Abstract
Although most Myxozoa species of the genera Ceratomyxa and Ellipsomyxa have been described in marine hosts worldwide, an increasing diversity has been reported infecting South American freshwater fish, mainly in Amazonian waters. The present study deals with two species of myxozoan ceratomyxids parasitizing the gallbladder of Amazonian ornamental cichlids fish: Ceratomyxa amazonensis is identified from a new host—Geophagus altifrons; while Ellipsomyxa santarenensis n. sp. is described infecting Satanoperca jurupari. Morphological (light microscopy and transmission electron microscopy), molecular (small ribosomal subunit DNA—SSU-rDNA sequencing) and phylogenetic analyses were used to characterize both species. Ceratomyxa amazonensis showed a prevalence of 64.2%, with plasmodia showing a vermiform shape and motility. For E. santarenensis n. sp., the prevalence was 33.3%. Ultrastructural analysis revealed that the vermiform C. amazonensis plasmodia were composed of an outer cytoplasmic region and a large vacuole occupying the inner area. In E. santarenensis n. sp., cytoplasmic expansions were observed in pseudoplasmodia originating pseudopodia. SSU rDNA sequencing-based genetic distance analysis revealed a very small difference between C. amazonensis, parasite of G. altifrons, and C. amazonensis, parasite of S. discus—host of the original description, thus showing that they are the same species occurring in a new host. For Ellipsomyxa santarenensis n. sp., molecular data revealed a difference of 1.6% for Ellipsomyxa amazonensis and Ellipsomyxa paraensis. The phylogenetic analysis revealed the grouping of E. santarenensis n. sp. together with the other freshwater Ellipsomyxa species of the Amazonian region, and associated with the morphological data, it was possible to identify it as a new taxon within the genus Ellipsomyxa.
1. Introduction
Myxozoa Grassé, 1970 is a subphylum of endoparasite cnidarians comprising the Class Malacosporea Canning, Curry, Feist, Longshaw & Okamura, 2000 and the Class Myxosporea Bütschli, 1881 [1,2]. The myxosporean family Ceratomyxidae Doflein, 1899 contains the genus Ceratomyxa Thélohan, 1892, which is currently composed of approximately 270 species. Most of these species have been isolated from the gallbladders of marine fish, but a surprising diversity has been reported from freshwater hosts in South America [3,4,5,6,7,8,9,10,11,12,13,14]. These freshwater South American Ceratomyxa are morphologically distinct from marine species, having vermiform plasmodia that are uniquely motile [8,9,15]. Ceratomyxidae also harbors the genus Ellipsomyxa Køie, 2003, which is less speciose than Ceratomyxa and, to date, comprises 16 species. Ellipsomyxa spp. have been mainly isolated from the gallbladders of fish caught in marine, brackish and estuarine waters, with some species having recently been described infecting freshwater fish from the Amazon Basin [16,17,18,19].
Cichlids are commonly found in Amazonian waters and are important in the ornamental fish trade [20,21]. In this study, aiming to increase the knowledge about South American myxozoan diversity, two species were isolated from the gallbladder of some cichlid fish commonly found in Amazonian waters. Gross morphological features provided the initial identification of these myxozoans as a species of Ceratomyxa infecting Geophagus altifrons Heckel, 1840, and an Ellipsomyxa species parasitizing Satanoperca jurupari (Heckel, 1840). Herein, further morphological and ultrastructure characteristics of these two myxozoans are described, together with analyses of small ribosomal subunit DNA-SSU rDNA sequences.
2. Materials and Methods
2.1. Field Collections and Parasite Sampling
Fourteen specimens of G. altifrons and six of S. jurupari were captured in waters where the Tapajós and Amazon rivers merge in the municipality of Santarém in Pará State, Brazil (2°23′49.79″ S 54°43′53.33″ W) during October 2021, January 2022 and August 2022. The fish were caught using a gill net and transported alive to the field laboratory. Capture authorization and access to genetic data were authorized by the Brazilian Ministry of the Environment SISBIO n° 67616-2 and SisGen n° A33CB83; the method of euthanasia was approved by the Ethics Committee of the Federal University of São Paulo CEUA n° 6549290920. All organs were examined under a Zeiss Stemi 2000 stereomicroscope. Confirmation of infections was done using a Zeiss Primo Star light microscope at 400×. Infected tissues or body fluids were fixed in 2.5% glutaraldehyde for morphological and ultrastructural analysis and in 100% ethanol for DNA amplicon sequencing.
2.2. Morphological Analyses
Myxozoa fixed in 2.5% glutaraldehyde were examined using a Carl Zeiss Axio Imager A2 differential interference contrast (DIC) microscope equipped with an Axiocam camera. Measurements were made on 30 mature myxospores using AxioVision 4.8 software, following criteria previously described [22,23,24,25,26]. For ultra-structural analysis, the glutaraldehyde fixed materials were washed, post-fixed in 2% OsO4 for 2 h at room temperature, dehydrated in increasing volumes of ethanol + propylene oxide and then embedded in EMbed 812 resin (EMS, Hatfield, PA, USA). After the resin hardened, semi-thin sections (~2 µm) were cut and then stained with toluidine blue. The quality of these sections was assessed by light microscopy. Ultrathin sections were next cut, and these were stained for contrast using uranyl acetate and lead citrate. The stained sections were viewed by transmission electron microscopy (Morgagni 268D) at the Advanced Center for Image Diagnosis (CADI) of the School of Veterinary Medicine and Animal Science of the University of São Paulo-FMVZ/USP.
