Pleistocene Glacial Refugia in the Don River Basin: Witness from the Endangered Depressed River Mussel
Abstract
1. Introduction
2. Materials and Methods
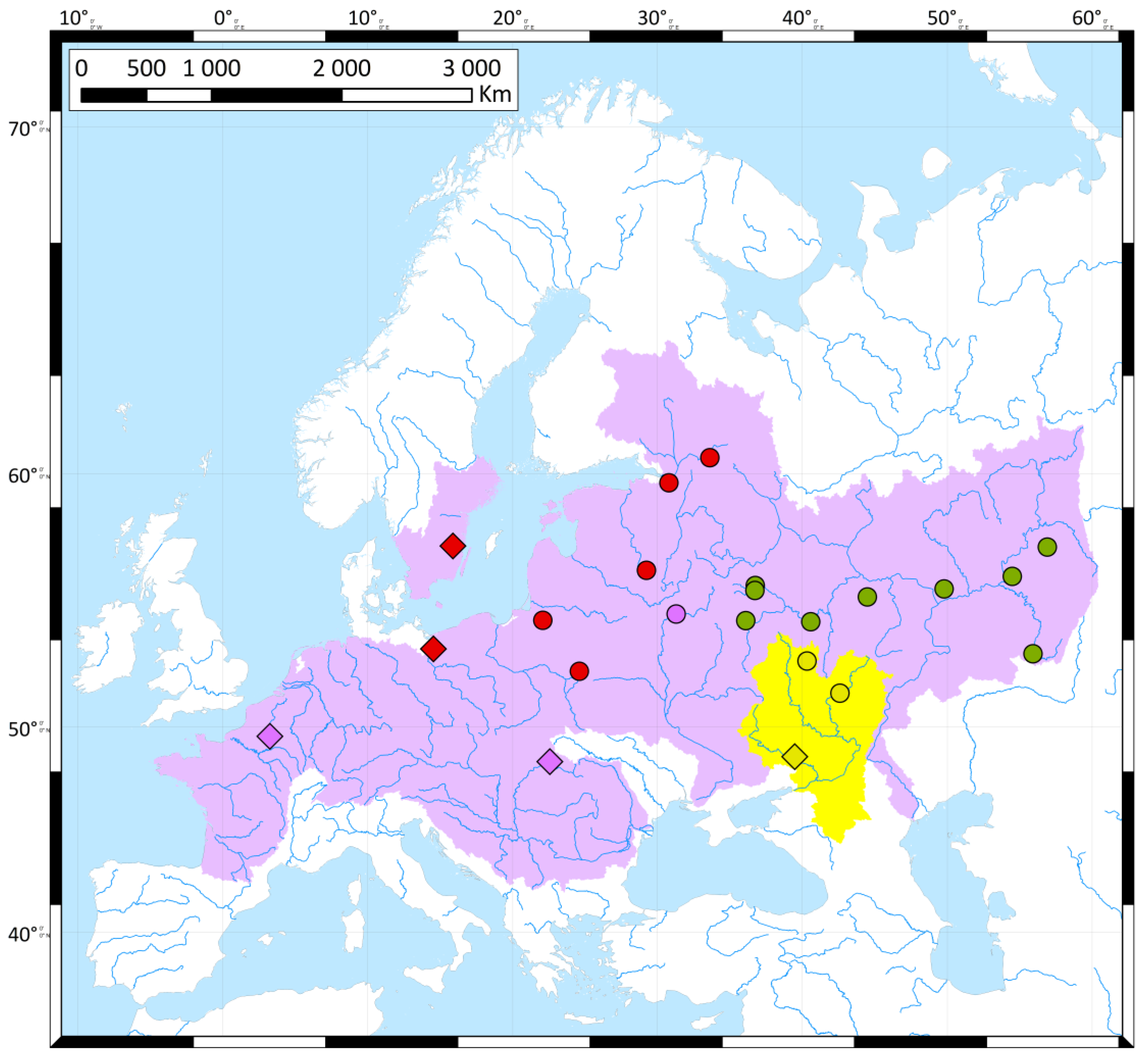
2.1. Sample Collection, DNA Extraction, PCR and Sequencing
2.2. Genetic Diversity, Genetic Differentiation, and Demographic History
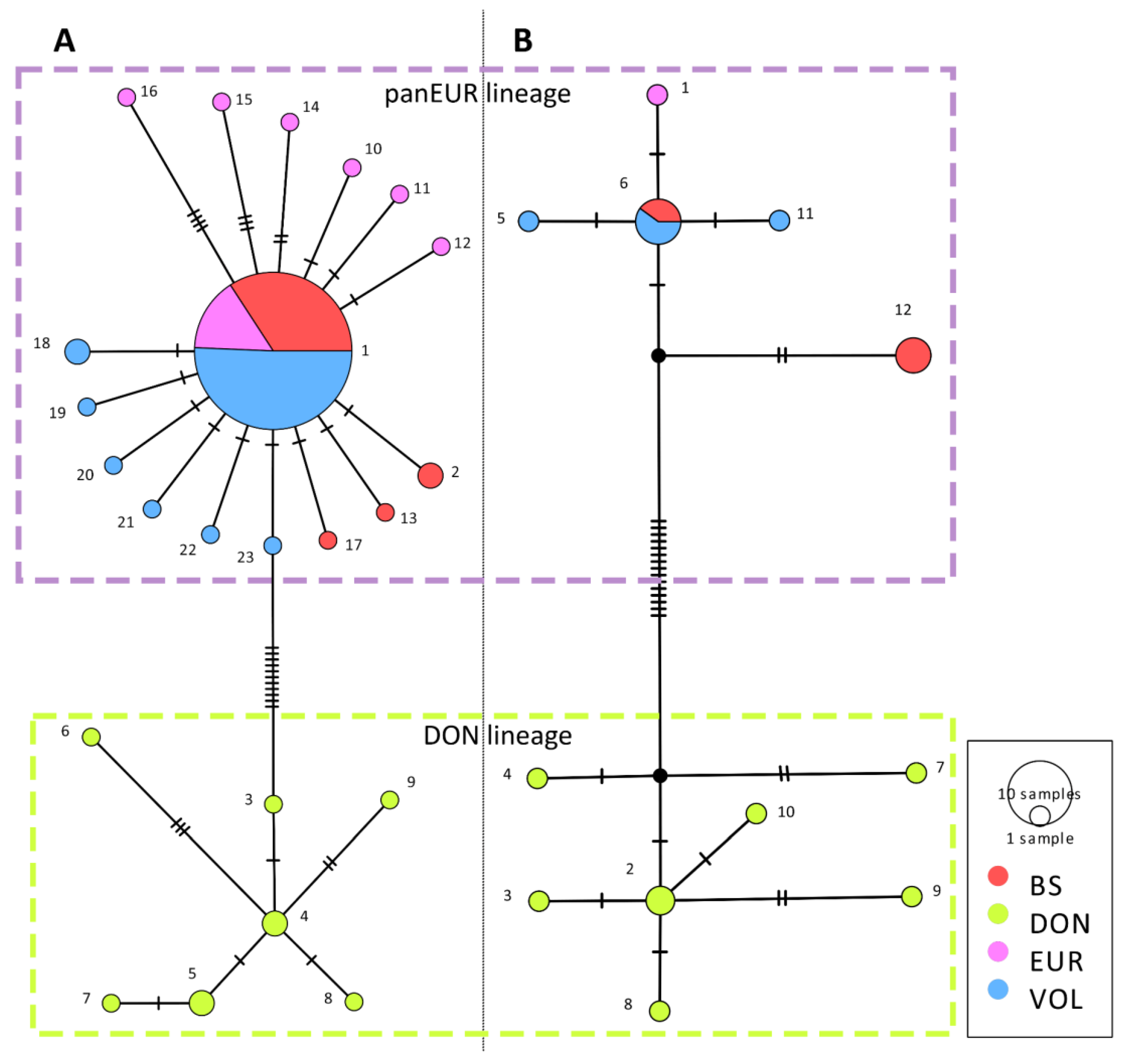
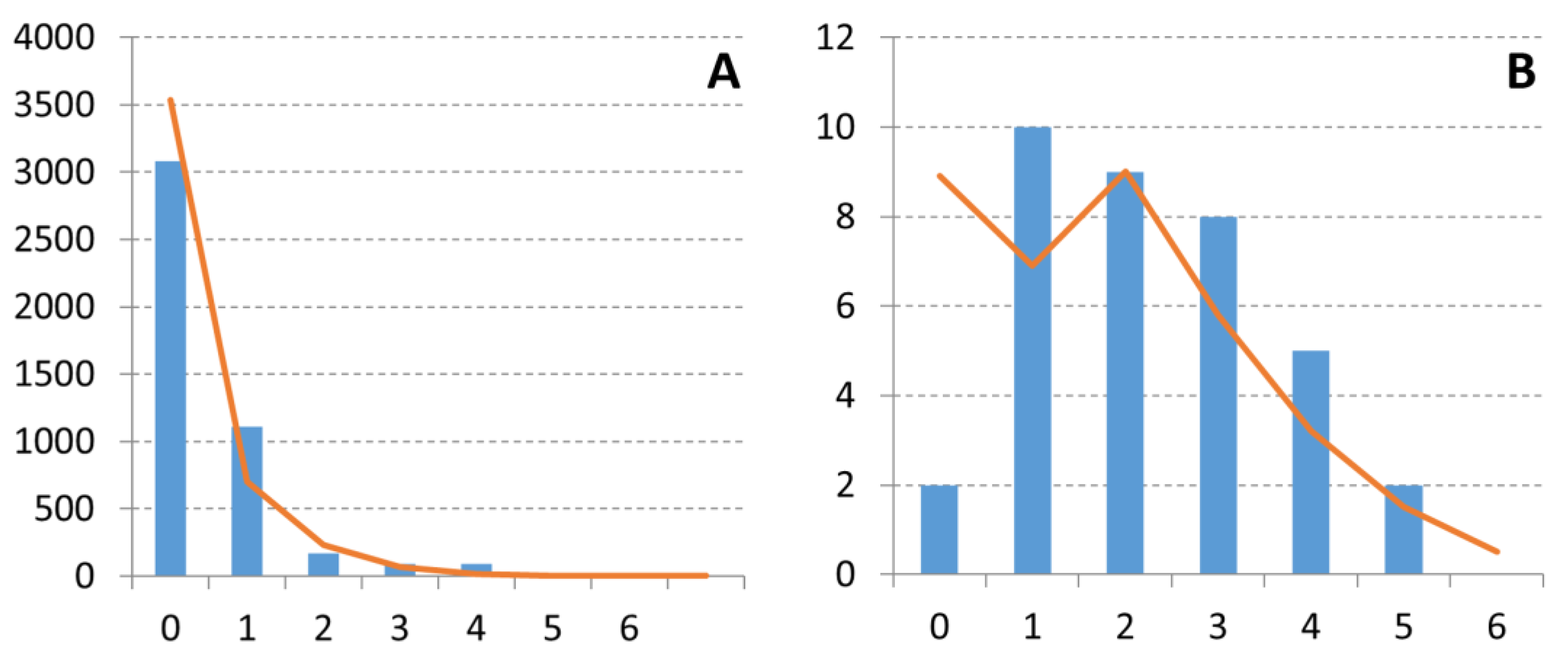
3. Results
3.1. Genetic Diversity
3.2. Genetic Differentiation and Genetic Structure
3.3. Demographic Trends
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hewitt, G. The Genetic Legacy of the Quaternary Ice Ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Consuegra, S.; García De Leániz, C.; Serdio, A.; González Morales, M.; Straus, L.G.; Knox, D.; Verspoor, E. Mitochondrial DNA Variation in Pleistocene and Modern Atlantic Salmon from the Iberian Glacial Refugium. Mol. Ecol. 2002, 11, 2037–2048. [Google Scholar] [CrossRef] [PubMed]
- Svenning, J.C.; Skov, F. Could the Tree Diversity Pattern in Europe Be Generated by Postglacial Dispersal Limitation? Ecol. Lett. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Hewitt, G.M. The Structure of Biodiversity—Insights from Molecular Phylogeography. Front. Zool. 2004, 1, 234. [Google Scholar] [CrossRef]
