Cryptic Clitellata: Molecular Species Delimitation of Clitellate Worms (Annelida): An Overview
Abstract
1. Introduction
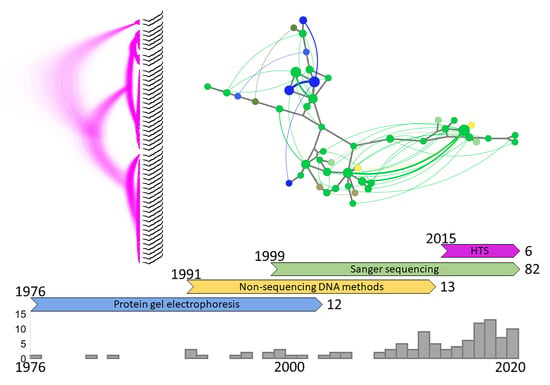
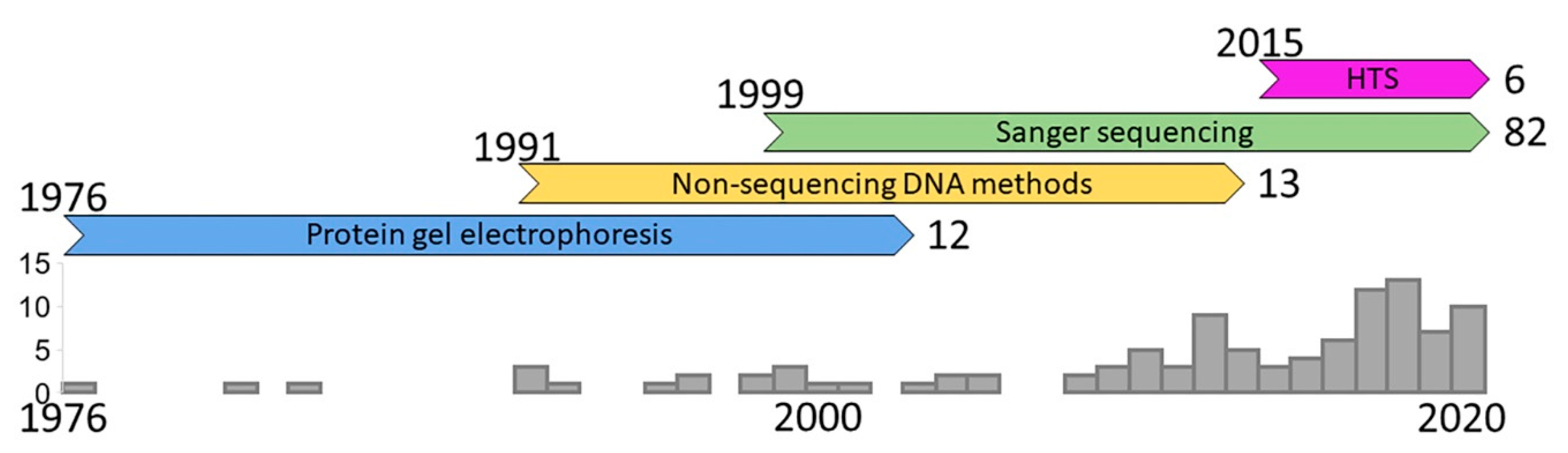
2. History of the Field
2.1. Protein Gel Electrophoresis
2.2. Non-Sequencing DNA Methods
2.3. Sanger Sequencing
2.4. High-Throughput Sequencing (HTS)
3. Taxonomical Treatment of Delimited Species
4. Future Development of the Field
5. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sites, J.W.; Marshall, J.C. Delimiting species: A Renaissance issue in systematic biology. Trends Ecol. Evol. 2003, 18, 462–470. [Google Scholar] [CrossRef]
- Fujita, M.K.; Leache, A.D.; Burbrink, F.T.; McGuire, J.A.; Moritz, C. Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol. Evol. 2012, 27, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Pfenninger, M.; Schwenk, K. Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evol. Biol. 2007, 7, 121. [Google Scholar] [CrossRef]
- Perez-Ponce de Leon, G.; Poulin, R. Taxonomic distribution of cryptic diversity among metazoans: Not so homogeneous after all. Biol. Lett. 2016, 12. [Google Scholar] [CrossRef]
- Erséus, C.; Gustafsson, D. Cryptic speciation in clitellate model organisms. In Annelids in Modern Biology; Shain, D.H., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; pp. 31–46. [Google Scholar] [CrossRef]
- Nygren, A. Cryptic polychaete diversity: A review. Zool. Scr. 2014, 43, 172–183. [Google Scholar] [CrossRef]
- Feckler, A.; Schulz, R.; Bundschuh, M. Cryptic lineages-same but different? Integr. Environ. Assess. Manag. 2013, 9, 172–173. [Google Scholar] [CrossRef]
- Marchán, D.F.; Díaz Cosín, D.J.; Novo, M. Why are we blind to cryptic species? Lessons from the eyeless. Eur. J. Soil Biol. 2018, 86, 49–51. [Google Scholar] [CrossRef]
- Halanych, K.M.; Borda, E. Developing models for Lophotrochozoan and annelid Biology. In Annelids in Modern Biology; Shain, D.H., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2009; pp. 1–12. [Google Scholar]
- Römbke, J.; Egeler, P. Oligochaete worms for ecotoxicological assessment of soils and sediments. In Annelids in Modern Biology; Shain, D.H., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2009; pp. 228–241. [Google Scholar] [CrossRef]
- Erséus, C.; Klinth, M.J.; Rota, E.; De Wit, P.; Gustafsson, D.R.; Martinsson, S. The popular model annelid Enchytraeus albidus is only one species in a complex of seashore white worms (Clitellata, Enchytraeidae). Org. Divers. Evol. 2019, 19, 105–133. [Google Scholar] [CrossRef]
- Gustafsson, D.R.; Price, D.A.; Erséus, C. Genetic variation in the popular lab worm Lumbriculus variegatus (Annelida: Clitellata: Lumbriculidae) reveals cryptic speciation. Mol. Phylogenet Evol. 2009, 51, 182–189. [Google Scholar] [CrossRef]
- Kille, P.; Andre, J.; Anderson, C.; Ang, H.N.; Bruford, M.W.; Bundy, J.G.; Donnelly, R.; Hodson, M.E.; Juma, G.; Lahive, E.; et al. DNA sequence variation and methylation in an arsenic tolerant earthworm population. Soil Biol. Biochem. 2013, 57, 524–532. [Google Scholar] [CrossRef]
- Römbke, J.; Aira, M.; Backeljau, T.; Breugelmans, K.; Dominguez, J.; Funke, E.; Graf, N.; Hajibabaei, M.; Perez-Losada, M.; Porto, P.G.; et al. DNA barcoding of earthworms (Eisenia fetida/andrei complex) from 28 ecotoxicological test laboratories. Appl. Soil Ecol. 2016, 104, 3–11. [Google Scholar] [CrossRef]
- Trontelj, P.; Utevsky, S.Y. Celebrity with a neglected taxonomy: Molecular systematics of the medicinal leech (genus Hirudo). Mol. Phylogenet Evol. 2005, 34, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Struck, T.H.; Golombek, A.; Weigert, A.; Franke, F.A.; Westheide, W.; Purschke, G.; Bleidorn, C.; Halanych, K.M. The evolution of annelids reveals two adaptive routes to the interstitial realm. Curr. Biol. 2015, 25, 1993–1999. [Google Scholar] [CrossRef] [PubMed]
- Struck, T.H.; Paul, C.; Hill, N.; Hartmann, S.; Hosel, C.; Kube, M.; Lieb, B.; Meyer, A.; Tiedemann, R.; Purschke, G.; et al. Phylogenomic analyses unravel annelid evolution. Nature 2011, 471, 95–98. [Google Scholar] [CrossRef]
- Erséus, C.; Williams, B.W.; Horn, K.M.; Halanych, K.M.; Santos, S.R.; James, S.W.; des Chatelliers, M.C.; Anderson, F.E. Phylogenomic analyses reveal a Palaeozoic radiation and support a freshwater origin for clitellate annelids. Zool. Scr. 2020, 49, 614–640. [Google Scholar] [CrossRef]
- Timm, T. Life forms in Oligochaeta: A literature review. Zool. Middle East 2012, 58, 71–82. [Google Scholar] [CrossRef]
- Kuo, D.H. The polychaete-to-clitellate transition: An EvoDevo perspective. Dev. Biol. 2017. [Google Scholar] [CrossRef]
- Brinkhurst, R.O.; Jamieson, B.G.M. Aquatic Oligochaeta of the World; Oliver and Boyd: Edinburgh, UK, 1971; p. 860. [Google Scholar]
- Cerca, J.; Meyer, C.; Purschke, G.; Struck, T.H. Delimitation of cryptic species drastically reduces the geographical ranges of marine interstitial ghost-worms (Stygocapitella; Annelida, Sedentaria). Mol. Phylogenet Evol. 2019. [Google Scholar] [CrossRef]
