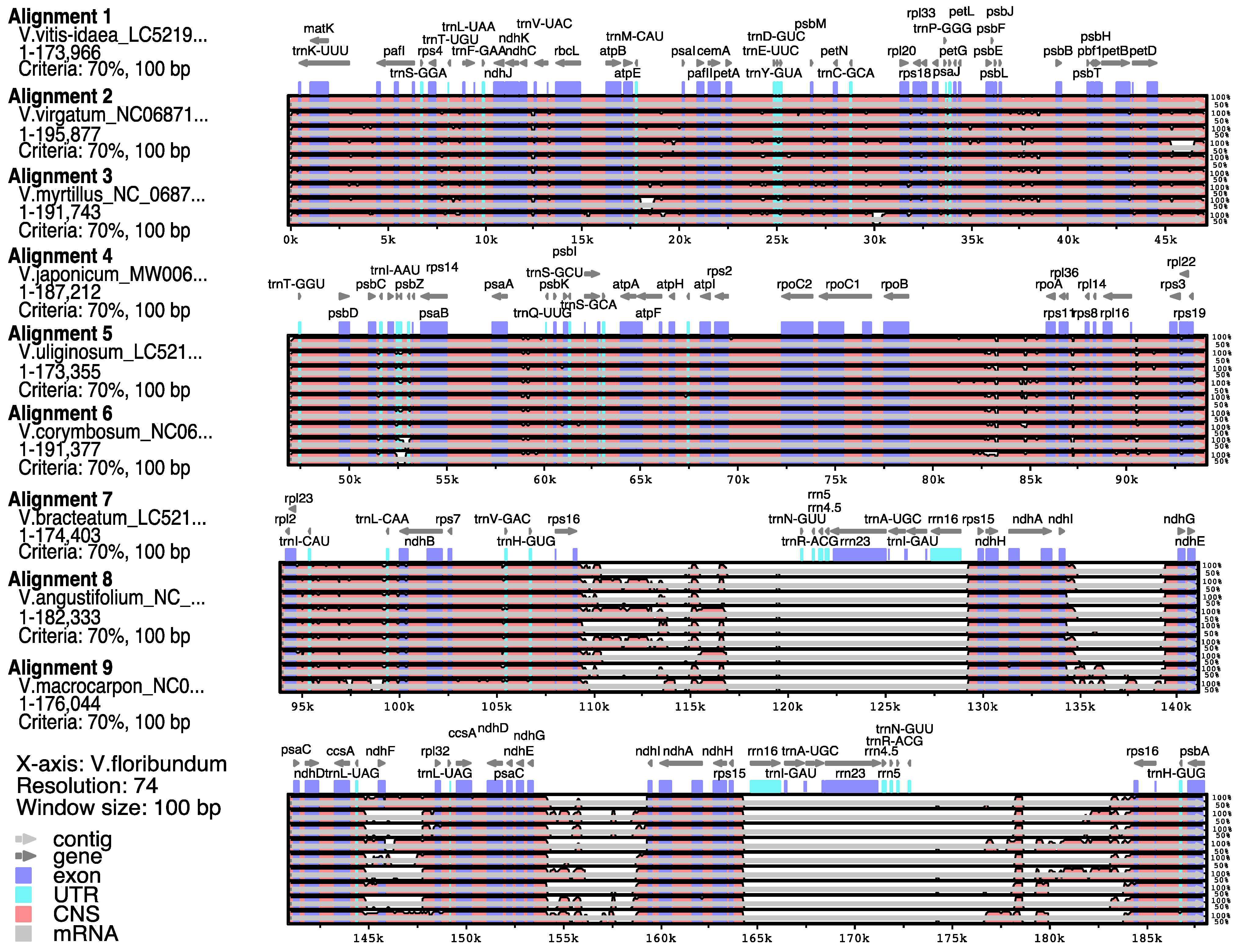
3.3. Comparison of Sequence Divergence between the cp Genome of V. floribundum and Other Vaccinium Species
3.4. Contraction and Expansion Analysis of the V. floribundum cp Genome Borders versus Other Vaccinium Species
3.5. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abreu, O.; Barreto, G.; Prieto, S. Vaccinium (Ericaceae): Ethnobotany and pharmacological potential. Emir. J. Food Agric. 2014, 26, 577–591. [Google Scholar] [CrossRef]
- Tong, Y.-H.; Xia, N.-H. Vaccinium damingshanense sp. nov. (Ericaceae) from Guangxi, China. Nord. J. Bot. 2014, 33, 74–78. [Google Scholar] [CrossRef]
- Meneses, L.; Morillo, L.; Vásquez-Castillo, W. In vitro propagation of Vaccinium floribundum Kunth from seeds: Promissory technology for mortiño accelerated production. Can. J. Plant Sci. 2022, 102, 216–224. [Google Scholar] [CrossRef]
- Fahrenkrog, A.M.; Matsumoto, G.O.; Toth, K.; Jokipii-Lukkari, S.; Salo, H.M.; Häggman, H.; Benevenuto, J.; Munoz, P.R. Chloroplast genome assemblies and comparative analyses of commercially important Vaccinium berry crops. Sci. Rep. 2022, 12, 21600. [Google Scholar] [CrossRef] [PubMed]
- Kron, K.A.; Powell, E.A.; Luteyn, J.L. Phylogenetic relationships within the blueberry tribe (Vaccinieae, Ericaceae) based on sequence data from MATK and nuclear ribosomal ITS regions, with comments on the placement of Satyria. Am. J. Bot. 2002, 89, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Shin, J.; Oh, D.-R.; Kim, D.-W.; Lee, H.-S.; Choi, C. Complete chloroplast genome sequences of Vaccinium bracteatum Thunb., V. vitisidaea L., and V. uliginosum L. (Ericaceae). Mitochondrial DNA Part B 2020, 5, 1843–1844. [Google Scholar] [CrossRef]
- Kim, Y.; Shin, J.; Oh, D.-R.; Kim, A.-Y.; Choi, C. Comparative analysis of complete chloroplast genome sequences and insertion-deletion (indel) polymorphisms to distinguish five Vaccinium species. Forests 2020, 11, 927. [Google Scholar] [CrossRef]
- Schreckinger, M.E.; Wang, J.; Yousef, G.; Lila, M.A.; de Mejia, E.G. Antioxidant capacity and in vitro inhibition of adipogenesis and inflammation by phenolic extracts of Vaccinium floribundum and Aristotelia chilensis. J. Agric. Food Chem. 2010, 58, 8966–8976. [Google Scholar] [CrossRef]
- Coba-Santamaría, P.; Coronel, D.; Verdugo, K.; Paredes, M.; Yugsi, E.; Huachi, L. Ethnobotanical study of the Mortiño (Vaccinium floribundum) as ancestral and potentially functional meal. LA GRANJA Rev. De Cienc. De La Vida 2012, 16, 5–13. [Google Scholar] [CrossRef]