- Makhrov, A.A.; Bolotov, I.N. Dispersal Routes and Species Identification of Freshwater Animals in Northern Europe: A Review of Molecular Evidence. Russ. J. Genet. 2006, 42, 1101–1115. [Google Scholar] [CrossRef]
- Schmitt, T. Molecular Biogeography of Europe: Pleistocene Cycles and Postglacial Trends. Front. Zool. 2007, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sworobowicz, L.; Mamos, T.; Grabowski, M.; Wysocka, A. Lasting through the Ice Age: The Role of the Proglacial Refugia in the Maintenance of Genetic Diversity, Population Growth, and High Dispersal Rate in a Widespread Freshwater Crustacean. Freshw. Biol. 2020, 65, 1028–1046. [Google Scholar] [CrossRef]
- Lyubas, A.A.; Tomilova, A.A.; Kondakov, A.V.; Konopleva, E.S.; Vikhrev, I.V.; Gofarov, M.Y.; Eliseeva, T.A.; Aksenova, O.V.; Bovykina, G.V.; Kryuk, D.V.; et al. Phylogeography and Genetic Diversity of Duck Mussel Anodonta Anatina (Bivalvia: Unionidae) in Eurasia. Diversity 2023, 15, 260. [Google Scholar] [CrossRef]
- Tomilova, A.A.; Lyubas, A.A.; Kondakov, A.V.; Vikhrev, I.V.; Gofarov, M.Y.; Kolosova, Y.S.; Vinarski, M.V.; Palatov, D.M.; Bolotov, I.N. Evidence for Plio-Pleistocene Duck Mussel Refugia in the Azov Sea River Basins. Diversity 2020, 12, 118. [Google Scholar] [CrossRef]