- Carstens, B.C.; Pelletier, T.A.; Reid, N.M.; Satler, J.D. How to fail at species delimitation. Mol. Ecol. 2013, 22, 4369–4383. [Google Scholar] [CrossRef]
- Mann, D.G.; Evans, K.M. The species concept and cryptic diversity. In Proceedings of the 12th International Conference on Harmful Algae, Copenhagen, Denmark, 4–8 September 2006; Moestrup, Ø., Doucette, G., Enevoldsen, H., Godhe, A., Hallegraeff, G., Luckas, B., Lundholm, N., Lewis, J., Rengefors, K., Sellner, K., et al., Eds.; International Society for the Study of Harmful Algae and Intergovernmental Oceanographic Commission of UNESCO: Copenhagen, Denmark, 2008. [Google Scholar]
- Christensen, B.; Jelnes, J. Sibling species in the oligochaete worm Lumbricillus rivalis (Enchytraeidae) revealed by enzyme polymorphisms and breeding experiments. Hereditas 1976, 83, 237–243. [Google Scholar] [CrossRef]
- Øien, N.; Stenersen, J. Esterases of earthworms—III. Electrophoresis reveals that Eisenia fetida (Savigny) is two species. Comp. Biochem. Physiol. Part C Comp. Pharmacol. 1984, 78, 277–282. [Google Scholar] [CrossRef]
- Jaenike, J. “Eisenia foetida” is two biological species. Megadrilogica 1982, 4, 6–8. [Google Scholar]
- Milbrink, G.; Nyman, L. Protein Taxonomy of Aquatic Oligochaetes and Its Ecological Applications. Oikos 1973, 24, 473. [Google Scholar] [CrossRef]
- Brockmeyer, V. Isozymes and general protein patterns for use in discrimination and identification of Enchytraeus species (Annelida, Oligochaeta). Z. Zool. Syst. Evol. 1991, 29, 343–361. [Google Scholar] [CrossRef]
- Christensen, B.; Hvilsom, M.; Pedersen, B.V. Genetic variation in coexisting sexual diploid and parthenogenetic triploid forms of Fridericia galba (Enchytraeidae, Oligochaeta) in a heterogeneous environment. Hereditas 1992, 117, 153–162. [Google Scholar] [CrossRef]
- Holmstrup, M.; Simonsen, V. Genetic and physiological differences between two morphs of the lumbricid earthworm Dendrodrilus rubidus (Savigny, 1826). Soil Biol. Biochem. 1996, 28, 1105–1107. [Google Scholar] [CrossRef]
- Schmelz, R. Separation of sympatric Fridericia species (Enchytraeidae, Oligochaeta) with isozyme and general protein patterns. Newsl. Enchytraeidae 1995, 4, 97–104. [Google Scholar]
- Collado, R.; Hass-Cordes, E.; Schmelz, R.M. Microtaxonomy of fragmenting Enchytraeus species using molecular markers, with a comment on species complexes in enchytraeids. Turk. J. Zool. 2012, 36, 85–94. [Google Scholar] [CrossRef]
- Schmelz, R.M.; Collado, R.; Myohara, M. A Taxonomic Study of Enchytraeus japonensis (Enchytraeidae, Oligochaeta): Morphological and Biochemical Comparisons with E. bigeminus. Zool. Sci. 2000, 17, 505–516. [Google Scholar] [CrossRef]
- Westheide, W.; Brockmeyer, V. Suggestions for an index of enchytraeid species (Oligochaeta) based on general protein patterns. Z. Zool. Syst. Evol. 1992, 30, 89–99. [Google Scholar] [CrossRef]
- Schmelz, R.M. Species separation and identification in the Enchytraeidae (Oligochaeta, Annelida): Combining morphology with general protein data. Hydrobiologia 1996, 334, 31–36. [Google Scholar] [CrossRef]
- Schmelz, R.M. Taxonomy of Fridericia (Oligochaeta, Enchytraeidae). Revision of species with morphological and biochemical methods. Abh. Nat. Ver. Hambg. (Neue Folge) 2003, 38, 1-415, 73 figs. [Google Scholar]
- Gabrich, A.; Jaros, P.P.; Brockmayer, V. Application of immunological methods for the taxonomic study of two selected animal taxa: Tisbe (Crustacea, Copepoda) and Enchytraeus (Annelida, Oligochaeta). Z. Zool. Syst. Evol. 1991, 29, 381–392. [Google Scholar] [CrossRef]
- Avise, J.; Lansman, R.; Shade, R. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics 1979, 92, 279–295. [Google Scholar]
- Schlegel, M.; Steinbrück, G.; Kramer, M.; Brockmeyer, V. Restriction fragment patterns as molecular markers for species identification and phylogenetic analysis in the genus Enchytraeus (Oligochaeta). Z. Zool. Syst. Evol. 1991, 29, 362–372. [Google Scholar] [CrossRef]
- Welsh, J.; McClelland, M. Fingerprinting Genomes Using PCR with Arbitrary Primers. Nucleic Acids Res. 1990, 18, 7213–7218. [Google Scholar] [CrossRef]
- Koperski, P.; Milanowski, R.; Krzyk, A. Searching for cryptic species in Erpobdella octoculata (L.) (Hirudinea: Clitellata): Discordance between the results of genetic analysis and cross-breeding experiments. Contrib. Zool. 2011, 80, 85–94. [Google Scholar] [CrossRef]
- Williams, J.G.K.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA Polymorphisms Amplified by Arbitrary Primers Are Useful as Genetic-Markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef]
- Bielecki, A.; Polok, K. Genetic variation and species identification among selected leeches (Hirudinea) revealed by RAPD markers. Biologia 2012, 67. [Google Scholar] [CrossRef]
- Schirmacher, A.; Schmidt, H.; Westheide, W. RAPD-PCR investigations on sibling species of terrestrial Enchytraeus (Annelida: Oligochaeta). Biochem. Syst. Ecol. 1998, 26, 35–44. [Google Scholar] [CrossRef]