2.3. DNA-SSU rDNA Sequencing
DNA extraction of parasite-infected bile fluids was performed using the DNeasy® Blood and Tissue Kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. Initial amplification for Ceratomyxa was performed using the universal primers 18E (CTGGTTGATTCTGCCAGT; [27]) and 18R (CTACGCAAACCTTGTTACG; [24]) primers, followed by a second round with Ceratomyxa-specific primers CerSAZ.1f (GTCCCTTCGATCGTAGTACCAC; [15]) and CerSAZ.3r (CTATCCCCACAGCCTGAAAACT; [15]), CerSAZ.4f (GTTGGTTAGTTTCCACGCGAAA; [15]) with 18R. For Ellipsomyxa, amplification was performed with ERIB1 (ACCTGGTTGATCCTGCCAG; [28]) and ERIB10 (CTTCCGCAGGT TCACCTACGG; [28]) primers, followed by a second round with ERIB1 and Ellipso.3r (CCAACAACTGAACCTTGTCATGG; [17]), Ellipso.4f (AACCACTCGTGCATTTAATCGTG; [17]) with ERIB10. Quantification and size estimation were determined by running the PCR products against a 1 Kb Plus DNA Ladder (Thermo Scientific, Carlsbad, CA, USA) through a 1.5% agarose gelin TBE buffer (0.045 M Tris-borate, 0.001 M EDTA pH 8.0). The fragments were stained with Sybr Safe (Thermo Scientific, Carlsbad, CA, USA) and visualized using a MiniBis Pro transilluminator. Amplicons were then purified using the QIAquick PCR Purification kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions and directly sequenced using the same PCR primers at 3.2 pmol plus MC5 (CCTGAGAAACGGCTACCACATCCA, [29]) and MC3 (GATTAGCCTGACAGATCACTCCACGA; [29]), using a BigDye 102 Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) in an ABI 3730 DNA 103 Analyser (Applied Biosystems) at the Central Laboratory of High Performance Technologies (LaCTAD), at the State University of Campinas—UNICAMP.
2.4. Phylogenetic Analyses
Amplicon sequences were aligned with all SSU rDNA sequences of Ellipsomyxa species available plus representative sequences of other genera based on the groupings obtained by Zatti et al. [17] using the default parameters of the Muscle algorithm [30] run on Geneious 7.1.3 [31]. The DNA-SSU rDNA sequences for Buddenbrockia plumatellae KF731698 and Tetracapsuloides bryosalmonae KF731712 were chosen as outgroups. The sequence of Ellipsomyxa tucujuensis MN999871 and Ellipsomyxa ariusi MN892549 generated inconsistent alignments, which led to high genetic differences and long branches in the phylogenetic tree. To avoid any further bias, we performed two phylogenetic analyses, one without these inconsistent sequences, which is presented in the body of the article, and another containing these sequences, which is presented as supplementary data (Figure S1). To assess replacement saturation, the Iss index for the alignment was estimated using the DAMBE 5 software package [32]. The number of base substitutions between sequences per site was calculated. Standard error was estimated using a bootstrap procedure with 2000 replicates. Analyses were conducted using the 2-parameter Kimura model using MEGA11 [33]. The best-fit model for nucleotide evolution in the resulting arrays was determined using the Akaike information criterion in the jModelTest software package [34], with GTR + I + G indicated for the SSU Gene Dataset. Phylogenetic analyses were performed using Bayesian Inference (BI) and Maximum Likelihood Inference (ML) using resources available in the CIPRES Science Gateway [35]. Bayesian analysis employed the following nucleotide substitution model settings for both data sets: lset nst = 6, rates = invariant, ncat = 4, shape = guess, inferrates = yes, and basefreq = empirical. For the Markov Monte Carlo chain (MCMC), chains with 10,000,000 generations were run, saving one tree every 1500 generations. The first 25% of the generations were discarded and the consensus tree (majority rule) was estimated using the remaining topologies, with only nodes with posterior probabilities greater than 90% were considered well supported. Maximum probability (ML) inference was implemented using RAxML [36] with bootstrap support values of 1000 repetitions and only nodes with bootstrap values greater than 70% were considered well supported. Trees were visualized using FigTree v.1.3.1 [37]. The genetic distance between the selected freshwater Ceratomyxa and Ellipssomyxa species was estimated using the p-distance method in MEGA11 with default parameters.
3. Results
Free plasmodia, which morphologically resembled Ceratomyxa, were observed in the gallbladder of G. altifrons (Figure 1, Figure 2 and Figure 3; Video S1). Free plasmodia with the general morphological characteristics of Ellipsomyxa were also observed in the gallbladder of S. jurupari (Figure 4 and Figure 5; Video S2). The prevalence of Ceratomyxa plasmodia in the gallbladder of G. altifrons was 64.2% (9/14), while for Ellipsomyxa, the infection prevalence in S. jurupari was less, only 33.3% (2/6). Morphological and molecular data are provided for each of these myxozoans.
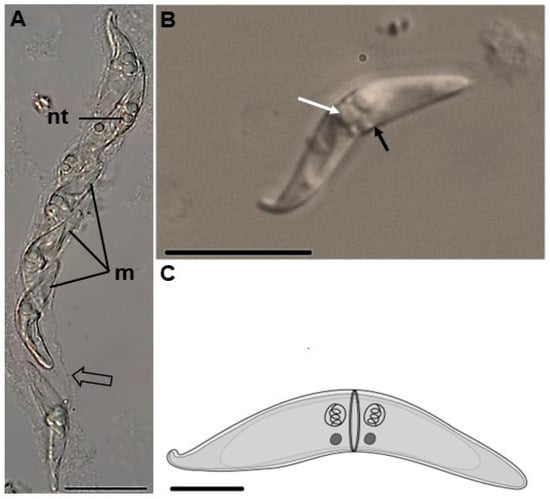
Figure 1.
Ceratomyxa amazonensis parasite of the gallbladder of Geophagus altifrons. (A,B) Differential interference contrast (DIC) photomicrograph. (A) Vermiform plasmodium (arrow) with multiple mature myxospores (m). Note the nematocysts (nt). Bar: 20 µm. (B) Mature spore showing nematocysts (white arrow) and a sutural line (black arrow). Bar: 20 µm. (C) Schematic representation of the myxospore. Bar: 10 µm.
