Morphological and Molecular Characterization of Anisakid Nematode Larvae (Nematoda: Anisakidae) in the Black Cusk eel Genypterus maculatus from the Southeastern Pacific Ocean off Peru
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and Morphological Analyses
2.2. DNA Extraction, PCR Amplification and DNA Sequencing
2.3. Molecular Analyses
| Access | Species | Host | Country | Stage | Reference |
|---|---|---|---|---|---|
| DQ116432 | Skrjabinisakis physeteris | Physeter macrocephalus | Mediterranean Sea | A | [33] |
| AB592801 | Skrjabinisakis physeteris | Beryx splendens | Japan | L | [34] |
| OR192868 | Skrjabinisakis physeteris (Sphy1) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| OR192869 | Skrjabinisakis physeteris (Sphy2) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| OR192870 | Skrjabinisakis physeteris (Sphy3) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| OR192871 | Skrjabinisakis physeteris (Sphy4) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| OR192872 | Skrjabinisakis physeteris (Sphy5) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| OR192873 | Skrjabinisakis physeteris (Sphy6) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| MH669506 | Skrjabinisakis brevispiculata | Diaphus sp. | Indian Ocean | L | [35] |
| DQ116433 | Skrjabinisakis brevispiculata | Kogia breviceps | No registred | A | [33] |
| OR192874 | Skrjabinisakis brevispiculata (Sbre1) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| OR192875 | Skrjabinisakis brevispiculata (Sbre2) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| OR192876 | Skrjabinisakis brevispiculata (Sbre3) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| OR192877 | Skrjabinisakis brevispiculata (Sbre4) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| DQ116434 | Skrjabinisakis paggiae | Kogia breviceps | West Atlantic Ocean (Florida coast) | A | [33] |
| AB592807 | Skrjabinisakis paggiae | Beryx splendens | Japan | L | [34] |
| DQ116430 | Anisakis ziphidarum | Mesoplodon layardii | Southeast Atlantic Ocean (South African coast) | A | [33] |
| AB517573 | Anisakis ziphidarum | Scomber japonicus | Japan | L | [36] |
| DQ116431 | Anisakis nascetti | Mesoplodon miros | Southeast Atlantic Ocean (South African coast) | A | [33] |
| GQ118167 | Anisakis nascetti | Mesoplodon grayi | From off New Zealand | A | [37] |
| DQ116427 | Anisakis typica | Delphinidae | Western North Atlantic Ocean | A | [33] |
| KC928266 | Anisakis typica | Katsuwonus pelamis | Southern Makassar Strait, Indonesia | L | [38] |
| KC810003 | Anisakis simplex | Balaenoptera acutorostrata | Northeastern Atlantic Ocean (Norwegian coast) | A | [39] |
| DQ116426 | Anisakis simplex | Delphinidae | Northeast Pacific coast | A | [33] |
| MZ546440 | Anisakis pegreffii | Seriolella violacea | Southeastern Pacific Ocean | L | [20] |
| DQ116428 | Anisakis pegreffii | Delphinus delphis | Northeast Atlantic Ocean (Spanish coast) | A | [33] |
| OR192866 | Anisakis pegreffii (Apeg1) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| OR192867 | Anisakis pegreffii (Apeg2) | Genypterus maculatus | Southeastern Pacific Ocean | L | Present study |
| MN385245 | Anisakis berlandi | Globicephala melas | New Zealand | A | [40] |
| KC810000 | Anisakis berlandi | Globicephala melas | New Zealand | A | [39] |
| JQ934891 | Hysterothylacium aduncum * | Trachurus trachurus | Croatia | L | [41] |
3. Results
3.1. Systematics and Morphological Characteristics of the Anisakid Larvae
- Class Chromadorea Inglis, 1983.
- Order Rhabditida Chitwood, 1933.
- Anisakidae Railliet and Henry, 1912.
3.1.1. Anisakis pegreffii Campana-Rouget and Biocca, 1955 (Figure 1A,B and Figure 2A,B)
- Site in host: body cavity.