- Trontelj, P.; Sotler, M.; Verovnik, R. Genetic differentiation between two species of the medicinal leech, Hirudo medicinalis and the neglected H. verbana, based on random-amplified polymorphic DNA. Parasitol. Res. 2004, 94, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Verovnik, R.; Trontelj, P.; Sket, B. Genetic differentiation and species status within the snail leech Glossiphonia complanata aggregate (Hirudinea: Glossiphoniidae) revealed by RAPD analysis. Arch. Hydrobiol. 1999, 144, 327–338. [Google Scholar] [CrossRef]
- Dyer, A.R.; Fowler, J.C.S.; Baker, G.H. Detecting genetic variation in exotic earthworms, Aporrectodea spp. (Lumbricidae), in Australian soils using RAPD markers. Soil Biol. Biochem. 1998, 30, 159–165. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- King, R.A.; Tibble, A.L.; Symondson, W.O. Opening a can of worms: Unprecedented sympatric cryptic diversity within British lumbricid earthworms. Mol. Ecol. 2008, 17, 4684–4698. [Google Scholar] [CrossRef]
- Andre, J.; King, R.A.; Sturzenbaum, S.R.; Kille, P.; Hodson, M.E.; Morgan, A.J. Molecular genetic differentiation in earthworms inhabiting a heterogeneous Pb-polluted landscape. Environ. Pollut. 2010, 158, 883–890. [Google Scholar] [CrossRef]
- Govedich, F.R.; Blinn, D.W.; Hevly, R.H.; Keim, P.S. Cryptic radiation in erpobdellid leeches in xeric landscapes: A molecular analysis of population differentiation. Can. J. Zool. 1999, 77, 52–57. [Google Scholar] [CrossRef]
- Spritz, R.A. Duplication/deletion polymorphism 5′-to the human beta globin gene. Nucleic Acids Res. 1981, 9, 5037–5047. [Google Scholar] [CrossRef]
- Tautz, D. Hypervariability of Simple Sequences as a General Source for Polymorphic DNA Markers. Nucleic Acids Res. 1989, 17, 6463–6471. [Google Scholar] [CrossRef]
- Weber, J.L.; May, P.E. Abundant Class of Human DNA Polymorphisms Which Can Be Typed Using the Polymerase Chain-Reaction. Am. J. Hum. Genet. 1989, 44, 388–396. [Google Scholar] [PubMed]
- Litt, M.; Luty, J.A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet. 1989, 44, 397–401. [Google Scholar] [PubMed]
- Donnelly, R.K.; Harper, G.L.; Morgan, A.J.; Orozco-Terwengel, P.; Pinto-Juma, G.A.; Bruford, M.W. Nuclear DNA recapitulates the cryptic mitochondrial lineages of Lumbricus rubellus and suggests the existence of cryptic species in an ecotoxological soil sentinel. Biol. J. Linn. Soc. 2013, 110, 780–795. [Google Scholar] [CrossRef][Green Version]
- Dupont, L.; Lazrek, F.; Porco, D.; King, R.A.; Rougerie, R.; Symondson, W.O.C.; Livet, A.; Richard, B.; Decaëns, T.; Butt, K.R.; et al. New insight into the genetic structure of the Allolobophora chlorotica aggregate in Europe using microsatellite and mitochondrial data. Pedobiologia 2011, 54, 217–224. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Coulson, A.R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975, 94, 441–448. [Google Scholar] [CrossRef]
- Beauchamp, K.A.; Kathman, R.D.; McDowell, T.S.; Hedrick, R.P. Molecular phylogeny of tubificid oligochaetes with special emphasis on Tubifex tubifex (Tubificidae). Mol. Phylogenet Evol. 2001, 19, 216–224. [Google Scholar] [CrossRef]
- Heethoff, M.; Etzold, K.; Scheu, S. Mitochondrial COII sequences indicate that the parthenogenetic earthworm Octolasion tyrtaeum (Savigny 1826) constitutes of two lineages differing in body size and genotype. Pedobiologia 2004, 48, 9–13. [Google Scholar] [CrossRef]
- Sturmbauer, C.; Opadiya, G.B.; Niederstätter, H.; Riedmann, A.; Dallinger, R. Mitochondrial DNA reveals cryptic oligochaete species differing in cadmium resistance. Mol. Biol. Evol. 1999, 16, 967–974. [Google Scholar] [CrossRef]
- Cech, G.; Boros, G.; Dózsa-Farkas, K. Revision of Bryodrilus glandulosus (Dózsa-Farkas, 1990) and Mesenchytraeus kuehnelti Dózsa-Farkas, 1991 (Oligochaeta: Enchytraeidae) using morphological and molecular data. Zool. Anz. 2012, 251, 253–262. [Google Scholar] [CrossRef]
- Dózsa-Farkas, K.; Porco, D.; Boros, G. Are Bryodrilus parvus Nurminen, 1970 and Bryodrilus librus (Nielsen and Christensen, 1959) (Annelida: Enchytraeidae) really different species? A revision based on DNA barcodes and morphological data. Zootaxa 2012, 3276, 38–50. [Google Scholar] [CrossRef]
- Rota, E.; Martinsson, S.; Erséus, C.; Petushkov, V.N.; Rodionova, N.S.; Omodeo, P. Green light to an integrative view of Microscolex phosphoreus (Dugès, 1837) (Annelida: Clitellata: Acanthodrilidae). Zootaxa 2018, 4496, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Martinsson, S.; Naveed, M.I. On the identity and phylogenetic position of Dero indica (Clitellata: Naididae). Biologia 2020, 75, 1685–1689. [Google Scholar] [CrossRef]
- Chang, C.H.; Lin, S.M.; Chen, J.H. Molecular systematics and phylogeography of the gigantic earthworms of the Metaphire formosae species group (Clitellata, Megascolecidae). Mol. Phylogenet Evol. 2008, 49, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Novo, M.; Almodóvar, A.; Díaz-Cosín, D.J. High genetic divergence of hormogastrid earthworms (Annelida, Oligochaeta) in the central Iberian Peninsula: Evolutionary and demographic implications. Zool. Scr. 2009, 38, 537–552. [Google Scholar] [CrossRef]