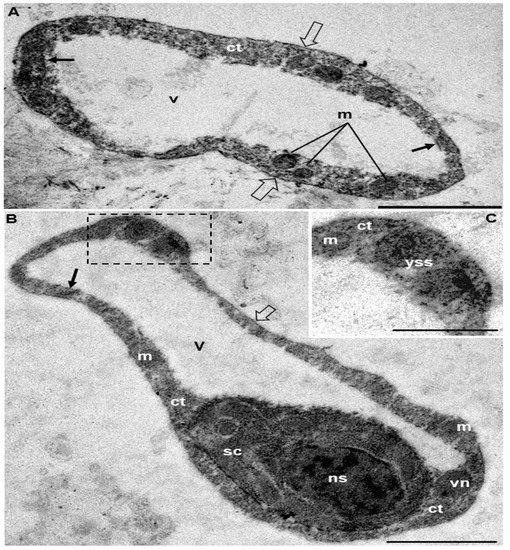
Figure 2.
Electron micrographs showing transversal sections of plasmodia of the Ceratomyxa amazonensis parasite of the gallbladder of Geophagus altifrons. (A) Plasmodium showing the plasmodial membrane (empty arrow), cytoplasm (ct), mitochondria (m), vacuolar membrane (black arrow) and vacuole (v). Bar: 1 μm. (B) Plasmodium showing plasmodial membrane (empty arrow), cytoplasm (ct), secondary cell (sc) and its nucleus (ns), mitochondria (m), vegetative nucleus (vn), vacuole membrane (black arrow), and extensive vacuolar area (v). Bar: 1 μm. (C) Magnification of (B) showing the region of cytoplasm (ct) with mitochondria (m) and early stages of sporogony development (yss). Bar: 0.5 μm.
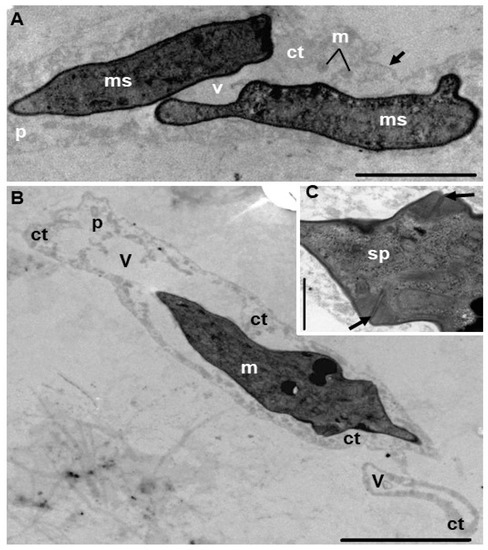
Figure 3.
Electron micrographs of vermiform plasmodia of the Ceratomyxa amazonensis parasite of the gallbladder of Geophagus altifrons. (A) Longitudinal section of a plasmodium (p) with two myxospores (ms) and mitochondria (m) in the cytoplasm (ct), vacuole region (v), and detail of the plasmodial membrane (black arrow). (B) Longitudinal section of a plasmodium (p) showing a thin layer of cytoplasm (ct) with mature myxospore (m) and extensive vacuole (v). Bar: 5 μm. (C) Magnification of the region of (B) showing the junction of the valves (black arrows) and the sporoplasm (sp) of a mature myxospore. Bar: 1 μm.
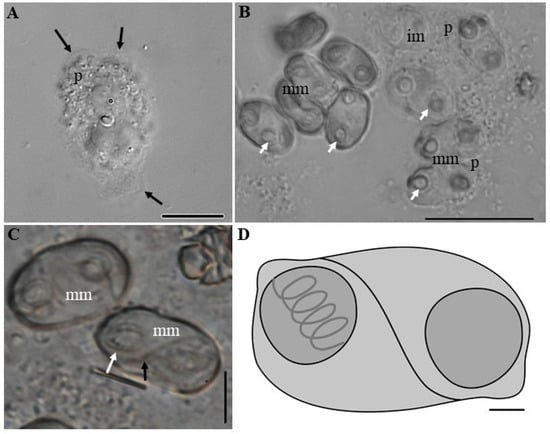
Figure 4.
Photomicrographs of Ellipsomyxa santarenensis n. sp., parasite of the gallbladder of Satanoperca jurupari. (A) Early developmental stage of pseudoplasmodium (p) showing pseudopod-like cytoplasmic expansions (black arrows). Bar: 10 µm. (B) Mature (mm) and immature (im) myxopores with nematocysts. Note diasporic pseudoplasmoduim (p) and nematocysts (white arrows). Bar: 20 µm. (C) Two mature myxospores (mm). Note the sultural line (black arrow) and nematocyst with tubules (white arrow). Bar: 5 µm. (D) Schematic drawing of a myxospore. Bar: 10 μm.
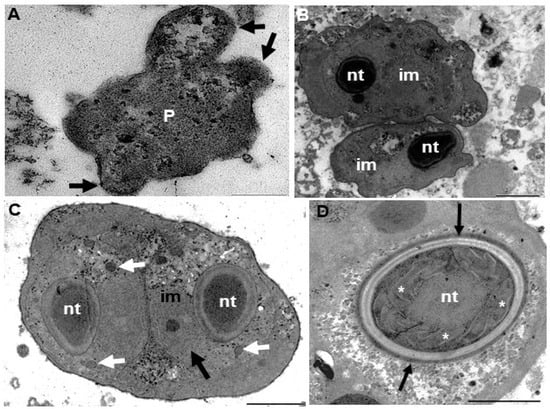
Figure 5.