- Cobo, M.M.; Gutiérrez, B.; Torres, A.F.; Torres, M.D.L. Preliminary analysis of the genetic diversity and population structure of mortiño (Vaccinium floribundum Kunth). Biochem. Syst. Ecol. 2016, 64, 14–21. [Google Scholar] [CrossRef]
- Racines-Oliva, A.M.; Hidalgo-Verdezoto, M.R.; Vasquez-Castillo, W.A. Domesticación de Mortiño (Vaccinium floribundum Kunth.): Frutal con gran potencial para la industria alimenticia. Agron. Colomb. 2016, 34, 51–53. [Google Scholar] [CrossRef]
- Pérez, B.P.; Endara, A.B.; Garrido, J.A.; Cárdenas, L.D.L.R. Extraction of anthocyanins from Mortiño (Vaccinium floribundum) and determination of their antioxidant capacity. Rev. Fac. Nac. De Agron. Medellin 2021, 74, 9453–9460. [Google Scholar] [CrossRef]
- Vega-Polo, P.; Cobo, M.M.; Argudo, A.; Gutierrez, B.; Rowntree, J.; Torres, M.D.L. Characterizing the genetic diversity of the Andean blueberry (Vaccinium floribundum Kunth.) across the Ecuadorian highlands. PLoS ONE 2021, 15, e0243420. [Google Scholar] [CrossRef] [PubMed]
- Magnitskiy, S.; Ligarreto, G.; Lancheros, H.O. Rooting of two types of cuttings of fruit crops Vaccinium floribundum Kunth and Disterigma alaternoides (Kunth) Niedenzu (Ericaceae). Agron. Colomb. 2011, 29, 361–371. [Google Scholar]
- Guevara-Terán, M.; Padilla-Arias, K.; Beltrán-Novoa, A.; González-Paramás, A.M.; Giampieri, F.; Battino, M.; Vásquez-Castillo, W.; Fernandez-Soto, P.; Tejera, E.; Alvarez-Suarez, J.M. Influence of altitudes and development stages on the chemical composition, antioxidant, and antimicrobial capacity of the wild andean blueberry (Vaccinium floribundum Kunth). Molecules 2022, 27, 7525. [Google Scholar] [CrossRef]
- Llivisaca, S.; Manzano, P.; Ruales, J.; Flores, J.; Mendoza, J.; Peralta, E.; Cevallos-Cevallos, J.M. Chemical, antimicrobial, and molecular characterization of mortiño (Vaccinium floribundum Kunth) fruits and leaves. Food Sci. Nutr. 2018, 6, 934–942. [Google Scholar] [CrossRef]
- Cerrato, A.; Piovesana, S.; Aita, S.E.; Cavaliere, C.; Felletti, S.; Laganà, A.; Montone, C.M.; Vargas-De-La-Cruz, C.; Capriotti, A.L. Detailed investigation of the composition and transformations of phenolic compounds in fresh and fermented Vaccinium floribundum berry extracts by high-resolution mass spectrometry and bioinformatics. Phytochem. Anal. 2022, 33, 507–516. [Google Scholar] [CrossRef]
- Chiluisa-Utreras, V.; Vela, D.; Vaca, I.; Acurio, R.; Chicaiza, J.; Peñaherrera, S. Expression of the ANS, CHS and DFR genes involved in the biosynthesis of anthocyanins in Vaccinium floribundum Kunth from ecuador, using RT-qPCR. Bionatura 2021, 6, 2180–2186. [Google Scholar] [CrossRef]