- Froufe, E.; Alekseyev, S.; Alexandrino, P.; Weiss, S. The Evolutionary History of Sharp- and Blunt-Snouted Lenok (Brachymystax Lenok (Pallas, 1773)) and Its Implications for the Paleo-Hydrological History of Siberia. BMC Evol. Biol. 2008, 8, 40. [Google Scholar] [CrossRef]
- Bernatchez, L. The Evolutionary History of Brown Trout (Salmo trutta L.) Inferred from Phylogeographic, Nested Clade, and Mismatch Analyses of Mitochondrial DNA Variation. Evolution 2001, 55, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Böhm, M.; Dewhurst-Richman, N.I.; Seddon, M.; Ledger, S.E.H.; Albrecht, C.; Allen, D.; Bogan, A.E.; Cordeiro, J.; Cummings, K.S.; Cuttelod, A.; et al. The Conservation Status of the World’s Freshwater Molluscs. Hydrobiologia 2021, 848, 3231–3254. [Google Scholar] [CrossRef]
- Vaughn, C.C.; Nichols, S.J.; Spooner, D.E. Community and Foodweb Ecology of Freshwater Mussels. J. N. Am. Benthol. Soc. 2008, 27, 409–423. [Google Scholar] [CrossRef]
- Vaughn, C.C. Ecosystem Services Provided by Freshwater Mussels. Hydrobiologia 2017, 810, 15–27. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Kondakov, A.V.; Konopleva, E.S.; Vikhrev, I.V.; Aksenova, O.V.; Aksenov, A.S.; Bespalaya, Y.V.; Borovskoy, A.V.; Danilov, P.P.; Dvoryankin, G.A.; et al. Integrative Taxonomy, Biogeography and Conservation of Freshwater Mussels (Unionidae) in Russia. Sci. Rep. 2020, 10, 59867. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Lima, M.; Sousa, R.; Geist, J.; Aldridge, D.C.; Araujo, R.; Bergengren, J.; Bespalaya, Y.; Bódis, E.; Burlakova, L.; Van Damme, D.; et al. Conservation Status of Freshwater Mussels in Europe: State of the Art and Future Challenges. Biol. Rev. 2017, 92, 572–607. [Google Scholar] [CrossRef]
- Mcivor, A.L.; Aldridge, D.C. The Reproductive Biology of the Depressed River Mussel, Pseudanodonta Complanata (Bivalvia: Unionidae), with Implications for Its Conservation. J. Molluscan Stud. 2007, 73, 259–266. [Google Scholar] [CrossRef]
- Ćmiel, A.M.; Zając, K.; Lipińska, A.M.; Zając, T. Is Pseudanodonta Complanata the Most Vulnerable of Widespread European Species of Unionids? An Intense Stress Test Leading to a Massive Die-Off. Aquat. Conserv. Mar. Freshw. Ecosyst. 2019, 29, 2185–2192. [Google Scholar] [CrossRef]
- Ożgo, M.; Urbańska, M.; Hoos, P.; Imhof, H.K.; Kirschenstein, M.; Mayr, J.; Michl, F.; Tobiasz, R.; Von Wesendonk, M.; Zimmermann, S.; et al. Invasive Zebra Mussel (Dreissena Polymorpha) Threatens an Exceptionally Large Population of the Depressed River Mussel (Pseudanodonta Complanata) in a Postglacial Lake. Ecol. Evol. 2020, 10, 4918–4927. [Google Scholar] [CrossRef]
- Van Damme, D. Pseudanodonta complanata. Available online: https://doi.org/10.2305/IUCN.UK.2011-2.RLTS.T18446A8279278.en (accessed on 20 April 2023).