- James, S.W.; Porco, D.; Decaens, T.; Richard, B.; Rougerie, R.; Erséus, C. DNA barcoding reveals cryptic diversity in Lumbricus terrestris L., 1758 (Clitellata): Resurrection of L. herculeus (Savigny, 1826). PLoS ONE 2010, 5, e15629. [Google Scholar] [CrossRef]
- Paoletti, M.G.; Blakemore, R.J.; Csuzdi, C.; Dorigo, L.; Dreon, A.L.; Gavinelli, F.; Lazzarini, F.; Manno, N.; Moretto, E.; Porco, D.; et al. Barcoding Eophila crodabepis sp. nov. (Annelida, Oligochaeta, Lumbricidae), a Large Stripy Earthworm from Alpine Foothills of Northeastern Italy Similar to Eophila tellinii (Rosa, 1888). PLoS ONE 2016, 11, e0151799. [Google Scholar] [CrossRef]
- Smythe, A.B.; Forgrave, K.; Patti, A.; Hochberg, R.; Litvaitis, M.K. Untangling the Ecology, Taxonomy, and Evolution of Chaetogaster limnaei (Oligochaeta: Naididae) Species Complex. J. Parasitol. 2015, 101, 320–326. [Google Scholar] [CrossRef]
- Szederjesi, T.; Pop, V.V.; Márton, O.; Krízsik, V.; Csuzdi, C. The Allolobophora sturanyi species group revisited: Integrated taxonomy and new taxa (Clitellata: Megadrili). Opusc. Zool. (Budap.) 2016, 47, 87–92. [Google Scholar] [CrossRef]
- Csuzdi, C.; Szederjesi, T.; Marchán, D.F.; Sosa, I.; Gavinelli, F.; Dorigo, L.; Pamio, A.; Dreon, A.L.; Fusaro, S.; Moretto, E.; et al. DNA barcoding of the Italian anecic Octodrilus species in rural (vineyard) and forested areas with description of Octodrilus zicsiniello sp. nov. (Clitellata, Megadrili). Zootaxa 2018, 4496, 43–64. [Google Scholar] [CrossRef]
- Seesamut, T.; Sutcharit, C.; Jirapatrasilp, P.; Chanabun, R.; Panha, S. Morphological and molecular evidence reveal a new species of the earthworm genus Pontodrilus Perrier, 1874 (Clitellata, Megascolecidae) from Thailand and Peninsular Malaysia. Zootaxa 2018, 4496, 218–237. [Google Scholar] [CrossRef]
- Nxele, T.C.; Plisko, J.D.; Mwabvu, T.; Zishiri, O.T. Molecular phylogeny of Kazimierzus Plisko, 2006 (Clitellata, Kazimierzidae) from the Western and Northern Cape Province inferred from mitochondrial DNA sequences. Afr. Invertebr. 2020, 61, 83–92. [Google Scholar] [CrossRef]
- Prantoni, A.L.; Belmonte-Lopes, R.; Lana, P.C.; Erséus, C. Genetic diversity of marine oligochaetous clitellates in selected areas of the South Atlantic as revealed by DNA barcoding. Invertebr. Syst. 2018, 32, 524–532. [Google Scholar] [CrossRef]
- Saglam, N.; Kutschera, U.; Saunders, R.; Saidel, W.M.; Balombini, K.L.W.; Shain, D.H. Phylogenetic and morphological resolution of the Helobdella stagnalis species-complex (Annelida: Clitellata: Hirudinea). Zootaxa 2018, 4403. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, N.; Lone, A.R.; Thakur, S.S.; Yadav, S. Interrogation of earthworm (Clitellata: Haplotaxida) taxonomy and the DNA sequence database. J. Asia-Pac. Biodivers. 2020. [Google Scholar] [CrossRef]
- Iwama, R.E.; Oceguera-Figueroa, A.; De Carle, D.; Manglicmot, C.; Erseus, C.; Miles, N.M.; Siddall, M.E.; Kvist, S. Broad geographic sampling and DNA barcoding do not support the presence of Helobdella stagnalis (Linnaeus, 1758) (Clitellata: Glossiphoniidae) in North America. Zootaxa 2019, 4671, 1–25. [Google Scholar] [CrossRef]
- Meyer, C.P.; Paulay, G. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef]
- Perez-Losada, M.; Ricoy, M.; Marshall, J.C.; Dominguez, J. Phylogenetic assessment of the earthworm Aporrectodea caliginosa species complex (Oligochaeta: Lumbricidae) based on mitochondrial and nuclear DNA sequences. Mol. Phylogenet Evol. 2009, 52, 293–302. [Google Scholar] [CrossRef]
- Buckley, T.R.; James, S.; Allwood, J.; Bartlam, S.; Howitt, R.; Prada, D. Phylogenetic analysis of New Zealand earthworms (Oligochaeta: Megascolecidae) reveals ancient clades and cryptic taxonomic diversity. Mol. Phylogenet Evol. 2011, 58, 85–96. [Google Scholar] [CrossRef]
- Kvist, S.; Sarkar, I.N.; Erséus, C. Genetic variation and phylogeny of the cosmopolitan marine genus Tubificoides (Annelida: Clitellata: Naididae: Tubificinae). Mol. Phylogenet Evol. 2010, 57, 687–702. [Google Scholar] [CrossRef]
- Matamoros, L.; Rota, E.; Erséus, C. Cryptic diversity among the achaetous Marionina (Annelida, Clitellata, Enchytraeidae). Syst. Biodivers. 2012, 10, 509–525. [Google Scholar] [CrossRef]
- Marchán, D.F.; Fernández, R.; Novo, M.; Díaz Cosín, D. New light into the hormogastrid riddle: Morphological and molecular description of Hormogaster joseantonioi sp. n. (Annelida, Clitellata, Hormogastridae). ZooKeys 2014, 414, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Saglam, N.; Saunders, R.; Lang, S.A.; Shain, D.H. A new species of Hirudo (Annelida: Hirudinidae): Historical biogeography of Eurasian medicinal leeches. BMC Zool. 2016, 1, 5. [Google Scholar] [CrossRef]