- Torres, M.L.; Trujillo, D.; Arahana, V. Cultivo in vitro del mortiño (Vaccinium floribundum Kunth). ACI Av. En Cienc. E Ing. 2010, 2, 9–15. [Google Scholar] [CrossRef]
- García, A.; Ruales, J. Study the effect of pre-treatment of drying ‘mortiño’ (Vaccinium floribundum Kunth) with reference to drying rate and total content of soluble polyphenols and anthocyanins. Rev. Politécnica 2018, 40, 47–57. [Google Scholar]
- Caranqui-Aldaz, J.M.; Romero-Saltos, H.; Hernández, F.; Martínez, R. Reproductive phenology of Vaccinium floribundum Kunth (Ericaceae) and codification according to the BBCH scale based on evidence from the volcano Chimborazo paramo (Ecuador). Sci. Hortic. 2022, 303, 111207. [Google Scholar] [CrossRef]
- Llivisaca-Contreras, S.A.; León-Tamariz, F.; Manzano-Santana, P.; Ruales, J.; Naranjo-Morán, J.; Serrano-Mena, L.; Chica-Martínez, E.; Cevallos-Cevallos, J.M. Mortiño (Vaccinium floribundum Kunth): An underutilized superplant from the andes. Horticulturae 2022, 8, 358. [Google Scholar] [CrossRef]
- Camacho, M. Los páramos ecuatorianos: Caracterización y consideraciones para su conservación y aprovechamiento sostenible. ANALES De La Univ. Cent. Del Ecuad. 2014, 372, 77–92. [Google Scholar] [CrossRef]
- Alarcón-Barrera, K.S.; Armijos-Montesinos, D.S.; García-Tenesaca, M.; Iturralde, G.; Jaramilo-Vivanco, T.; Granda-Albuja, M.G.; Giampieri, F.; Alvarez-Suarez, J.M. Wild Andean blackberry (Rubus glaucus Benth) and Andean blueberry (Vaccinium floribundum Kunth) from the highlands of Ecuador: Nutritional composition and protective effect on human dermal fibroblasts against cytotoxic oxidative damage. J. Berry Res. 2018, 8, 223–236. [Google Scholar] [CrossRef]
- Flores-López, F.; Galaitsi, S.; Escobar, M.; Purkey, D. Modeling of andean páramo ecosystems’ hydrological response to environmental change. Water 2016, 8, 94. [Google Scholar] [CrossRef]
- Kim, S.-C.; Lee, J.-W.; Choi, B.-K. Seven complete chloroplast genomes from Symplocos: Genome organization and comparative analysis. Forests 2021, 12, 608. [Google Scholar] [CrossRef]
- Fajardo, D.; Senalik, D.; Ames, M.; Zhu, H.; Steffan, S.; Harbut, R.; Polashock, J.; Vorsa, N.; Gillespie, E.; Kron, K.; et al. Complete plastid genome sequence of Vaccinium macrocarpon: Structure, gene content, and rearrangements revealed by next generation sequencing. Tree Genet. Genomes 2012, 9, 489–498. [Google Scholar] [CrossRef]
- Szczecińska, M.; Gomolińska, A.; Szkudlarz, P.; Sawicki, J. Plastid and nuclear genomic resources of a relict and endangered plant species: Chamaedaphne calyculata (L.) Moench (Ericaceae). Turk. J. Bot. 2014, 38, 1229–1238. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Q.; Guo, W.; Wei, H.; Wang, J.; Zhu, D.; Tan, Y. The complete chloroplast genome of Vaccinium duclouxii, an endemic species in China. Mitochondrial DNA Part B 2019, 4, 2215–2216. [Google Scholar] [CrossRef]