- Zając, K. Pseudanodonta Complanata (Rossmässler, 1835). Available online: http://www.iop.krakow.pl/pckz/opis6113-2.html?id=129&je=pl (accessed on 20 April 2023).
- Bonk, M. A New Locality of the Depressed River Mussel Pseudanodonta Complanata (Rossmässler, 1835) (Bivalvia: Unionidae) in the Wisłok River (Carpathian Mountains). Folia Malacol. 2019, 27, 71–74. [Google Scholar] [CrossRef]
- Ollard, I.; Aldridge, D.C. Declines in Freshwater Mussel Density, Size and Productivity in the River Thames over the Past Half Century. J. Anim. Ecol. 2023, 92, 112–123. [Google Scholar] [CrossRef]
- Geist, J. Strategies for the Conservation of Endangered Freshwater Pearl Mussels (Margaritifera margaritifera L.): A Synthesis of Conservation Genetics and Ecology. Hydrobiologia 2010, 644, 69–88. [Google Scholar] [CrossRef]
- Geist, J.; Kuehn, R. Genetic Diversity and Differentiation of Central European Freshwater Pearl Mussel (Margaritifera margaritifera L.) Populations: Implications for Conservation and Management. Mol. Ecol. 2005, 14, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.J. Population Genetics, Ecology and Waterway Management in the Conservation of Pseudanodonta Complanata (Rossmassler). Ph.D. Thesis, University of Cambridge, Cambridge, UK, 1999. [Google Scholar]
- Källersjö, M.; Von Proschwitz, T.; Lundberg, S.; Eldenäs, P.; Erséus, C. Evaluation of ITS RDNA as a Complement to Mitochondrial Gene Sequences for Phylogenetic Studies in Freshwater Mussels: An Example Using Unionidae from North-Western Europe. Zool. Scr. 2005, 34, 415–424. [Google Scholar] [CrossRef]
- Skidmore, R.; Leach, C.; Hoffman, J.; Amos, W.; Aldridge, D. Conservation Genetics of the Endangered Depressed River Mussel, Pseudanodonta Complanata, Using Amplified Fragment Length Polymorphism (AFLP) Markers. Aquat. Conserv. 2010, 20, 560–567. [Google Scholar] [CrossRef]
- Karlsson, S.; Larsen, B.M.; Eriksen, L.; Hagen, M. Four Methods of Nondestructive DNA Sampling from Freshwater Pearl Mussels Margaritifera margaritifera L. (Bivalvia:Unionoida). Freshw. Sci. 2013, 32, 525–530. [Google Scholar] [CrossRef]
- Berg, D.J.; Haag, W.R.; Guttman, S.I.; Sickel, J.B. Mantle Biopsy: A Technique for Nondestructive Tissue-Sampling of Freshwater Mussels. J. N. Am. Benthol. Soc. 1995, 14, 577–581. [Google Scholar] [CrossRef]
- Lehner, B.; Verdin, K.; Jarvis, A. New Global Hydrography Derived from Spaceborne Elevation Data. Eos Trans. AGU 2008, 89, 93–94. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Serb, J.M.; Buhay, J.E.; Lydeard, C. Molecular Systematics of the North American Freshwater Bivalve Genus Quadrula (Unionidae: Ambleminae) Based on Mitochondrial ND1 Sequences. Mol. Phylogenet. Evol. 2003, 28, 1–11. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Kondakov, A.V.; Vikhrev, I.V.; Aksenova, O.V.; Bespalaya, Y.V.; Gofarov, M.Y.; Kolosova, Y.S.; Konopleva, E.S.; Spitsyn, V.M.; Tanmuangpak, K.; et al. Ancient River Inference Explains Exceptional Oriental Freshwater Mussel Radiations. Sci. Rep. 2017, 7, 2312. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Snell, Q.; Walke, P.; Posada, D.; Crandall, K. TCS: Estimating Gene Genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium, Kyoto, Japan, 15–19 April 2002; p. 7. [Google Scholar]