- De Sosa, I.; Marchán, D.F.; Novo, M.; Almodóvar, A.; Díaz Cosín, D.J. Bless this phylogeographic mess–Comparative study of Eiseniella tetraedra (Annelida, Oligochaeta) between an Atlantic area and a continental Mediterranean area in Spain. Eur. J. Soil Biol. 2017, 78, 50–56. [Google Scholar] [CrossRef]
- Marchán, D.F.; Fernández, R.; de Sosa, I.; Díaz Cosín, D.J.; Novo, M. Pinpointing cryptic borders: Fine-scale phylogeography and genetic landscape analysis of the Hormogaster elisae complex (Oligochaeta, Hormogastridae). Mol. Phylogenet Evol. 2017, 112, 185–193. [Google Scholar] [CrossRef]
- Anderson, K.; Braoudakis, G.; Kvist, S. Genetic variation, pseudocryptic diversity, and phylogeny of Erpobdella (Annelida: Hirudinida: Erpobdelliformes), with emphasis on Canadian species. Mol. Phylogenet Evol. 2019. [Google Scholar] [CrossRef]
- Timm, T.; Arslan, N.; Rüzgar, M.; Martinsson, S.; Erséus, C. Oligochaeta (Annelida) of the profundal of Lake Hazar (Turkey), with description of Potamothrix alatus hazaricus n. ssp. Zootaxa 2013, 3716, 144–156. [Google Scholar] [CrossRef]
- De Wit, P.; Erséus, C. Genetic variation and phylogeny of Scandinavian species of Grania (Annelida: Clitellata: Enchytraeidae), with the discovery of a cryptic species. J. Zool. Syst. Evol. Res. 2010, 48, 285–293. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Eiroa, J.; Mato, S.; Domínguez, J. Phylogenetic species delimitation of the earthworms Eisenia fetida (Savigny, 1826) and Eisenia andrei Bouché, 1972 (Oligochaeta, Lumbricidae) based on mitochondrial and nuclear DNA sequences. Pedobiologia 2005, 49, 317–324. [Google Scholar] [CrossRef]
- Martinsson, S.; Erséus, C. Cryptic diversity in the well-studied terrestrial worm Cognettia sphagnetorum (Clitellata: Enchytraeidae). Pedobiologia 2014, 57, 27–35. [Google Scholar] [CrossRef]
- Achurra, A.; Rodriguez, P.; Erséus, C. Pseudo-cryptic speciation in the subterranean medium: A new species of Stylodrilus Claparède, 1862, with a revision of the status of Bichaeta Bretscher, 1900 (Annelida, Clitellata, Lumbriculidae). Zool. Anz. 2015, 257, 71–86. [Google Scholar] [CrossRef]
- Envall, I.; Gustavsson, L.M.; Erseus, C. Genetic and chaetal variation in Nais worms (Annelida, Clitellata, Naididae). Zool. J. Linn. Soc. 2012, 165, 495–520. [Google Scholar] [CrossRef]
- Achurra, A.; Erséus, C. DNA barcoding and species delimitation: The Stylodrilus heringianus case (Annelida : Clitellata : Lumbriculidae). Invertebr. Syst. 2013, 27, 118–128. [Google Scholar] [CrossRef]
- Dozsa-Farkas, K.; Felföldi, T. Unexpected occurrence of Hemifridericia bivesiculata Christensen & Dozsa-Farkas, 2006 in Hungary, a species presumed to be endemic to Devon Island, Canada, and its comparative analysis with H. parva Nielsen & Christensen, 1959 (Enchytraeidae, Oligochaeta). Zootaxa 2015, 3914, 185–194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Prantoni, A.L.; De Wit, P.; Erséus, C. First reports of Grania (Clitellata: Enchytraeidae) from Africa and South America: Molecular phylogeny and descriptions of nine new species. Zool. J. Linn. Soc. 2016, 176, 485–510. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Berman, D.I.; Bazarova, N.E.; Bulakhova, N.A.; Porco, D.; Peltek, S.E. Cryptic genetic lineages in Eisenia nordenskioldi pallida (Oligochaeta, Lumbricidae). Eur. J. Soil. Biol. 2016, 75, 151–156. [Google Scholar] [CrossRef]
- Dózsa-Farkas, K.; Csitári, B.; Felföldi, T. A new Cernosvitoviella species (Clitellata: Enchytraeidae) and its comparison with other Cernosvitoviella species from Sphagnum mires in Hungary. Zootaxa 2017, 4254, 322. [Google Scholar] [CrossRef]
- Dozsa-Farkas, K.; Felfoldi, T. Comparative morphological and molecular taxonomic study of six Achaeta species (Clitellata: Enchytraeidae) with the description of a new Achaeta species from Koszeg Mountains, Hungary. Zootaxa 2017, 4273, 177–194. [Google Scholar] [CrossRef]
- Martinsson, S.; Klinth, M.; Erséus, C. A new Scandinavian Chamaedrilus species (Clitellata: Enchytraeidae), with additional notes on others. Zootaxa 2018, 4521, 417–429. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Golovanova, E.V.; Peltek, S.E. Genetic diversity of the earthworm Octolasion tyrtaeum (Lumbricidae, Annelida). Pedobiologia 2014, 57, 245–250. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Rapoport, I.B.; Poluboyarova, T.V.; Geraskina, A.P.; Golovanova, E.V.; Peltek, S.E. Morphotypes and genetic diversity of Dendrobaena schmidti (Lumbricidae, Annelida). Vavilov J. Genet. Breed. 2020, 24, 48–54. [Google Scholar] [CrossRef]
- Nagy, H.; Felföldi, T.; Dózsa-Farkas, K. Morphological and molecular distinction of two Fridericia species (Clitellata, Enchytraeidae) having same spermatheca type. Zootaxa 2018, 4496, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Rota, E.; Martinsson, S.; Erséus, C. Two new bioluminescent Henlea from Siberia and lack of molecular support for Hepatogaster (Annelida, Clitellata, Enchytraeidae). Org. Divers. Evol. 2018, 18, 291–312. [Google Scholar] [CrossRef]