- Diaz-Garcia, L.; Rodriguez-Bonilla, L.; Rohde, J.; Smith, T.; Zalapa, J. Pacbio sequencing reveals identical organelle genomes between American cranberry (Vaccinium macrocarpon Ait.) and a wild relative. Genes 2019, 10, 291. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Luo, L.; Huang, Y.; Li, G.; Wang, X.; Cheng, T.; Sun, Z.; Wang, F.; Zhang, L.; Li, W. The complete chloroplast genome of Vaccinium fragile (Vacciniaceae), a shrub endemic to China. Mitochondrial DNA Part B 2019, 4, 2310–2311. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-C.; Baek, S.-H.; Lee, J.-W.; Hyun, H.J. Complete chloroplast genome of Vaccinium oldhamii and phylogenetic analysis. Mitochondrial DNA Part B 2019, 4, 902–903. [Google Scholar] [CrossRef]
- Cho, W.-B.; Han, E.-K.; Choi, I.-S.; Son, D.C.; Chung, G.Y.; Lee, J.-H. The complete plastid genome sequence of Vaccinium japonicum (Ericales: Ericaceae), a deciduous broad-leaved shrub endemic to east Asia. Mitochondrial DNA Part B 2021, 6, 1926–1928. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.-R.; Chen, Q.-X.; Niu, J.-Q.; Guo, Y.-P. The complete chloroplast genome of highbush blueberry (Vaccinium corymbosum). Mitochondrial DNA Part B 2021, 7, 87–88. [Google Scholar] [CrossRef] [PubMed]
- Saldaña, C.L.; Chávez-Galarza, J.C.; De la Cruz, G.; Jhoncon, J.H.; Guerrero-Abad, J.C.; Vásquez, H.V.; Maicelo, J.L.; Arbizu, C.I. Revealing the complete chloroplast genome of an Andean horticultural crop, sweet cucumber (Solanum muricatum), and its comparison with other Solanaceae species. Data 2022, 7, 123. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Chaisson, M.J.; Tesler, G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinform. 2012, 13, 238. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Zhou, W.; Armijos, C.E.; Lee, C.; Lu, R.; Wang, J.; Tracey, A.; Jansen, R.K.; Jones, A.M.; Jones, C.D. Plastid genome assembly using long-read data (PtGAUL). bioRxiv 2022, 1–39. [Google Scholar] [CrossRef]
- Firtina, C.; Kim, J.S.; Alser, M.; Cali, D.S.; Cicek, A.E.; Alkan, C.; Mutlu, O. Apollo: A sequencing-technology-independent, scalable and accurate assembly polishing algorithm. Bioinformatics 2020, 36, 3669–3679. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; The R Foundation for Statistical Computing: Indianapolis, IN, USA, 2013; Volume 201, Available online: http://www.R-project.org/ (accessed on 29 November 2022).