- Froufe, E.; Sobral, C.; Teixeira, A.; Sousa, R.; Varandas, S.; Aldridge, D.C.; Lopes-Lima, M. Genetic Diversity of the Pan-European Freshwater Mussel Anodonta Anatina (Bivalvia: Unionoida) Based on CO1: New Phylogenetic Insights and Implications for Conservation. Aquat. Conserv. 2014, 24, 561–574. [Google Scholar] [CrossRef]
- Vavilov, N.I. The Origin, Variation, Immunity and Breeding of Cultivated Plants; Chester, K.S., Translator; Ronald Press: New York, NY, USA, 1951. [Google Scholar]
- Hewitt, G.M. Some Genetic Consequences of Ice Ages, and Their Role in Divergence and Speciation. Biol. J. Linn. Soc. 1996, 58, 247–276. [Google Scholar] [CrossRef]
- Jones, E.L. What Is a Refugium? Questions for the Middle–Upper Palaeolithic Transition in Peninsular Southern Europe. J. Quat. Sci. 2022, 37, 136–141. [Google Scholar] [CrossRef]
- Bennett, K.D.; Provan, J. What Do We Mean by “Refugia”? Quat. Sci. Rev. 2008, 27, 2449–2455. [Google Scholar] [CrossRef]
- Artamonova, V.S.; Bolotov, I.N.; Vinarski, M.V.; Makhrov, A.A. Fresh-and Brackish-Water Cold-Tolerant Species of Southern Europe: Migrants from the Paratethys That Colonized the Arctic. Water 2021, 13, 1161. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Konopleva, E.S.; Chan, N.; Lunn, Z.; Win, T.; Gofarov, M.Y.; Kondakov, A.V.; Tomilova, A.A.; Vikhrev, I.V. A Riverine Biodiversity Hotspot in Northern Myanmar Supports Three New and Narrowly Endemic Freshwater Mussel Species. Aquat. Conserv. 2022, 23, 3850. [Google Scholar] [CrossRef]
- Mangerud, J.; Jakobsson, M.; Alexanderson, H.; Astakhov, V.; Clarke, G.K.C.; Henriksen, M.; Hjort, C.; Krinner, G.; Lunkka, J.P.; Möller, P.; et al. Ice-Dammed Lakes and Rerouting of the Drainage of Northern Eurasia during the Last Glaciation. Quat. Sci. Rev. 2004, 23, 1313–1332. [Google Scholar] [CrossRef]
- Son, M.O.; Prokin, A.A.; Dubov, P.G.; Konopacka, A.; Grabowski, M.; Macneil, C.; Panov, V.E. Caspian Invaders vs. Ponto-Caspian Locals—Range Expansion of Invasive Macroinvertebrates from the Volga Basin Results in High Biological Pollution of the Lower Don River. Manag. Biol. Invasions 2020, 11, 178–200. [Google Scholar] [CrossRef]
- Artaev, O.; Pashkov, A.; Vekhov, D.; Saprykin, M.; Shapovalov, M.; Levina, M.; Levin, B. Fish Occurrence in the Kuban River Basin (Russia). Biodivers. Data J. 2021, 9, e76701. [Google Scholar] [CrossRef]
- Barinova, S.; Wink, M.; Makhrov, A.A.; Artamonova, V.S.; Sun, Y.-H.; Fang, Y.; Pashkov, A.N.; Reshetnikov, A.N. New Records of the Alien Chinese Ricefish (Oryzias Sinensis) and Its Dispersal History across Eurasia. Diversity 2023, 15, 317. [Google Scholar] [CrossRef]
- Mezhzherin, S.V.; Yanovich, L.M.; Zhalay, E.I.; Vasilieva, L.A.; Pampura, M.M. Genetic and morphological variability and differentiation of freshwater mussels (Bivavia, Unionidae, Anodontinae) in Ukraine. Vestn. Zool. 2014, 48, 99. [Google Scholar] [CrossRef]
- Prié, V.; Puillandre, N. Molecular phylogeny, taxonomy, and distribution of French Unio species (Bivalvia, Unionidae). Hydrobiologia 2014, 735, 95–110. [Google Scholar] [CrossRef]