- Nagy, H.; Dózsa-Farkas, K.; Hong, Y.; Felföldi, T. Extending the geographic distribution of Bryodrilus ehlersi (Annelida, Enchytraeidae): Morphological and molecular comparison of korean and european specimens. Acta Zool. Acad. Sci. Hung. 2020, 66, 345–360. [Google Scholar] [CrossRef]
- Novo, M.; Almodovar, A.; Fernandez, R.; Trigo, D.; Diaz Cosin, D.J. Cryptic speciation of hormogastrid earthworms revealed by mitochondrial and nuclear data. Mol. Phylogenet Evol. 2010, 56, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Shekhovtsov, S.V.; Golovanova, E.V.; Peltek, S.E. Cryptic diversity within the Nordenskiold’s earthworm, Eisenia nordenskioldi subsp. nordenskioldi (Lumbricidae, Annelida). Eur. J. Soil. Biol. 2013, 58, 13–18. [Google Scholar] [CrossRef]
- Latif, R.; Malek, M.; Aminjan, A.R.; Pasantes, J.J.; Briones, M.J.I.; Csuzdi, C. Integrative taxonomy of some Iranian peregrine earthworm species using morphology and barcoding (Annelida: Megadrili). Zootaxa 2020, 4877, 163–173. [Google Scholar] [CrossRef]
- Liu, Y.; Fend, S.V.; Martinsson, S.; Erséus, C. Extensive cryptic diversity in the cosmopolitan sludge worm Limnodrilus hoffmeisteri (Clitellata, Naididae). Org. Divers. Evol. 2017, 17, 477–495. [Google Scholar] [CrossRef]
- Torii, T.; Erséus, C.; Martinsson, S.; Ito, M. Morphological and genetic characterization of the first species of Thalassodrilides (Annelida: Clitellata: Naididae: Limnodriloidinae) from Japan. Species Divers. 2016, 21, 117–125. [Google Scholar] [CrossRef]
- Felföldi, T.; Dózsa-Farkas, K.; Nagy, H.; Hong, Y. Three new enchytraeid species (Enchytraeidae, Annelida) from mountain soils of Korea and ten species new for the country. Zootaxa 2020, 4896, 1–45. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.; Barraclough, T.G.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.P.; Hazell, S.; Kamoun, S.; Sumlin, W.D.; Vogler, A.P. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Jeratthitikul, E.; Bantaowong, U.; Panha, S. DNA barcoding of the Thai species of terrestrial earthworms in the genera Amynthas and Metaphire (Haplotaxida: Megascolecidae). Eur. J. Soil Biol. 2017, 81, 39–47. [Google Scholar] [CrossRef]
- Klinth, M.J.; Martinsson, S.; Erséus, C. Phylogeny and species delimitation of North European Lumbricillus (Clitellata, Enchytraeidae). Zool. Scr. 2017, 46, 96–110. [Google Scholar] [CrossRef]
- Latif, R.; Malek, M.; Csuzdi, C. When morphology and DNA are discordant: Integrated taxonomic studies on the Eisenia fetida/andrei complex from different parts of Iran (Annelida, Clitellata: Megadrili). Eur. J. Soil Biol. 2017, 81, 55–63. [Google Scholar] [CrossRef]
- Martinsson, S.; Achurra, A.; Svensson, M.; Erséus, C. Integrative taxonomy of the freshwater worm Rhyacodrilus falciformis s.l. (Clitellata: Naididae), with the description of a new species. Zool. Scr. 2013, 42, 612–622. [Google Scholar] [CrossRef]
- Martinsson, S.; Klinth, M.; Erséus, C. Testing species hypotheses for Fridericia magna, an enchytraeid worm (Annelida: Clitellata) with great mitochondrial variation. BMC Evol. Biol. 2020, 20, 1–12. [Google Scholar] [CrossRef]
- Martinsson, S.; Rhodén, C.; Erséus, C. Barcoding gap, but no support for cryptic speciation in the earthworm Aporrectodea longa (Clitellata: Lumbricidae). Mitochondrial DNA 2017, 28, 147–155. [Google Scholar] [CrossRef]
- Martin, P.; Martinsson, S.; Wuillot, J.; Erséus, C. Integrative species delimitation and phylogeny of the branchiate worm Branchiodrilus (Clitellata, Naididae). Zool. Scr. 2018, 47, 727–742. [Google Scholar] [CrossRef]
- Martinsson, S.; Erséus, C. Cryptic diversity in supposedly species-poor genera of Enchytraeidae (Annelida: Clitellata). Zool. J. Linn. Soc. 2018, 183, 749–762. [Google Scholar] [CrossRef]
- Martin, P.; Sonet, G.; Smitz, N.; Backeljau, T. Phylogenetic analysis of the Baikalodrilus species flock (Annelida: Clitellata: Naididae), an endemic genus to Lake Baikal (Russia). Zool. J. Linn. Soc. 2019, 187, 987–1015. [Google Scholar] [CrossRef]
- Reid, N.M.; Carstens, B.C. Phylogenetic estimation error can decrease the accuracy of species delimitation: A Bayesian implementation of the general mixed Yule-coalescent model. BMC Evol. Biol. 2012, 12, 196. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Almodóvar, A.; Novo, M.; Simancas, B.; Díaz Cosín, D.J. Adding complexity to the complex: New insights into the phylogeny, diversification and origin of parthenogenesis in the Aporrectodea caliginosa species complex (Oligochaeta, Lumbricidae). Mol. Phylogenet Evol. 2012, 64, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Novo, M.; Almodovar, A.; Fernandez, R.; Trigo, D.; Diaz-Cosin, D.J.; Giribet, G. Appearances can be deceptive: Different diversification patterns within a group of Mediterranean earthworms (Oligochaeta, Hormogastridae). Mol. Ecol. 2012, 21, 3776–3793. [Google Scholar] [CrossRef] [PubMed]