- Specimens deposited: Hologenophore (MUSM-HEL 5141).
- Representative DNA sequence: Sequences were deposited in GenBank under the accession numbers OR192866 and OR192867 for the mtDNA cox2.
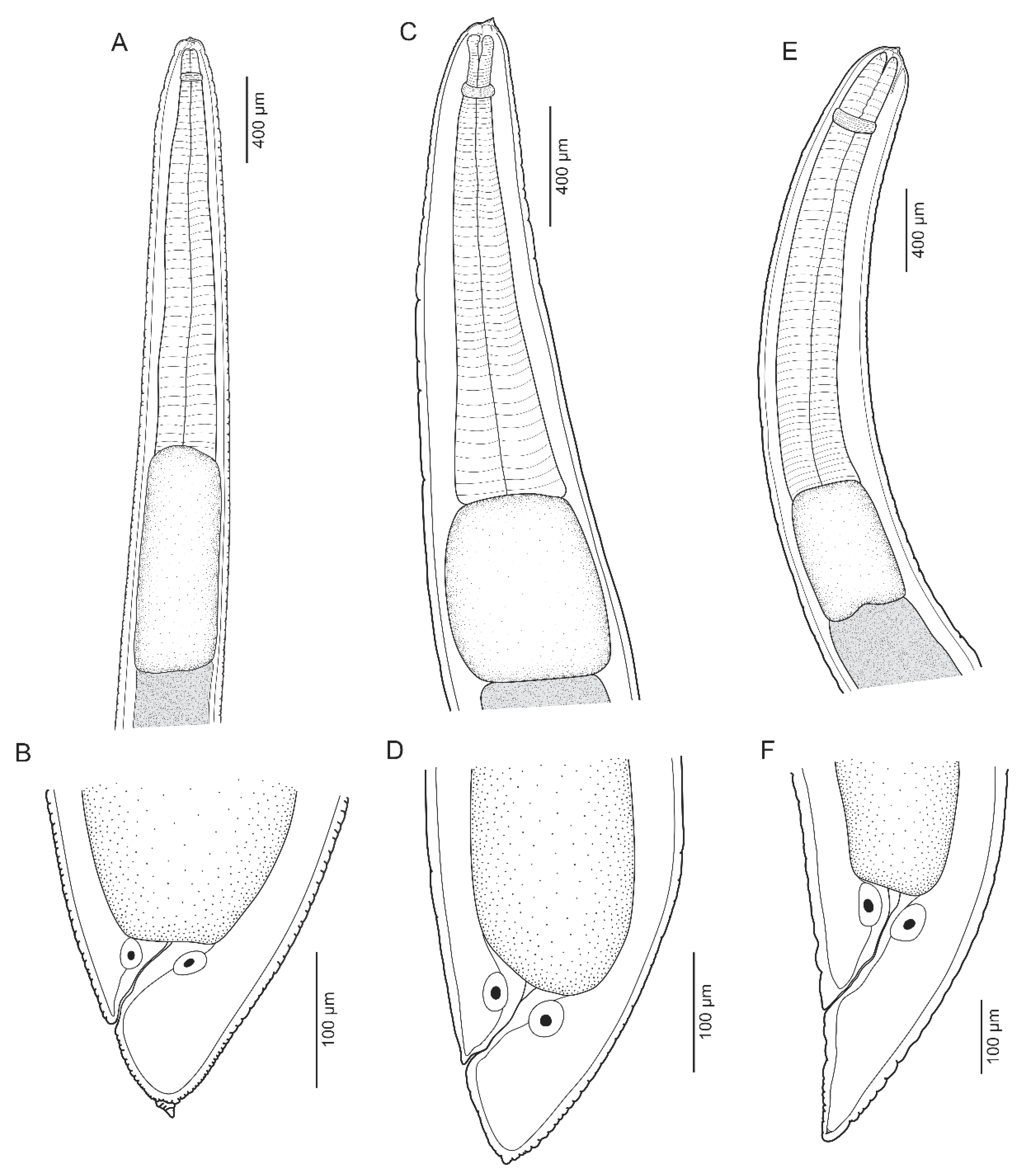
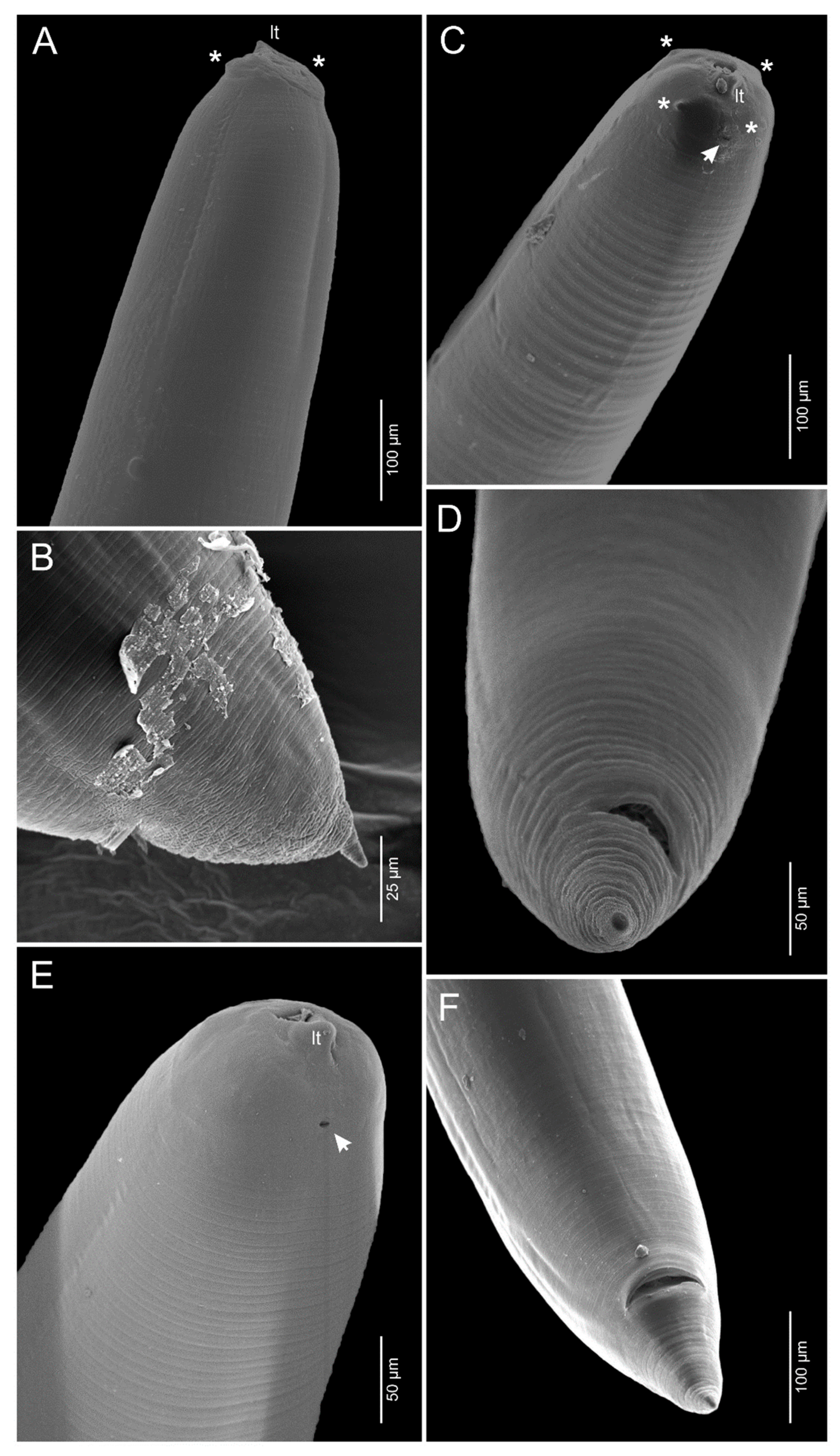
3.1.2. Skrjabinisakis brevispiculata (Dollfus, 1966) Safonova, Voronova and Vainutis, 2021 (Figure 1C,D and Figure 2C,D)
- Site in host: body cavity.