Electromicrographs of Ellipsomyxa santarenensis n. sp., parasite of the gallbladder of Satanoperca jurupari. (A) Immature pseudoplasmodium (P). Note expansions of the membrane (black arrows). Bar: 0.5 μm. (B) Dysporic pseudoplasmodium with immature myxospores (im) and their nematocysts (nt). Bar: 5 μm. (C) Immature myxospore showing two nematocysts (nt) with the polar filaments still exteriorized (white arrows). Note the presence of a cnidocyte nucleus (black arrow). Bar: 2 μm. (D) Nematocyst in cross-section with details of the tubule (*) and double layer of the nematocyst wall (black arrows). Bar: 0.5 µm.
Taxonomic summary
Phylum Cnidaria Verril, 1865.
Subphylum Endocnidozoa Schuchert, 1996.
Class Myxozoa Grassé, 1970.
Subclasse Myxosporea Bütschli, 1881.
Order Bivalvulida Shulman, 1959.
Family Ceratomyxidade Doflein, 1899.
3.1. Myxozoa Parasite of Geophagus altifrons Heckel, 1840
Genus: Ceratomyxa Thélohan, 1892.
Species: Ceratomyxa amazonensis Mathews, Naldoni; Maia & Adriano, 2016 (Figure 1, Figure 2 and Figure 3).
Locality: Tapajós River, near the city of Santarém, Pará State, Brazil (2°23′49.79″ S 54°43′53.33″ W).
Specimens deposited: one glass slide with Giemsa-stained myxospores (paratype) was deposited in the Myxozoa collection of the Museum of Biological Diversity), University of Campinas (UNICAMP), São Paulo, Brazil (accession number ZUEC MYX 115).
Prevalence: 64.2% (9/14).
Site of infection: Gallblader—free plasmodia swimming in the bile.
Representative DNA sequences: SSU rDNA (1527 and 1408 bp) was deposited in the GenBank database. (accession number OR142123).
3.1.1. Morphological Data
Plasmodia present in the gallbladder of G. altifrons exhibited a vermiform shape with undulatory motility (Video S1), measuring 106.9 ± 52.1 (67.3–202.6) μm in length and 7.6 ± 1.7 (4.9–11.1) µm in width (n = 10). Mature myxospores displayed smooth, equal-sized, slightly curved valves with blunted ends, measuring 4.9 ± 1.4 (3.1–6.3) μm in length, 23.8 ± 5.9 (18, 8–28.6) µm in thickness, and a posterior angle of 159.7° ± 10.6° (174.9°–144.7°). Sub-spheric nematocysts were equal in size and measured 2.4 ± 0.8 (1.2–5.6) μm in length and 1.9 ± 0.3 (1.5–2.8) μm in width and were located one on each side of the sutural line. Tubules could be observed within the nematocysts and were coiled with 3–4 turns (Figure 1; Table 1). Transmission electron microscopy suggested that the vermiform plasmodia were organized as follows: an outer cytoplasmic region, which contained organelles and different stages of sporogony development, and a large vacuole occupying the inner area (Figure 2 and Figure 3).

Table 1.
Morphological comparison of characteristics of Ceratomyxa amazonensis isolated in this study with other Ceratomyxa species parasitizing the gallbladders of freshwater fish in South America. The measurements (in μm) were obtained in accordance with Gunter et al. [25].
3.1.2. SSU rDNA Data
Amplification and sequencing of SSU rDNA generated a 1527 nucleotide read. The BLAST search against the GenBank database showed a 99.7% sequence similarity hit with the SSU rDNA sequence of C. amazonensis. When sequence similarity was compared with the twelve freshwater Ceratomyxa species for which SSU rDNA data were available, not surprisingly the highest similarity for the query sequence was to C. amazonensis (0.6%), while the greatest sequence divergence (8.3%) was with Ceratomyxa mandi Araújo, Adriano, Franzolin, Zatti & Naldoni, 2022 (Table 2).

Table 2.
Dissimilarity matrix of SSU rDNA sequences from selected freshwater Ceratomyxa species.
3.2. Myxozoa Parasite of Satanoperca jurupari (Heckel, 1840)
Genus: Ellipsomyxa Køie, 2003.
Locality: Tapajós River, near the city of Santarém, Pará State, Brazil (2°23′49.79″ S 54°43′53.33 W).
Specimens deposited: one glass slide with Giemsa-stained myxospores (syntype) was deposited in the Myxozoa collection of the Museum of Biological Diversity), University of Campinas (UNICAMP), São Paulo, Brazil (accession number ZUEC MYX 114).
Prevalence: 33.3% (2/6).
Site of infection: Gallbladder, free in bile
Representative DNA sequences: SSU rDNA (1584 bp) was deposited in the GenBank database. (accession number OR142132).
ZooBank registration: The Life Science Identifier for E. santarenesis n. sp. is urn:lsid:zoobank.org:pub:CEBA6D8E-2530-496A-A7FB-24E2D6DC7168.
Etymology: The specific epithet “santarenensis” derives from the type locality, Santarém municipality.
3.2.1. Morphological Data
Diasporic amorphous pseudoplasmodia of E. santarenensis n. sp. showed cytoplasmic pseudopod-like expansions (Video S2) and measured 27.9 μm (20.6–35.8 μm; n = 4) in length and 23.4 μm (20–31.4 μm; n = 4) in width (Figure 4a and Figure 5a). Mature myxospores were ellipsoidal to the plane of the suture line, with smooth valves measuring 12.0 ± 3.2 (10.7–13.7) μm in length and 7.6 ± 1.5 (5.6–8.2) µm in width. The transverse suture line was curved in an “S” shape. Spherical nematocysts of equal size were located at opposite ends of myxospores and measured 2.8 ± 0.4 (2.0–3.6) μm in diameter. A tubule with 4–5 coils was arranged perpendicularly to the longitudinal axis of each nematocyst (Table 3). In ultrastructure analysis, cytoplasmic expansions were observed in the pseudoplasmodia, forming structures similar to pseudopods (Figure 5).

Table 3.