- Novo, M.; Fernandez, R.; Marchan, D.F.; Monica, G.; Cosin, D.J. Compilation of morphological and molecular data, a necessity for taxonomy: The case of Hormogaster abbatissae sp. n. (Annelida, Clitellata, Hormogastridae). ZooKeys 2012, 242, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Jirapatrasilp, P.; Backeljau, T.; Prasankok, P.; Chanabun, R.; Panha, S. Untangling a mess of worms: Species delimitations reveal morphological crypsis and variability in Southeast Asian semi-aquatic earthworms (Almidae, Glyphidrilus). Mol. Phylogenet Evol. 2019, 139, 106531. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef]
- De Carle, D.; Oceguera-Figueroa, A.; Tessler, M.; Siddall, M.E.; Kvist, S. Phylogenetic analysis of Placobdella (Hirudinea: Rhynchobdellida: Glossiphoniidae) with consideration of COI variation. Mol. Phylogenet Evol. 2017, 114, 234–248. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D. A DNA-based registry for all animal species: The barcode index number (BIN) system. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef]
- Doyle, J.J. The Irrelevance of Allele Tree Topologies for Species Delimitation, and a Non-Topological Alternative. Syst. Bot. 1995, 20, 574–588. [Google Scholar] [CrossRef]
- Flot, J.F.; Couloux, A.; Tillier, S. Haplowebs as a graphical tool for delimiting species: A revival of Doyle’s “field for recombination” approach and its application to the coral genus Pocillopora in Clipperton. BMC Evol. Biol. 2010, 10, 372. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. Statistical tests for taxonomic distinctiveness from observations of monophyly. Evolution 2007, 61, 317–323. [Google Scholar] [CrossRef]
- Rodrigo, A.; Bertels, F.; Heled, J.; Noder, R.; Shearman, H.; Tsai, P. The perils of plenty: What are we going to do with all these genes? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 3893–3902. [Google Scholar] [CrossRef] [PubMed]
- Masters, B.C.; Fan, V.; Ross, H.A. Species Delimitation—a Geneious plugin for the exploration of species boundaries. Mol. Ecol. Resour. 2011, 11, 154–157. [Google Scholar] [CrossRef]
- Rannala, B.; Yang, Z. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 2003, 164, 1645–1656. [Google Scholar]
- Rannala, B. The art and science of species delimitation. Curr. Zool. 2015, 61, 846–853. [Google Scholar] [CrossRef]
- Yang, Z. The BPP program for species tree estimation and species delimitation. Curr. Zool. 2015, 61, 854–865. [Google Scholar] [CrossRef]
- Yang, Z.; Rannala, B. Bayesian species delimitation using multilocus sequence data. Proc. Natl. Acad. Sci. USA 2010, 107, 9264–9269. [Google Scholar] [CrossRef]
- Martinsson, S.; Erseus, C. Cryptic speciation and limited hybridization within Lumbricus earthworms (Clitellata: Lumbricidae). Mol. Phylogenet Evol. 2017, 106, 18–27. [Google Scholar] [CrossRef]
- Martinsson, S.; Erséus, C. Hybridisation and species delimitation of Scandinavian Eisenia spp. (Clitellata: Lumbricidae). Eur. J. Soil Biol. 2018, 88, 41–47. [Google Scholar] [CrossRef]
- Taheri, S.; James, S.; Roy, V.; Decaens, T.; Williams, B.W.; Anderson, F.; Rougerie, R.; Chang, C.H.; Brown, G.; Cunha, L.; et al. Complex taxonomy of the ‘brush tail’ peregrine earthworm Pontoscolex corethrurus. Mol. Phylogenet Evol. 2018, 124, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Aydin, Z.; Oxelman, B. DISSECT: An assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics 2015, 31, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Miller, M.R.; Dunham, J.P.; Amores, A.; Cresko, W.A.; Johnson, E.A. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007, 17, 240–248. [Google Scholar] [CrossRef]
- Davey, J.W.; Blaxter, M.L. RADSeq: Next-generation population genetics. Brief. Funct. Genom. 2010, 9, 416–423. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Giska, I.; Sechi, P.; Babik, W. Deeply divergent sympatric mitochondrial lineages of the earthworm Lumbricus rubellus are not reproductively isolated. BMC Evol. Biol. 2015, 15, 217. [Google Scholar] [CrossRef]
- Anderson, C.; Cunha, L.; Sechi, P.; Kille, P.; Spurgeon, D. Genetic variation in populations of the earthworm, Lumbricus rubellus, across contaminated mine sites. BMC Genet. 2017, 18, 97. [Google Scholar] [CrossRef]
- Marchán, D.F.; Novo, M.; Sanchez, N.; Dominguez, J.; Diaz Cosin, D.J.; Fernández, R. Local adaptation fuels cryptic speciation in terrestrial annelids. Mol. Phylogenet Evol. 2020, 146, 106767. [Google Scholar] [CrossRef] [PubMed]
- Shekhovtsov, S.V.; Ershov, N.I.; Vasiliev, G.V.; Peltek, S.E. Transcriptomic analysis confirms differences among nuclear genomes of cryptic earthworm lineages living in sympatry. BMC Evol. Biol. 2019, 19. [Google Scholar] [CrossRef] [PubMed]
- Shekhovtsov, S.V.; Shipova, A.A.; Poluboyarova, T.V.; Vasiliev, G.V.; Golovanova, E.V.; Geraskina, A.P.; Bulakhova, N.A.; Szederjesi, T.; Peltek, S.E. Species Delimitation of the Eisenia nordenskioldi Complex (Oligochaeta, Lumbricidae) Using Transcriptomic Data. Front. Genet. 2020, 11, 01508. [Google Scholar] [CrossRef] [PubMed]