- Specimens deposited: Hologenophore (MUSM-HEL 5142).
- Representative DNA sequence: Sequences were deposited in GenBank under the accession numbers OR192874–OR192877 for the mtDNA cox2.
3.1.3. Skrjabinisakis physeteris (Baylis, 1923) Safonova, Voronova and Vainutis, 2021 (Figure 1E,F and Figure 2E,F)
- Site in host: body cavity.
- Specimens deposited: Hologenophore (MUSM-HEL 5143).
- Representative DNA sequence: Sequences were deposited in GenBank under the accession numbers OR192868–OR192873 for the mtDNA cox2.
3.2. Phylogenetic Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Froese, R.; Pauly, D. FishBase. Available online: https://www.fishbase.se/search.php (accessed on 24 May 2023).
- Reyes, P.; Hüne, M. Peces del sur de Chile; Ocho Libros: Santiago de Chile, Chile, 2012; p. 497. [Google Scholar]
- SERNAPESCA. Anuario Estadístico de Pesca. Servicio Nacional de Pesca. Ministerio de Economía, Fomento y Reconstrucción. Chile. Available online: http://www.sernapesca.cl/informacion-utilidad/anuarios-estadisticos-de-pesca-y-acuicultura (accessed on 24 May 2023).
- Bahamonde, N.N.; Zavala, P.F. Contenidos gástricos en Genypterus maculatus (Tschudi) y Genypterus blacodes (Schneider) capturados en Chile entre 31° y 37° S. Boletín Del Mus. Nac. De História Nat. Chile 1981, 38, 53–59. [Google Scholar]
- Chirichigno, N.; Vélez, M. Clave para identificar los peces marinos del Perú. In Publicación Especial del Instituto del Mar, 2nd ed.; Instituto del Mar del Perú: Callao, Peru, 1998; p. 500. [Google Scholar]
- Shamsi, S. The occurrence of Anisakis spp. in Australian waters: Past, present, and future trends. Parasitol. Res. 2021, 120, 3007–3033. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, N.A.; Abdel-Ghaffar, F.; Fayed, H.O.; Hassan, A.A. Morphological and molecular identification of third-stage larvae of Anisakis typica (Nematoda: Anisakidae) from Red Sea coral trout. Plectropomus Areolatus. Parasitol. Res. 2023, 122, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.W.; Wootten, R. Anisakis and anisakiasis. Adv. Parasitol. 1978, 16, 93–163. [Google Scholar]
- Fiorenza, E.A.; Wendt, C.A.; Dobkowski, K.A.; King, T.L.; Pappaionou, M.; Rabinowitz, P.; Samhouri, J.F.; Wood, C.L. It’s a wormy world: Meta-analysis reveals several decades of change in the global abundance of the parasitic nematodes Anisakis spp. and Pseudoterranova spp. in marine fishes and invertebrates. Glob. Chang. Biol. 2020, 26, 2854–2866. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, S. Parasite loss or parasite gain? Story of Contracaecum nematodes in antipodean waters. Parasite Epidemiol. Control 2019, 4, e00087. [Google Scholar] [CrossRef] [PubMed]
- Mattiucci, S.; Nascetti, G. Advances and trends in the molecular systematics of anisakid nematodes, with implications for their evolutionary ecology and host-parasite co-evolutionary processes. Adv. Parasitol. 2008, 66, 47–148. [Google Scholar]
- Bao, M.; Pierce, G.J.; Pascual, S.; González-Muñoz, M.; Mattiucci, S.; Mladineo, I.; Cipriani, P.; Bušelić, I.; Strachan, N.J. Assessing the risk of an emerging zoonosis of worldwide concern: Anisakiasis. Sci. Rep. 2017, 7, 43699. [Google Scholar] [CrossRef]