Comparison of the myxospore dimensions of all Ellipsomyxa species described parasitizing freshwater fish hosts. Dimensions are given in micrometers.
3.2.2. SSU rDNA Data
Sequencing of the SSU rDNA amplicon provided a 1584 bp sequence for E. santarenensis n. sp. BLAST searches revealed that the greatest similarity of the sequence of E. santarenensis n. sp. was for that of Ellipsomyxa amazonensis Zatti, Atkinson, Maia, Corrêa, Bartholomew & Adriano, 2018, with 99.6%. An analysis of the genetic distances between Ellipsomyxa species from other freshwater environments (Table 4) indicated that the smallest distance between the E. santarenensis n. sp. isolated herein from S. jurupari and E. amazonensis and Ellipsomyxa paraensis Zatti, Maia & Adriano, 2020 was 1.6%. The greatest divergence was 8.8% between E. santarenensis n. sp. and Ellipsomyxa tucujuensis Ferreira, Silva Carvalho, Bittencourt, Hamoy, Matos &Videira, 2021.

Table 4.
Dissimilarity matrix of SSU rDNA sequences of freshwater Ellipsomyxa species.
3.3. Phylogenetic Analysis
Both ML and BL phylogenetic analyses were in accordance and showed Ellipsomyxa species to be monophyletic. However, freshwater Ellipsomyxa species were not monophyletic. Ellipsomyxa santarenensis n. sp. parasite of S. jurupari reported herein clustered together with other Amazonian freshwater species, but Ellipsomyxa plagioscioni Zatti, Maia & Adriano, 2020, which is a parasite of the Amazonian Plagioscion squamosissimus, emerged in the marine Ellipsomyxa lineage (Figure 6 and Figure S1).
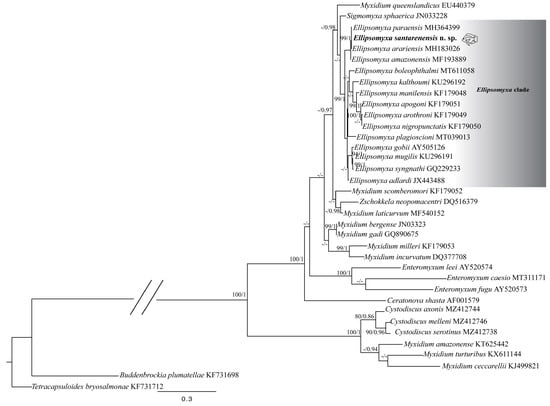
Figure 6.
Maximum Likelihood (ML) consensus phylogenetic tree obtained from the SSU-rDNA sequences from Ellipsomyxa santarenensis n. sp., parasites of Satanoperca jurupari. GenBank accession numbers are presented in front of the names of each species.
4. Discussion
Knowledge pertaining to the diversity and radiation of the myxozoan genera Ceratomyxa and Ellipsomyxa in freshwater environments is lacking, with data emerging from the South American continent as a result of studies over recent years. Taxonomic, morphological and molecular comparisons provided in this study were, therefore, based on freshwater species from South American species.
Regarding the C. amazonensis isolated in this study from G. altifrons caught in the Tapajós River, molecular data provided a genetic distance of only 0.6% to another C. amazonenses isolated (sequence no. KX236169) from Symphysodon discus Heckel, 1840 caught in the Rio Negro [7], which is in the same watershed as the Tapajós River. Symphysodon discus, such as G. altifrons, is a cichlid fish. However, despite the low genetic divergence, morphological data of C. amazonensis isolated from G. altifrons differed from those obtained in the original description from S. discus, being shorter, thicker and slightly curved, while the myxospores of C. amazonensis of S. discus were strongly arched (see Table 1 and Figure 1 of Mathews et al. [7]). Morphological data of the myxospores obtained in this study corroborate another characterization of C. amazonensis performed by Sousa et al. [39], who reported slightly curved myxospores in infections of S. discus specimens caught in the Unini River, which is a tributary of the Negro River (Table 1). The SSU rDNA genetic distances between the C. amazonensis parasite of G. altifrons and C. amazonensis isolated from S. discus from the Unini River were only 0.2% (sequence no. MN064752—Sousa et al., [39]). Thus, despite morphological discrepancies between the findings in this study and those of Mathews et al. [8], the morphological data provided herein are similar to those obtained by Sousa et al. [39]. The SSU rDNA data obtained from the three studies enable intraspecific genetic divergence to be inferred and supported G. altifrons captured in the Tapajós River as a new vertebrate host for C. amazonensis. The infection incidence of a species between different hosts has also been observed for Ceratomyxa tuniensis Thabet, Mansour, Omar, & Tlig-Zouari, 2016, which was isolated from the gallbladder of Caranax rhonchus Geoffroy Saint-Hilaire, 1817 and Trachurus trachurus Linnaeus, 1758 in the Gulf of Gabes, Tunisia [26]. The SSU rDNA sequence similarities between C. amazonensis isolated from G. altiforns and S. discus (Mathews et al., [7] and Sousa et al., [39]) confirmed that these were the same species. However, the contrast between the morphological features of myxospores presented in this study and those of Sousa et al. [39], compared to those provided in the original description by Mathews et al. [7], was intriguing. Extreme morphological plasticity in Ceratomyxa spp. myxospores has been previously observed [23] and is likely host-dependent, which makes species determination using morphology alone difficult without corroborating evidence afforded by molecular data. Zhai et al. [40] also reported that morphological plasticity was common in myxosporeans but with clear overlap within minimum and maximum size ranges. Thus, the discrepancies between the morphological features of myxospores presented in the original description of C. amazonensis [7] and those from subsequent studies (Sousa et al., [39] and this study) (Table 1) might be attributed to either morphological plasticity or some mistaken or unobserved data.