- Anderson, F.E.; Williams, B.W.; Horn, K.M.; Erseus, C.; Halanych, K.M.; Santos, S.R.; James, S.W. Phylogenomic analyses of Crassiclitellata support major Northern and Southern Hemisphere clades and a Pangaean origin for earthworms. BMC Evol. Biol. 2017, 17, 123. [Google Scholar] [CrossRef]
- Novo, M.; Fernández, R.; Andrade, S.C.S.; Marchán, D.F.; Cunha, L.; Díaz Cosín, D.J. Phylogenomic analyses of a Mediterranean earthworm family (Annelida: Hormogastridae). Mol. Phylogenet Evol. 2016, 94, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, A.R.; Emme, S.A.; Lemmon, E.M. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst. Biol. 2012, 61, 727–744. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.; Dornburg, A.; Zapfe, K.L.; Anderson, F.E.; James, S.W.; Erseus, C.; Moriarty Lemmon, E.; Lemmon, A.R.; Williams, B.W. Phylogenomic Analysis of a Putative Missing Link Sparks Reinterpretation of Leech Evolution. Genome Biol. Evol. 2019, 11, 3082–3093. [Google Scholar] [CrossRef] [PubMed]
- Kvist, S.; Manzano-Marin, A.; de Carle, D.; Trontelj, P.; Siddall, M.E. Draft genome of the European medicinal leech Hirudo medicinalis (Annelida, Clitellata, Hirudiniformes) with emphasis on anticoagulants. Sci. Rep. 2020, 10, 9885. [Google Scholar] [CrossRef] [PubMed]
- Zwarycz, A.S.; Nossa, C.W.; Putnam, N.H.; Ryan, J.F. Timing and Scope of Genomic Expansion within Annelida: Evidence from Homeoboxes in the Genome of the Earthworm Eisenia fetida. Genome Biol. Evol. 2015, 8, 271–281. [Google Scholar] [CrossRef]
- Simakov, O.; Marletaz, F.; Cho, S.J.; Edsinger-Gonzales, E.; Havlak, P.; Hellsten, U.; Kuo, D.H.; Larsson, T.; Lv, J.; Arendt, D.; et al. Insights into bilaterian evolution from three spiralian genomes. Nature 2013, 493, 526–531. [Google Scholar] [CrossRef]
- Dozsa-Farkas, K.; Felföldi, T.; Nagy, H.; Hong, Y. New enchytraeid species from Mount Hallasan (Jeju Island, Korea) (Enchytraeidae, Oligochaeta). Zootaxa 2018, 4496, 337–381. [Google Scholar] [CrossRef] [PubMed]
- Dózsa-Farkas, K.; Nagy, H.; Felföldi, T. Two new species of Fridericia (Annelida: Enchytraeidae) from Hungarian caves. Eur. J. Taxon. 2019, 553. [Google Scholar] [CrossRef]
- Klinth, M.J.; Rota, E.; Erséus, C. Taxonomy of North European Lumbricillus (Clitellata, Enchytraeidae). ZooKeys 2017, 703, 15–96. [Google Scholar] [CrossRef] [PubMed]
- Martinsson, S.; Rota, E.; Erséus, C. Revision of Cognettia (Clitellata, Enchytraeidae): Re-establishment of Chamaedrilus and description of cryptic species in the sphagnetorum complex. Syst. Biodivers 2015, 13, 257–277. [Google Scholar] [CrossRef]
- Martinsson, S.; Rota, E.; Erséus, C. On the identity of Chamaedrilus glandulosus (Michaelsen, 1888) (Clitellata, Enchytraeidae), with the description of a new species. ZooKeys 2015, 501, 1–14. [Google Scholar] [CrossRef]
- Marchán, D.F.; Fernández, R.; Domínguez, J.; Díaz Cosín, D.J.; Novo, M. Genome-informed integrative taxonomic description of three cryptic species in the earthworm genus Carpetania (Oligochaeta, Hormogastridae). Syst. Biodivers. 2020, 1–13. [Google Scholar] [CrossRef]
- Marchán, D.F.; Fernández, R.; Sánchez, N.; de Sosa, I.; Díaz Cosín, D.J.; Novo, M. Insights into the diversity of Hormogastridae (Annelida, Oligochaeta) with descriptions of six new species. Zootaxa 2018, 4496, 65–95. [Google Scholar] [CrossRef]
- Kvist, S.; Erséus, C. Two new European species of the marine genus Tubificoides (Annelida: Clitellata: Naididae) with notes on the morphology of T. pseudogaster (Dahl, 1960). Zootaxa 2018, 4433, 561. [Google Scholar] [CrossRef]
- Kvist, S.; Oceguera-Figueroa, A.; Siddall, M.E.; Erseus, C. Barcoding, types and the Hirudo files: Using information content to critically evaluate the identity of DNA barcodes. Mitochondrial DNA 2010, 21, 198–205. [Google Scholar] [CrossRef]
- Schmelz, R.M.; Beylich, A.; Boros, G.; Dózsa-Farkas, K.; Graefe, U.; Hong, Y.; Römbke, J.; Schlaghamersky, J.; Martinsson, S. How to deal with cryptic species in Enchytraeidae, with recommendations on taxonomical descriptions. Opusc. Zool. 2017, 48, 45–51. [Google Scholar] [CrossRef]
- Eberle, J.; Ahrens, D.; Mayer, C.; Niehuis, O.; Misof, B. A Plea for Standardized Nuclear Markers in Metazoan DNA Taxonomy. Trends Ecol. Evol. 2020. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinsson, S.; Erséus, C. Cryptic Clitellata: Molecular Species Delimitation of Clitellate Worms (Annelida): An Overview. Diversity 2021, 13, 36. https://doi.org/10.3390/d13020036
Martinsson S, Erséus C. Cryptic Clitellata: Molecular Species Delimitation of Clitellate Worms (Annelida): An Overview. Diversity. 2021; 13(2):36. https://doi.org/10.3390/d13020036
Chicago/Turabian StyleMartinsson, Svante, and Christer Erséus. 2021. "Cryptic Clitellata: Molecular Species Delimitation of Clitellate Worms (Annelida): An Overview" Diversity 13, no. 2: 36. https://doi.org/10.3390/d13020036
APA StyleMartinsson, S., & Erséus, C. (2021). Cryptic Clitellata: Molecular Species Delimitation of Clitellate Worms (Annelida): An Overview. Diversity, 13(2), 36. https://doi.org/10.3390/d13020036