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. ProgressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
- Cai, L.; Zhang, H.; Davis, C.C. PhyloHerb: A high-throughput phylogenomic pipeline for processing genome skimming data. Appl. Plant Sci. 2022, 10, 11475. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1.3.1. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 17 December 2022).
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Yao, X.; Tang, P.; Li, Z.; Li, D.; Liu, Y.; Huang, H. The first complete chloroplast genome sequences in Actinidiaceae: Genome structure and comparative analysis. PLoS ONE 2015, 10, e0129347. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D. Comparative organization of chloroplast genomes. Annu. Rev. Genet. 1985, 19, 325–354. [Google Scholar] [CrossRef]
- Logacheva, M.D.; Schelkunov, M.I.; Shtratnikova, V.Y.; Matveeva, M.V.; Penin, A.A. Comparative analysis of plastid genomes of non-photosynthetic Ericaceae and their photosynthetic relatives. Sci. Rep. 2016, 6, 30042. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Alberola, F.; Del Campo, E.M.; Lázaro-Gimeno, D.; Mezquita-Claramonte, S.; Molins, A.; Mateu-Andrés, I.; Pedrola-Monfort, J.; Casano, L.M.; Barreno, E. Balanced gene losses, duplications and intensive rearrangements led to an unusual regularly sized genome in arbutus unedo chloroplasts. PLoS ONE 2013, 8, e79685. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, T.; Zhang, Y.; Li, Y.; Gong, J.; Yi, Y. The complete chloroplast genome of Rhododendron delavayi (Ericaceae). Mitochondrial DNA Part B 2019, 5, 37–38. [Google Scholar] [CrossRef]
- Yang, J.-B.; Tang, M.; Li, H.-T.; Zhang, Z.-R.; Li, D.-Z. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013, 13, 84. [Google Scholar] [CrossRef]
- Könyves, K.; Bilsborrow, J.; Christodoulou, M.D.; Culham, A.; David, J. Comparative plastomics of amaryllidaceae: Inverted repeat expansion and the degradation of the ndh genes in Strumaria truncata Jacq. PeerJ 2021, 9, e12400. [Google Scholar] [CrossRef] [PubMed]
- Endo, T.; Shikanai, T.; Takabayashi, A.; Asada, K.; Sato, F. The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett. 1999, 457, 5–8. [Google Scholar] [CrossRef]