- Riccardi, N.; Froufe, E.; Bogan, A.E.; Zieritz, A.; Teixeira, A.; Vanetti, I.; Varandas, S.; Zaccara, S.; Nagel, K.O.; Lopes-Lima, M. Phylogeny of European Anodontini (Bivalvia: Unionidae) with a redescription of Anodonta exulcerata. Zool. J. Linn. Soc. 2020, 189, 745–761. [Google Scholar] [CrossRef]
- Soroka, M. Characteristics of mitochondrial DNA of unionid bivalves (Mollusca: Bivalvia: Unionidae). I. Detection and characteristics of doubly uniparental inheritance (DUI) of unionid mitochondrial DNA. Folia Malacol. 2010, 18, 147–188. [Google Scholar] [CrossRef]
| Population Group | n | NH | Hd ± SD | π ± SD | Fu’s F-Test | Tajima’s D-Test | MMD under Spatial Expansion Model | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FS | p-Value | D | p-Value | SSD | p-Value | r | p-Value | |||||
| BS | 31 | 4 | 0.24 ± 0.1 | 0.0004 ± 0.0005 | −2.77 | 0.003 | −1.54 | 0.027 | 0.000 | 0.057 | 0.324 | 0.705 |
| DON | 9 | 7 | 0.94 ± 0.07 | 0.004 ± 0.003 | −3.37 | 0.009 | −1.44 | 0.080 | 0.012 | 0.423 | 0.068 | 0.473 |
| EUR | 18 | 7 | 0.57 ± 0.14 | 0.002 ± 0.002 | −2.75 | 0.02 | −2.28 | 0.001 | 0.003 | 0.86 | 0.063 | 0.898 |
| VOL | 47 | 7 | 0.28 ± 0.09 | 0.001 ± 0.001 | −7.23 | 0.0001 | −2.02 | 0.003 | 0.000 | 0.015 | 0.273 | 0.690 |
| panEUR (BS + EUR + VOL) | 96 | 16 | 0.32 ± 0.06 | 0.001 ± 0.001 | −21.39 | 0.0001 | −2.59 | 0.0001 | 0.000 | 0.15 | 0.230 | 0.877 |
| Molecular Marker | Group of Populations | BS | DON | EUR | VOL |
|---|---|---|---|---|---|
| COI | BS | 0.023 ± 0.006 | 0.001 | 0 | |
| DON | 0.951 | 0.024 ± 0.006 | 0.023 ± 0.006 | ||
| EUR | 0.022 | 0.888 | 0.001 | ||
| VOL | 0.010 | 0.957 | 0.032 | ||
| ND1 | DON | n/a | 0.021 ± 0.005 | 0.021 ± 0.005 | |
| EUR | n/a | 0.86 | 0.002 ± 0.001 | ||
| VOL | n/a | 0.89 | −0.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vikhrev, I.V.; Yunitsyna, O.A.; Kondakov, A.V.; Pestova, E.P.; Bovykina, G.V.; Konopleva, E.S.; Kruk, D.V.; Lyubas, A.A.; Soboleva, A.A.; Bolotov, I.N. Pleistocene Glacial Refugia in the Don River Basin: Witness from the Endangered Depressed River Mussel. Diversity 2023, 15, 829. https://doi.org/10.3390/d15070829
Vikhrev IV, Yunitsyna OA, Kondakov AV, Pestova EP, Bovykina GV, Konopleva ES, Kruk DV, Lyubas AA, Soboleva AA, Bolotov IN. Pleistocene Glacial Refugia in the Don River Basin: Witness from the Endangered Depressed River Mussel. Diversity. 2023; 15(7):829. https://doi.org/10.3390/d15070829
Chicago/Turabian StyleVikhrev, Ilya V., Olesya A. Yunitsyna, Alexander V. Kondakov, Elizaveta P. Pestova, Galina V. Bovykina, Ekaterina S. Konopleva, Darya V. Kruk, Artem A. Lyubas, Alena A. Soboleva, and Ivan N. Bolotov. 2023. "Pleistocene Glacial Refugia in the Don River Basin: Witness from the Endangered Depressed River Mussel" Diversity 15, no. 7: 829. https://doi.org/10.3390/d15070829
APA StyleVikhrev, I. V., Yunitsyna, O. A., Kondakov, A. V., Pestova, E. P., Bovykina, G. V., Konopleva, E. S., Kruk, D. V., Lyubas, A. A., Soboleva, A. A., & Bolotov, I. N. (2023). Pleistocene Glacial Refugia in the Don River Basin: Witness from the Endangered Depressed River Mussel. Diversity, 15(7), 829. https://doi.org/10.3390/d15070829