Ultrastructural analyses revealed asynchronous development, with plasmodia from C. amazonensis containing early sporogonic developmental stages and immature and mature myxospores, which are all common features of the Ceratomyxa species with this kind of vermiform plasmodia [15]. Sporogonic stages, as well as cytoplasmic organelles, were arranged in an external cytoplasmic region, while a large vacuole occupied the central region of the plasmodia. These plasmodial anatomical appearances have all been reported previously in other South American freshwater Ceratomyxa species [8,15].
The myxospores of E. santarensensis n. sp. isolated in this study from the gallbladder of S. jurupari were similar in length and width to those of E. amazonensis, E. paraensis and Ellipsomyxa arariensis Silva; Matos, Lima, Furtado, Hamoy & Matos, 2020. However, the myxospores of the new species differed from those of the other three species, presenting spherical nematocysts with 2.8 µm and containing tubules with 4–5 turns, while for E. amazonensis the nematocysts had 3.8 × 3.1 µm and tubules with 2–3 turns, for E. paraensis the nematocysts measured 3.2 × 2.6 µm and also had tubules with 2–3 turns, and for E. arariensis the nematocysts presented 3.5 × 2.6 µm and tubules with 5–6 turns (Table 2). The SSU rDNA sequencing data suggested that among freshwater Ellipsomyxa species, E. amazonensis and E. paraensis were the species with the lowest genetic distance to E. santarensensis n. sp., with 1.6% divergence. Although there is no definition of the exact interspecific genetic distance to define species [23], careful analysis supported that the level of interspecific difference in the genus Ellipsomyxa can be as low as 0.5% between Ellipsomyxa gobii Køie, 2003 and Ellipsomyxa syngnathi Køie & Karlsbakk, 2009, 0.3% between E. syngnathi and Ellipsomyxa mugilis (Sitja-Bobadilla & Alvarez-Pellitero, 1993), and 0.2% between Ellipsomyxa nigropunctatis Heiniger & Adlard, 2014 and Ellipsomyxa arothroni Heiniger & Adlard, 2014 (see Table 3 in Zatti et al., [17]). Thus, based on morphological and molecular differences, E. santarensensis n. sp. is proposed herein as a new myxosporean species. It is the second species of the genus Ellipsomyxa to be isolated from S. jurupari, with an earlier description of Ellipsomyxa tucujuensis Ferreira, Silva, de Carvalho, Bittencourt, Hamoy, Matos, & Videira, 2021, also infecting this cichlid fish [19]. Currently, there are only 16 species in the genus Ellipsomyxa, with five species reported from fish captured in the Amazon River. The Amazon basin may, therefore, be an area of large and yet underexplored region of Ellipsomyxa species diversity.
This study corroborates the analyses carried out by Zatti et al. [17], who recovered E. plagioscioni, a parasite of the sciaenid, grouping in a marine lineage, showing that South American freshwater Ellipsomyxa species do not form a monophyletic group. However, the other freshwater species formed a single clade (Figure 6 and Figure S1). As suggested by Zatti et al. [17], this may reflect distinct change events between the marine and freshwater environments, possibly driven by vertebrate or invertebrate hosts.
In summary, the present study expands information on species diversity and host ranges of myxosporeans in the Amazon basin. Knowledge of the occurrence and distribution of parasitic fauna of ornamental fish, such as cichlids, has enormous importance to the local aquarium industry in the Amazon, since the ornamental fish trade has been considered an important route of myxozoan transmission [41,42,43,44], and therefore, preventive monitoring plans must be considered in the activity.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15070830/s1.
Author Contributions
R.T.A.F., M.I.M. and E.A.A.: conceived the initial idea for the study, undertook sampling and participated in the writing of the draft manuscript; R.T.A.F. and M.I.M.: performed morphological, molecular analysis and phylogenetic analyses; E.A.A. and M.I.M.: supervision; R.T.A.F. and E.A.A.: ultra structural analysis; P.F.L. and E.A.A.: project administration, funding acquisition, revision and organization of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Fundação de Amparo à Pesquisa do Estado de São Paulo. (FAPESP grant #2019/17427-3), and in-part by the Coordination for the Improvement of Higher Education Personnel (CAPES Finance Code 001). R.T.A. Figueredo was supported with a scholarship provided by CAPES (grant #88887.505484/2020-00). M.I. Muller was supported with a Postdoctoral fellowship granted by FAPESP (grant #2017/16546-3). E.A. Adriano received a research productivity grant from the Brazilian Fostering Agency CNPq (grant #304687/2020-0). P.F. Long was supported by the University of São Paulo (grant number 13.1.1502.9.8) and King’s College London.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional of Ethics Research Committee of the Federal University of São Paulo (CEUA # 6549290920-17/12/2020). Capture authorization and access to genetic data were authorized by the Brazilian Ministry of the Environment SISBIO n° 67616-2 and SisGen n° A33CB83; the method of euthanasia was approved by the Ethics Committee of the Federal University of São Paulo CEUA n° 6549290920.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors thank Lincoln Lima Corrêa for the logistical support of the fieldwork in Santarém, Frank Raynner Vasconcelos Ribeiro for identifying fish species, and the fishermen Fernando Dias de Souza, Francisco dos Santos Pinto, and Arlindo Teixeira Guimarães for their local knowledge of fish and providing study material from the Amazon and Tapajós rivers.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Okamura, B.; Hartigan, A.; Naldoni, J. Extensive uncharted biodiversity: The parasite dimension. Integr. Comp. Biol. 2018, 58, 1132–1145. [Google Scholar] [CrossRef] [PubMed]