- Chen, H.X.; Zhang, L.P.; Gibson, D.I.; Lu, L.; Xu, Z.; Li, H.T.; Ju, H.D.; Li, L. Detection of ascaridoid nematode parasites in the important marine food-fish Conger myriaster (Brevoort) (Anguilliformes: Congridae) from the Zhoushan Fishery, China. Parasites Vectors 2018, 11, 274. [Google Scholar] [CrossRef]
- Shamsi, S.; Chen, Y.; Poupa, A.; Ghadam, M.; Justine, J.L. Occurrence of anisakid parasites in marine fishes and whales off New Caledonia. Parasitol. Res. 2018, 117, 3195–3204. [Google Scholar] [CrossRef]
- Mattiucci, S.; Cipriani, P.; Levsen, A.; Paoletti, M.; Nascetti, G. Molecular epidemiology of Anisakis and anisakiasis: An ecological and evolutionary road map. Adv. Parasitol. 2018, 99, 93–263. [Google Scholar] [PubMed]
- Lin, R.Q.; Dong, S.J.; Nie, K.; Wang, C.R.; Li, A.X.; Song, H.Q.; Huang, W.Y.; Zhu, X.Q. Sequence analysis of the first internal transcribed spacer of rDNA supports the existence of an intermediate Fasciola between F. hepatica and F. gigantica in main land China. Parasitol. Res. 2007, 101, 813–817. [Google Scholar] [CrossRef]
- Tunya, R.; Wongsawad, C.; Wongsawad, P.; Chai, J.Y. Morphological and molecular characteristics of Anisakis typica larvae in two species of threadfin bream, Nemipterus hexodon and N. japonicus, from the Gulf of Thailand. Korean J. Parasitol. 2020, 58, 15–25. [Google Scholar] [CrossRef]
- Luque, J.L.; Cruces, C.; Chero, J.; Paschoal, F.; Alves, P.V.; Da Silva, A.C.; Sánchez, L.; Iannacone, J. Checklist of metazoan parasites of fishes from Peru. Neotrop. Helminthol. 2016, 10, 301–375. [Google Scholar]
- Aco Alburqueque, R.; Palomba, M.; Santoro, M.; Mattiucci, S. Molecular identification of zoonotic parasites of the genus Anisakis (nematoda: Anisakidae) from fish of the southeastern Pacific Ocean (off Peru coast). Pathogens 2020, 9, 910. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Rojas, R.; Mondragón-Martínez, A.; De los Santos, E.R.; Cruz-Neyra, L.; García-Candela, E.; Delgado-Escalante, A.; Sanchez-Venegas, J.R. Molecular identification and epidemiological data of Anisakis spp. (Nematoda: Anisakidae) larvae from Southeastern Pacific Ocean off Peru. Int. J. Parasitol. Parasites Wildl. 2021, 16, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Bush, A.O.; Lafferty, K.D.; Lotz, J.M.; Shostak, A.W. Parasitology meets ecology on its own terms: Margolis et al. revisited. J. Parasitol. 1997, 83, 575–583. [Google Scholar] [CrossRef]
- Nadler, S.A.; Hudspeth, D.S.S. Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology: Hypotheses of structural and sequence evolution. J. Parasitol. 2000, 86, 380–393. [Google Scholar] [CrossRef]
- Filatov, D.A. Proseq: A software for preparation and evolutionary analysis of DNA sequence data sets. Mol. Ecol. Notes 2002, 2, 621–624. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Santorum, J.M.; Darriba, D.; Taboada, G.L.; Posada, D. jmodeltest.org: Selection of nucleotide substitution models on the cloud. Bioinformatics 2014, 30, 1310–1311. [Google Scholar] [CrossRef] [PubMed]
- Akaike, H. A new look at the statistical model identification. Sel. Pap. Hirotugu Akaike. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.; Teslenko, M. Draft MrBayes Version 3.2 Manual: Tutorials and Model Summaries. 2011. Available online: https://bioweb.pasteur.fr/docs/modules/mrbayes/3.1.2/Manual_MrBayes_v3.2.0_draft.pdf (accessed on 2 April 2023).