- Sanderson, M.J.; Copetti, D.; Burquez, A.; Bustamante, E.; Charboneau, J.L.M.; Eguiarte, L.E.; Kumar, S.; Lee, H.O.; Lee, J.; McMahon, M.; et al. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): Loss of the ndh gene suite and inverted repeat. Am. J. Bot. 2015, 102, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Omelchenko, D.O.; Krinitsina, A.A.; Belenikin, M.S.; Konorov, E.A.; Kuptsov, S.V.; Logacheva, M.D.; Speranskaya, A.S. Complete plastome sequencing of Allium paradoxum reveals unusual rearrangements and the loss of the ndh genes as compared to Allium ursinum and other onions. Gene 2020, 726, 144154. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.; Lasso, E. Insights into the functional ecology of paramo plants in Colombia. Biotropica 2021, 53, 1415–1431. [Google Scholar] [CrossRef]
- Ramos, C.; Buitrago, S.P.; Pulido, K.L.; Vanegas, L.J. Environmental variability and physiological responses from Polylepis cuadrijuga (Rosaceae) in a fragmented environment in the Páramo de la Rusia (Colombia). Rev. De Biol. Trop. 2013, 61, 11134. [Google Scholar] [CrossRef]
- Madriñán, S.; Cortés, A.J.; Richardson, J.E. Páramo is the world’s fastest evolving and coolest biodiversity hotspot. Front. Genet. 2013, 4, 192. [Google Scholar] [CrossRef]
- Rahbek, C.; Borregaard, M.K.; Colwell, R.K.; Dalsgaard, B.; Holt, B.G.; Morueta-Holme, N.; Nogues-Bravo, D.; Whittaker, R.J.; Fjeldså, J. Humboldt’s enigma: What causes global patterns of mountain biodiversity? Science 2019, 365, 1108–1113. [Google Scholar] [CrossRef]
- Valencia, J.B.; Mesa, J.; León, J.G.; Madriñán, S.; Cortés, A.J. Climate vulnerability assessment of the espeletia complex on páramo sky islands in the northern Andes. Front. Ecol. Evol. 2020, 8, 565708. [Google Scholar] [CrossRef]
- Testolin, R.; Attorre, F.; Borchardt, P.; Brand, R.F.; Bruelheide, H.; Chytrý, M.; De Sanctis, M.; Dolezal, J.; Finckh, M.; Haider, S.; et al. Global patterns and drivers of alpine plant species richness. Glob. Ecol. Biogeogr. 2021, 30, 1218–1231. [Google Scholar] [CrossRef]
- Schley, R.J.; Twyford, A.D.; Pennington, R.T. Hybridization: A ‘double-edged sword’ for neotropical plant diversity. Bot. J. Linn. Soc. 2021, 199, 331–356. [Google Scholar] [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Song, W.; Chen, Z.; Shi, W.; Han, W.; Feng, Q.; Shi, C.; Engel, M.S.; Wang, S. Comparative analysis of complete chloroplast genomes of nine species of Litsea (Lauraceae): Hypervariable regions, positive selection, and phylogenetic relationships. Genes 2022, 13, 1550. [Google Scholar] [CrossRef]