- Okamura, B.; Gruhl, A.; Bartholomew, J.L. An Introduction to Myxozoan Evolution, Ecology and Development; Springer International Publisher: Cham, Switzerland, 2015; pp. 69–84. [Google Scholar]
- Eiras, J.C. Synopsis of the species of the genus Ceratomyxa Thelohan, 1892 (Myxozoa: Myxosporea: Ceratomyxidae). Syst. Parasitol. 2006, 65, 49–71. [Google Scholar] [CrossRef] [PubMed]
- Eiras, J.C.; Cruz, C.; Saraiva, A. Synopsis of the species of Ceratomyxa Thelohan, 1892 (Cnidaria, Myxosporea, Ceratomyxidae) described between 2007 and 2017. Syst. Parasitol. 2018, 95, 427–446. [Google Scholar] [CrossRef]
- Azevedo, C.; Rocha, S.; Casal, G.; São Clemente, C.S.; Matos, P.S.; Al-Quraishy, A.; Matos, E. Light and ultrastructural description of Meglitschia mylei n. sp. (Myxozoa) from Myleus rubripinnis (Teleostei: Serrasalmidae) in the Amazon River System. J. Eukaryot. Microbiol. 2011, 58, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Rocha, S.; Casal, G.; São Clemente, C.S.; Matos, P.S.; Al-Quraishy, A.; Matos, E. Ultrastructural description of Ceratomyxa microlepis sp. nov. (Phylum Myxozoa): A parasite infecting the gall bladder of Hemiodus microlepis, a freshwater teleost from the Amazon River. Mem. Inst. Oswaldo Cruz. 2013, 108, 150–154. [Google Scholar] [CrossRef]
- Mathews, P.D.; Naldoni, J.; Maia, A.A.M.; Adriano, E.A. Morphology and small subunit rDNA-based phylogeny of Ceratomyxa amazonensis n. sp. parasite of Symphysodon discus, an ornamental freshwater fish from Amazon. Parasitol. Res. 2016, 115, 4021–4025. [Google Scholar] [CrossRef]
- Adriano, E.A.; Okamura, B. Motility, morphology and phylogeny of the plasmodial worm, Ceratomyxa vermiformis n. sp. (Cnidaria: Myxozoa: Myxosporea). Parasitology 2017, 144, 158–168. [Google Scholar] [CrossRef]
- Zatti, S.A.; Atkinson, S.D.; Bartholomew, J.L.; Maia, A.A.M.; Adriano, E.A. Ceratomyxa gracillima n. sp. (Cnidaria: Myxosporea) provides evidence of panmixia and ceratomyxid radiation in the Amazon basin. Parasitology 2018, 145, 1137–1146. [Google Scholar] [CrossRef]
- Silva, M.S.; Carvalho, A.E.F.B.; Hamoy, I.; Matos, E.R. Coelozoic parasite of the family Ceratomyxidae (Myxozoa, Bivalvulida) described from motile vermiform plasmodia found in Hemiodus unimaculatus Bloch, 1794. Parasitol. Res. 2020, 119, 871–878. [Google Scholar] [CrossRef]
- Bittencourt, L.S.; Silva, D.T.; Hamoy, I.; Carvalho, A.A.; Silva, M.F.; Videira, M.; Carvalho, J.C.T.; Matos, E.R. Morphological and Phylogenetic Features of Ceratomyxa macapaensis n. sp. (Myxozoa: Ceratomyxidae) in Mesonauta festivus Heckel, 1840 (Cichliformes: Cichlidae) from the eastern Amazon region. Acta Parasitol. 2021, 67, 322–329. [Google Scholar] [CrossRef]
- Zatti, S.A.; Adriano, E.A.; Araújo, B.L.; Franzolin, G.N.; Maia, A.A.M. Expanding the geographic distribution of the freshwater parasite Ceratomyxa (Cnidaria: Myxozoa) with vermiform-type plasmodia. Microb. Pathog. 2022, 162, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Araújo, B.L.; Adriano, E.A.; Franzolin, G.N.; Zatti, S.A.; Naldoni, J. A novel Ceratomyxa species (Myxozoa: Cnidaria) infecting an Amazonian catfish. Parasitology 2022, 89, 102582. [Google Scholar] [CrossRef] [PubMed]
- Franzolin, G.N.; Araújo, B.L.; Zatti, S.A.; Naldoni, J.; Adriano, E.A. Occurrence of the host-parasite system Rhaphiodon vulpinus and Ceratomyxa barbata n. sp. in the two largest watersheds in South America. Parasitol. Int. 2022, 91, 102651. [Google Scholar] [CrossRef]
- Adriano, E.A.; Zatti, S.A.; Okamura, B. How to build single-celled cnidarians with worm-like motility: Lessons from Myxozoa. J. Anat. 2022, 240, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Zatti, S.A.; Atkinson, S.D.; Maia, A.A.; Corrêa, L.L.; Bartholomew, J.L.; Adriano, E.A. Novel Myxobolus and Ellipsomyxa species (Cnidaria: Myxozoa) parasiting Brachyplatystoma rousseauxii (Siluriformes: Pimelodidae) in the Amazon basin, Brazil. Parasitol. Int. 2018, 67, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Zatti, S.A.; Maia, A.A.; Adriano, E.A. Growing diversity supports radiation of an Ellipsomyxa lineage into the Amazon freshwater: Description of two novel species parasitizing fish from Tapajós and Amazon rivers. Acta Trop. 2020, 211, 105616. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.T.; Matos, P.S.; Lima, A.M.; Furtado, A.P.; Hamoy, I.; Matos, E.R. Ellipsomyxa arariensis n. sp. (Myxozoa: Ceratomyxidae), a new myxozoan parasite of Pygocentrus nattereri Kner, 1858 (Teleostei: Characidae) and Pimelodus ornatus Kner, 1858 (Teleostei: Pimelodidae) from Marajó Island, in the Brazilian Amazon region. Parasitol. Res. 2018, 117, 3537–3545. [Google Scholar] [CrossRef]
- Ferreira, R.L.S.; Silva, D.T.; Carvalho, A.A.; Bittencourt, L.S.; Hamoy, I.; Matos, E.; Videira, M. Ellipsomyxa tucujuensis n. sp. (Myxozoa: Ceratomyxidae), parasita de Satanoperca jurupari (Osteichthyes: Cichlidae) da Amazônia brasileira. Parasitol. Int. 2021, 83, 102332. [Google Scholar] [CrossRef]
- Silvano, R.A.; Nitschke, P.P.; Vieira, K.C.; Nagl, P.; Martínez, A.T.; Dutra, M.C.; Andrade, M.C. Atlas of Fish of Tapajós and Negro Rivers III: Perciformes and Other Fish Groups. In Fish and Fisheries in the Brazilian Amazon; Springer: Cham, Switzerland, 2020; pp. 321–414. [Google Scholar]
- Froese, R.; Pauly, D. World Wide Web Electronic Publication. FishBase. 2022. Available online: www.fishbase.org (accessed on 4 April 2023).