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES science gateway: A community resource for phylogenetic analyses. In Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, Salt Lake City, UT, USA, 18–21 July 2011; pp. 1–8. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kimura, M.A. Simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Valentini, A.; Mattiucci, S.; Bondanelli, P.; Webb, S.C.; Mignucci-Giannone, A.A.; Colom-Llavina, M.M.; Nascetti, G. Genetic relationships among Anisakis species (Nematoda: Anisakidae) inferred from mitochondrial cox-2 sequences, and comparison with allozyme data. J. Parasitol. 2006, 92, 156. [Google Scholar] [CrossRef]
- Murata, R.; Suzuki, J.; Sadamasu, K.; Kai, A. Morphological and molecular characterization of Anisakis larvae (Nematoda: Anisakidae) in Beryx splendens from Japanese waters. Parasitol. Int. 2011, 60, 193–198. [Google Scholar] [CrossRef]
- Cabrera-Gil, S.; Deshmukh, A.; Cervera-Estevan, C.; Fraija-Fernández, N.; Fernández, M.; Aznar, F.J. Anisakis infections in lantern fish (Myctophidae) from the Arabian Sea: A dual role for lantern fish in the life cycle of Anisakis brevispiculata? Deep Sea Res. Part I Oceanogr. Res. Pap. 2018, 141, 43–50. [Google Scholar] [CrossRef]
- Suzuki, J.; Murata, R.; Hosaka, M.; Araki, J. Risk factors for human Anisakis infection and association between the geographic origins of Scomber japonicus and anisakid nematodes. Int. J. Food Microbiol. 2010, 137, 88–93. [Google Scholar] [CrossRef]
- Mattiucci, S.; Paoletti, M.; Webb, S.C. Anisakis nascettii n. sp. (Nematoda: Anisakidae) from beaked whales of the southern hemisphere: Morphological description, genetic relationships between congeners and ecological data. Syst. Parasitol. 2009, 74, 199–217. [Google Scholar] [CrossRef] [PubMed]
- Anshary, H.; Sriwulan; Freeman, M.A.; Ogawa, K. Occurrence and molecular identification of Anisakis Dujardin, 1845 from marine fish in southern Makassar Strait, Indonesia. Korean J. Parasitol. 2014, 52, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Mattiucci, S.; Cipriani, P.; Webb, S.C.; Paoletti, M.; Marcer, F.; Bellisario, B.; Gibson, D.I.; Nascetti, G. Genetic and morphological approaches distinguish the three sibling species of the Anisakis simplex species complex, with a species designation as Anisakis berlandi sp. for A. simplex sp. C (Nematoda: Anisakidae). J. Parasitol. 2014, 100, 199–214. [Google Scholar] [CrossRef]
- Bello, E.; Paoletti, M.; Webb, S.C.; Nascetti, G.; Mattiucci, S. Cross-species utility of microsatellite loci for the genetic characterisation of Anisakis berlandi (Nematoda: Anisakidae). Parasite 2020, 27, 9. [Google Scholar] [CrossRef] [PubMed]
- Vardić Smrzlić, I.; Valic, D.; Kapetanovic, D.; Kurtovic, B.; Teskeredzic, E. Molecular characterisation of Anisakidae larvae from fish in Adriatic Sea. Parasitol. Res. 2012, 111, 2385–2391. [Google Scholar] [CrossRef]
- Nielsen, J.G.; Cohen, D.M.; Markle, D.F.; Robins, C.R. Ophidiiform fishes of the world (Order Ophidiiformes). An annotated and illustrated catalogue of pearlfishes, cusk-eels, brotulas and other ophidiiform fishes known to date. FAO Fish. Synop. 1999, 125, 178. [Google Scholar]