- Singh, N.V.; Patil, P.G.; Sowjanya, R.P.; Parashuram, S.; Natarajan, P.; Babu, K.D.; Pal, R.K.; Sharma, J.; Reddy, U.K. Chloroplast genome sequencing, comparative analysis, and discovery of unique cytoplasmic variants in pomegranate (Punica granatum L.). Front. Genet. 2021, 12, 704075. [Google Scholar] [CrossRef]
- Guo, Y.-Y.; Yang, J.-X.; Bai, M.-Z.; Zhang, G.-Q.; Liu, Z.-J. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 2021, 21, 248. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jiang, M.; Chen, H.; Liu, Y.; Liu, C.; Wu, W. Comparative genome analysis revealed gene inversions, boundary expansions and contractions, and gene loss in the Stemona sessilifolia (Miq.) Miq. chloroplast genome. PLoS ONE 2021, 16, e0247736. [Google Scholar] [CrossRef]
- Yan, M.; Zhao, X.; Zhou, J.; Huo, Y.; Ding, Y.; Yuan, Z. The Complete Chloroplast Genomes of Punica granatum and a Comparison with Other Species in Lythraceae. Int. J. Mol. Sci. 2019, 20, 2886. [Google Scholar] [CrossRef]
- Zhidkin, R.R.; Matveeva, T.V. Phylogeny problems of the genus Vaccinium L. and ways to solve them. Ecol. Genet. 2022, 20, 151–164. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
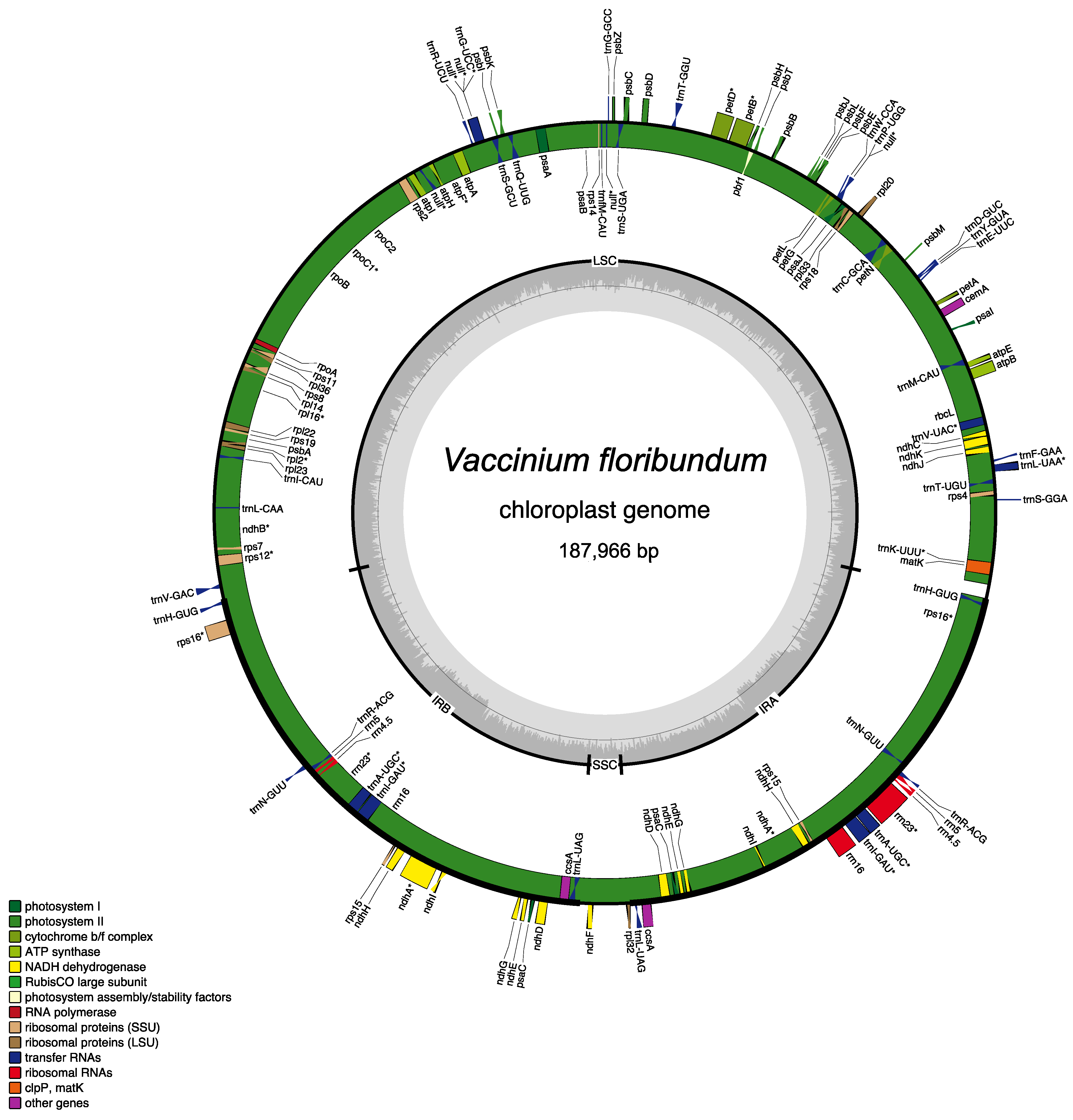


| Whole cp Genome Sequence | |
| Length (bp) | 187,966 |
| GC content (%) | 36.8 |
| Protein Coding Genes (PCGs) 1 | 84 |
| Unique Genes | 113 |
| Genes 1 | 134 |
| Transfer RNA (tRNA) genes 1 | 42 |
| Ribosomal RNA (rRNA) genes 1 | 8 |
| LSC region | |
| Length (bp) | 107,283 |
| PCGs 1 | 62 |
| Genes 1 | 92 |
| tRNA genes 1 | 30 |
| rRNA genes 1 | 0 |
| GC content (%) | 35.9 |
| IR regions | |
| Length (bp) | 38,421 |
| PCGs 1 | 10 |
| Genes 1 | 20 |
| tRNA genes 1 | 6 |
| rRNA genes 1 | 4 |
| GC content (%) | 38.3 |
| SSC region | |
| Length (bp) | 3841 |
| PCGs 1 | 2 |
| Genes 1 | 2 |
| tRNA genes 1 | 0 |
| tRNA genes 1 | 0 |
| GC content (%) | 29.5 |
| Gene Category | Gene Group | Gene Name |
|---|---|---|
| Photosynthesis | Subunits of ATP synthase | atpA, atpB, atpE, atpF2, atpH, atpI |
| Subunits of NADH-dehydrogenase | ndhA2, 3, 5, ndhB2, ndhC, ndhD3, 5, ndhE3, 5, ndhF1, ndhG3, 5, ndhH3, 5, ndhI3, 5, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB2, petD2, petG, petL, petN | |