- Lom, J.; Arthur, J. A guideline for the preparation of species descriptions in Myxosporea. J. Fish Dis. 1989, 12, 151–156. [Google Scholar] [CrossRef]
- Gunter, N.L.; Whipps, C.N.; Adlard, R.D. Ceratomyxa (Myxozoa: Bivalvulida): Robust taxon or genus of convenience. Int. J. Parasitol. 2009, 39, 1395–1405. [Google Scholar] [CrossRef]
- Whipps, C.M.; Adlard, R.D.; Bryant, M.S.; Lester, R.J.G.; Findlay, V.; Kent, M.L. First report of three Kudoa species from Eastern Australia: Kudoa thyrsites from Mahi mahi (Coryphaena hippurus), Kudoa amamiensis and Kudoa minithyrsites n. sp. from Sweeper (Pempheris ypsilychnus). J. Eukaryot. Microbiol. 2003, 50, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Heiniger, H.; Adlard, R.D. Relatedness of novel species of Myxidium Bütschli, 1882, Zschokkella Auerbach, 1910 and Ellipsomyxa Køie, 2003 (Myxosporea: Bivalvulida) from the gall bladders of marine fishes (Teleostei) from Australian waters. Syst. Parasitol. 2014, 87, 47–72. [Google Scholar] [CrossRef] [PubMed]
- Thabet, A.; Mansour, L.; Al Omar, S.Y.; Tlig-Zouari, S. Ceratomyxa tunisiensis n. sp. (Myxosporea: Bivalvulida) from the gallbladders of two carangid fish caught off the coast of Tunisia. J. Eukaryot. Microbiol. 2016, 63, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M.; Dixon, M.T. Ribosomal DNA: Molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991, 66, 411–453. [Google Scholar] [CrossRef] [PubMed]
- Barta, J.R.; Martin, D.S.; Liberator, P.A.; Dashkevicz, M.; Anderson, J.W.; Feighner, S.D.; Elbrecht, A.; Perkins-Barrow, A.; Jenkins, M.C.; Danforth, H.D.; et al. Phylogenetic relationships among eight Eimeria species infecting domestic fowl inferred using complete small subunit ribosomal DNA sequences. J. Parasitol. 1997, 83, 262–271. [Google Scholar] [CrossRef]
- Molnár, K.; Eszterbauer, E.; Székely, C.; Dan, A.; Harrach, B. Morphological and molecular biological studies on intramuscular Myxobolus spp. of cyprinid fish. J. Fish Dis. 2002, 25, 643–652. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Drummond, A. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Xia, X. DAMBE5: A comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2013, 30, 1720–1728. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, K. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 1–8. [Google Scholar]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. Molecular Evolution, Phylogenetics and Epidemiology: Fig-Tree. 2009. Available online: http.ac.uk/software/figtree/ (accessed on 10 November 2015).
- Zatti, S.A.; Atkinson, S.D.; Bartholomew, J.L.; Maia, A.A.M.; Adriano, E.A. Amazonian waters harbour an ancient freshwater Ceratomyxa lineage (Cnidaria: Myxosporea). Acta Trop. 2017, 169, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Sousa, F.B.; Milanin, T.; Morandini, A.C.; Espinoza, L.L.; Flores-Gonzales, A.; Gomes, A.L.S.; Mathews Delgado, P. Molecular diagnostic based on 18S rDNA and supplemental taxonomic data of the cnidarian coelozoic Ceratomyxa (Cnidaria, Myxosporea) and comments on the intraspecific morphological variation. Zoosystematics Evol. 2021, 97, 307–314. [Google Scholar] [CrossRef]
- Zhai, Y.; Whipps, C.M.; Gu, Z.; Guo, Q.; Wu, Z.; Wang, H.; Liu, Y. Intraspecific morphometric variation in myxosporeans. Folia Parasitol. 2016, 63, 001. [Google Scholar] [CrossRef]
- Mathews, P.D.; Mertins, O.; Pereira, J.O.L.; Maia, A.A.M.; Adriano, E.A. Morphology and 18S rDNA sequencing of Henneguya peruviensis n. sp. (Cnidaria: Myxosporea), a parasite of the Amazonian ornamental fish Hyphessobrycon loretoensis from Peru: A myxosporean dispersal approach. Acta Tropica. 2018, 187, 207–213. [Google Scholar] [CrossRef]
- Biosecurity, N.Z. Import Risk Analysis: Ornamental Fish; Ministry of Agriculture and Forestry: Wellington, New Zealand, 2005. [Google Scholar]
- Hallett, S.L.; Hartigan, A.; Atkinson, S.D. Myxozoans on the move: Dispersal modes, exotic species and emerging diseases. In Myxozoan Evolution, Ecology and Development; Springer International Publisher: Cham, Switzerland, 2015; pp. 343–362. [Google Scholar]
- Figueredo, R.T.A.; Müller, M.I.; Arana, S.; Long, P.F.; Adriano, E.A. Phylogenetic and host-parasite relationship analyses of Henneguya caquetaia sp. nov (Myxosporea: Myxobolidae) infecting an Amazonian cichlid fish. Microb. Pathog. 2023, 179, 106116. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).