- Berland, B. Nematodes from some Norwegian marine fishes. Sarsia 1961, 2, 1–50. [Google Scholar] [CrossRef]
- Safonova, A.E.; Voronova, A.N.; Vainutis, K.S. First report on molecular identification of Anisakis simplex in Oncorhynchus nerka from the fish market, with taxonomical issues within Anisakidae. J. Nematol. 2021, 53, 1–10. [Google Scholar] [CrossRef]
- Takano, T.; Sata, N. Multigene phylogenetic analysis reveals non-monophyly of Anisakis sl and Pseudoterranova (Nematoda: Anisakidae). Parasitol. Int. 2022, 91, 102631. [Google Scholar] [CrossRef]
- Bao, M.; Olsen, K.M.; Levsen, A.; Cipriani, P.; Giulietti, L.; Storesund, J.E.; García-Seoane, E.; Karlsbakk, E. Characterization of Pseudoterranova ceticola (Nematoda: Anisakidae) larvae from meso/bathypelagic fishes off Macaronesia (NW Africa waters). Sci. Rep. 2022, 12, 17695. [Google Scholar] [CrossRef]
- Cipriani, P.; Palomba, M.; Giulietti, L.; Marcer, F.; Mazzariol, S.; Santoro, M.; Mattiucci, S. Distribution and genetic diversity of Anisakis spp. in cetaceans from the Northeast Atlantic Ocean and the Mediterranean Sea. Sci. Rep. 2022, 12, 13664. [Google Scholar] [CrossRef] [PubMed]
- Mattiucci, S.; Paggi, L.; Nascetti, G.; Abollo, E.; Webb, S.C.; Pascual, S.; Cianchi, R.; Bullini, L. Genetic divergence and reproductive isolation between Anisakis brevispiculata and Anisakis physeteris (Nematoda: Anisakidae) s. Int. J. Parasitol. 2001, 31, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Quiazon, K.M.A.; Yoshinaga, T.; Santos, M.D.; Ogawa, K. Identification of larval Anisakis spp. (Nematoda: Anisakidae) in Alaska pollock (Theragra chalcogramma) in northern Japan using morphological and molecular markers. J. Parasitol. 2009, 95, 1227–1232. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chero, J.D.; Ñacari, L.; Cruces, C.L.; Lopez, D.F.; Cacique, E.; Severino, R.; Lopez, J.; Luque, J.L.; Saéz, G. Morphological and Molecular Characterization of Anisakid Nematode Larvae (Nematoda: Anisakidae) in the Black Cusk eel Genypterus maculatus from the Southeastern Pacific Ocean off Peru. Diversity 2023, 15, 820. https://doi.org/10.3390/d15070820
Chero JD, Ñacari L, Cruces CL, Lopez DF, Cacique E, Severino R, Lopez J, Luque JL, Saéz G. Morphological and Molecular Characterization of Anisakid Nematode Larvae (Nematoda: Anisakidae) in the Black Cusk eel Genypterus maculatus from the Southeastern Pacific Ocean off Peru. Diversity. 2023; 15(7):820. https://doi.org/10.3390/d15070820
Chicago/Turabian StyleChero, Jhon Darly, Luis Ñacari, Celso Luis Cruces, David Fermín Lopez, Edson Cacique, Ruperto Severino, Jorge Lopez, José Luis Luque, and Gloria Saéz. 2023. "Morphological and Molecular Characterization of Anisakid Nematode Larvae (Nematoda: Anisakidae) in the Black Cusk eel Genypterus maculatus from the Southeastern Pacific Ocean off Peru" Diversity 15, no. 7: 820. https://doi.org/10.3390/d15070820
APA StyleChero, J. D., Ñacari, L., Cruces, C. L., Lopez, D. F., Cacique, E., Severino, R., Lopez, J., Luque, J. L., & Saéz, G. (2023). Morphological and Molecular Characterization of Anisakid Nematode Larvae (Nematoda: Anisakidae) in the Black Cusk eel Genypterus maculatus from the Southeastern Pacific Ocean off Peru. Diversity, 15(7), 820. https://doi.org/10.3390/d15070820