| Subunits of photosystem I | psaA, psaB, psaC3, 5, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbT, psbZ | |
| Subunit of rubisco | rbcL | |
| rRNA | rrn163, 5, rrn232, 3, 5, rrn4.53, 5, rrn53, 5 | |
| tRNA | trnA-UGC2, 3, 5, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU3, 4, trnG-GCC, trnG-UCC2, trnH-GUG3, 5, trnI-AAU, trnI-CAU, trnI-GAU2, 3, 5, trnK-UUU2, trnL-CAA, trnL-UAA2, trnL-UAG3, 5, trnM-CAU, trnN-GUU3, 5, trnP-GGG, trnP-UGG, trnQ-UUG, trnR-ACG3, 5, trnR-UCU, trnS-CGA2, trnS-GCA2, trnS-GCU, trnS-GGA, trnS-UGA, trnT-CGU2, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC2, trnW-CCA, trnY-GUA | |
| Self-replication | The large subunit of the ribosome (LSU) | rpl14, rpl162, rpl2, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| The small subunit of the ribosome (SSU) | rpoA, rpoB, rpoC12, rpoC2 | |
| DNA-dependent RNA polymerase | rps11, rps12, rps14, rps153, 5, rps162, 3, 5, rps18, rps19, rps2, rps3, rps4, rps7, rps8 | |
| Other genes | c-type cytochrome synthesis | ccsA3, 5 |
| Envelop membrane protein | cemA | |
| Unknown | Hypothetical cp reading frame | pbfI (psbN), ycf2, ycf3 (pafI) 2, ycf4 (pafII) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López, K.E.R.; Armijos, C.E.; Parra, M.; Torres, M.d.L. The First Complete Chloroplast Genome Sequence of Mortiño (Vaccinium floribundum) and Comparative Analyses with Other Vaccinium Species. Horticulturae 2023, 9, 302. https://doi.org/10.3390/horticulturae9030302
López KER, Armijos CE, Parra M, Torres MdL. The First Complete Chloroplast Genome Sequence of Mortiño (Vaccinium floribundum) and Comparative Analyses with Other Vaccinium Species. Horticulturae. 2023; 9(3):302. https://doi.org/10.3390/horticulturae9030302
Chicago/Turabian StyleLópez, Karla E. Rojas, Carolina E. Armijos, Manuela Parra, and María de Lourdes Torres. 2023. "The First Complete Chloroplast Genome Sequence of Mortiño (Vaccinium floribundum) and Comparative Analyses with Other Vaccinium Species" Horticulturae 9, no. 3: 302. https://doi.org/10.3390/horticulturae9030302
APA StyleLópez, K. E. R., Armijos, C. E., Parra, M., & Torres, M. d. L. (2023). The First Complete Chloroplast Genome Sequence of Mortiño (Vaccinium floribundum) and Comparative Analyses with Other Vaccinium Species. Horticulturae, 9(3), 302. https://doi.org/10.3390/horticulturae9030302






