Soil Bacterial Community and Soil Enzyme Activity Depending on the Cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Area
2.2. Methodology of Microbiological Analyses
2.2.1. Bacterial Count
2.2.2. DNA Extraction and Bioinformatic Analysis of Specific Bacterial Taxa
2.3. Methodology of Biochemical Analyses
2.4. Methodology of Chemical and Physicochemical Analyses of Soil
2.5. Statistical Analysis
3. Results
3.1. Physicochemical and Chemical Properties of Soil
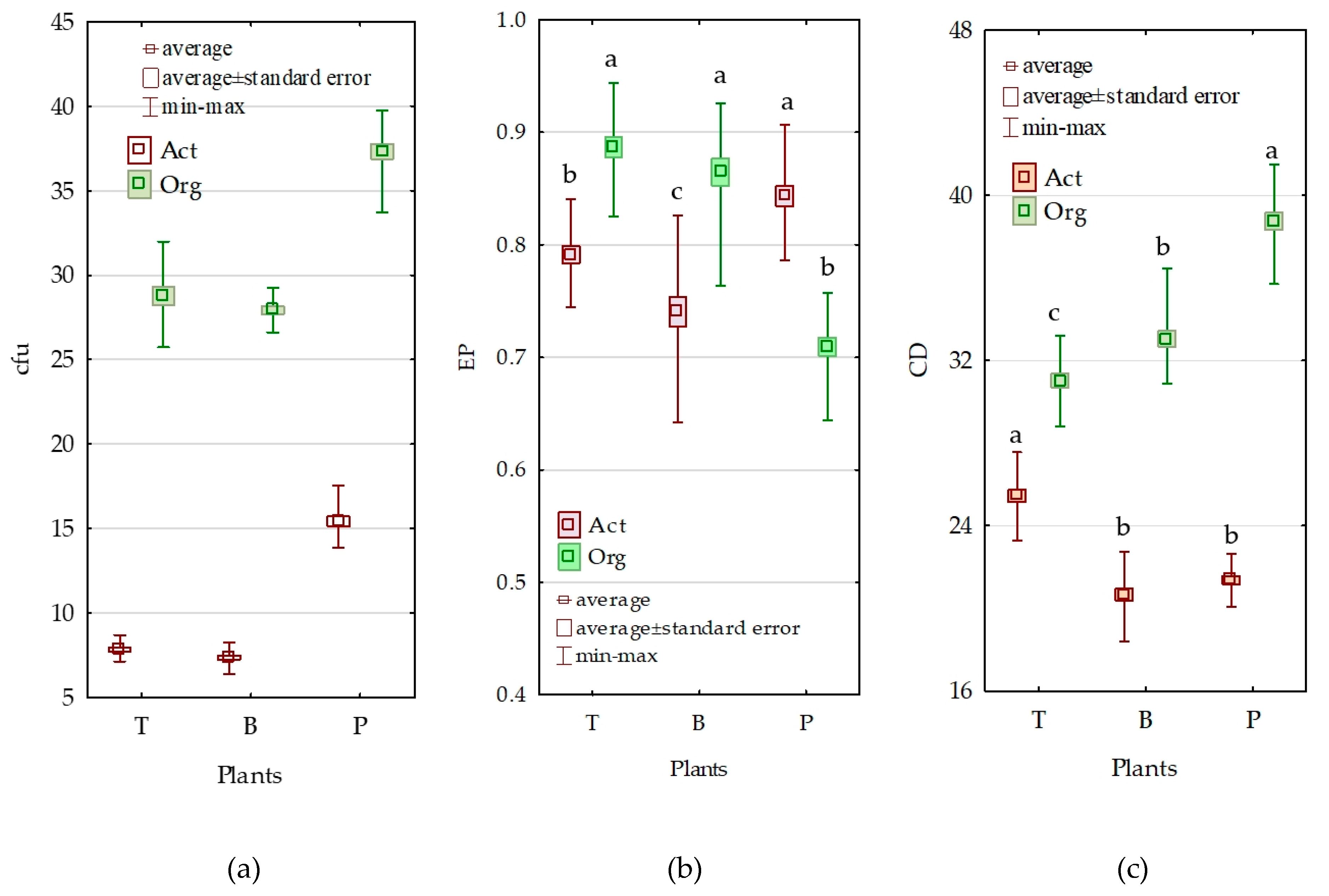
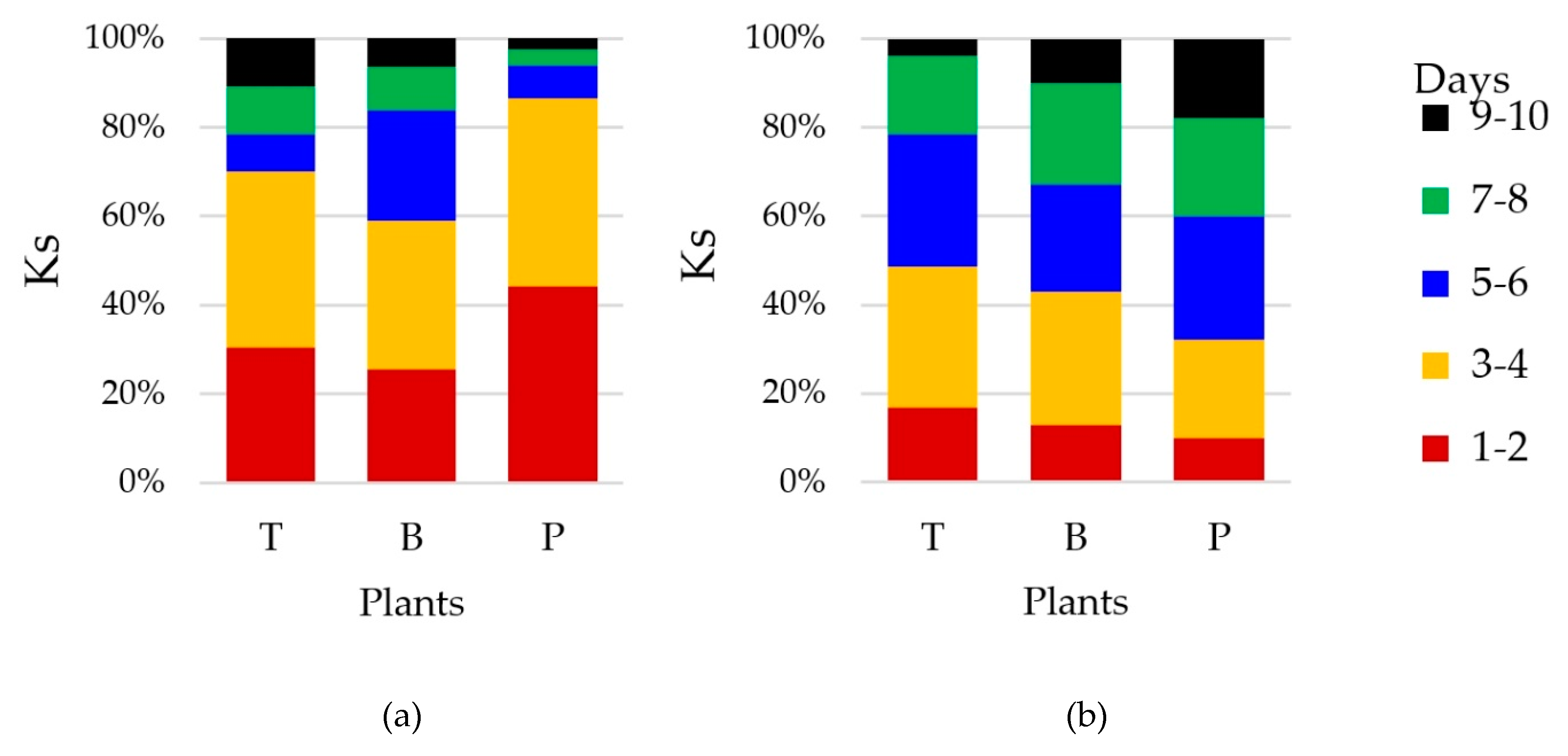
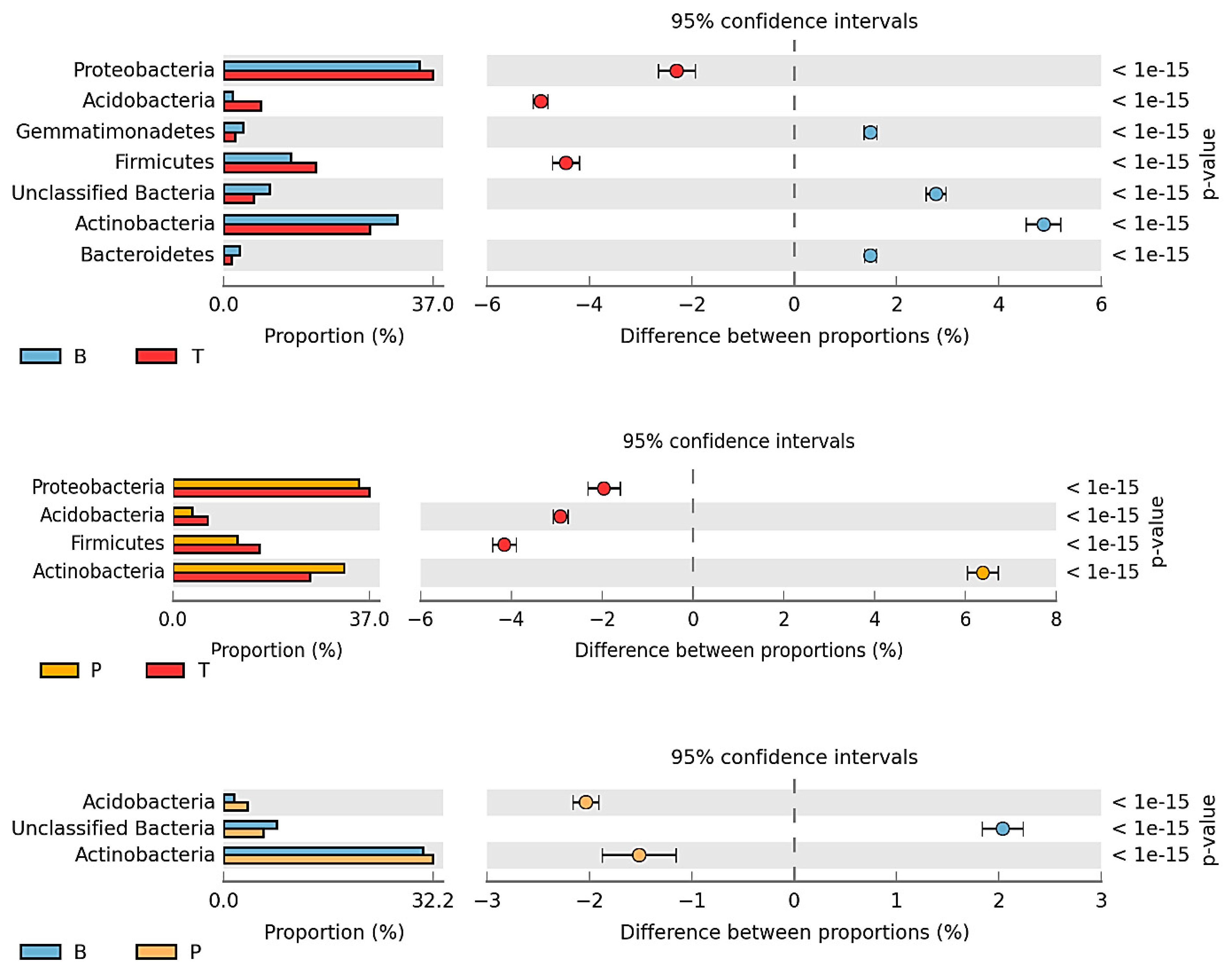
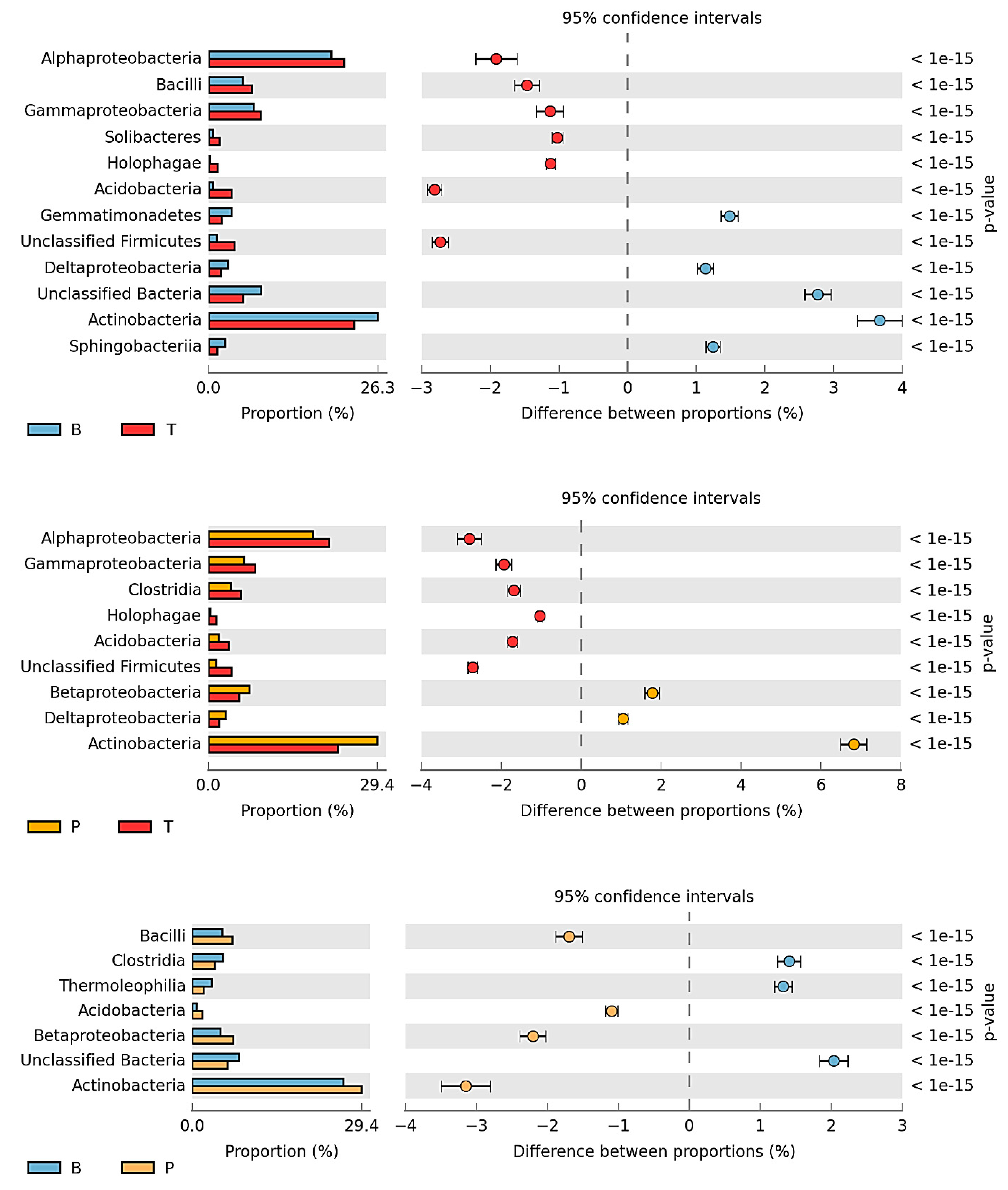
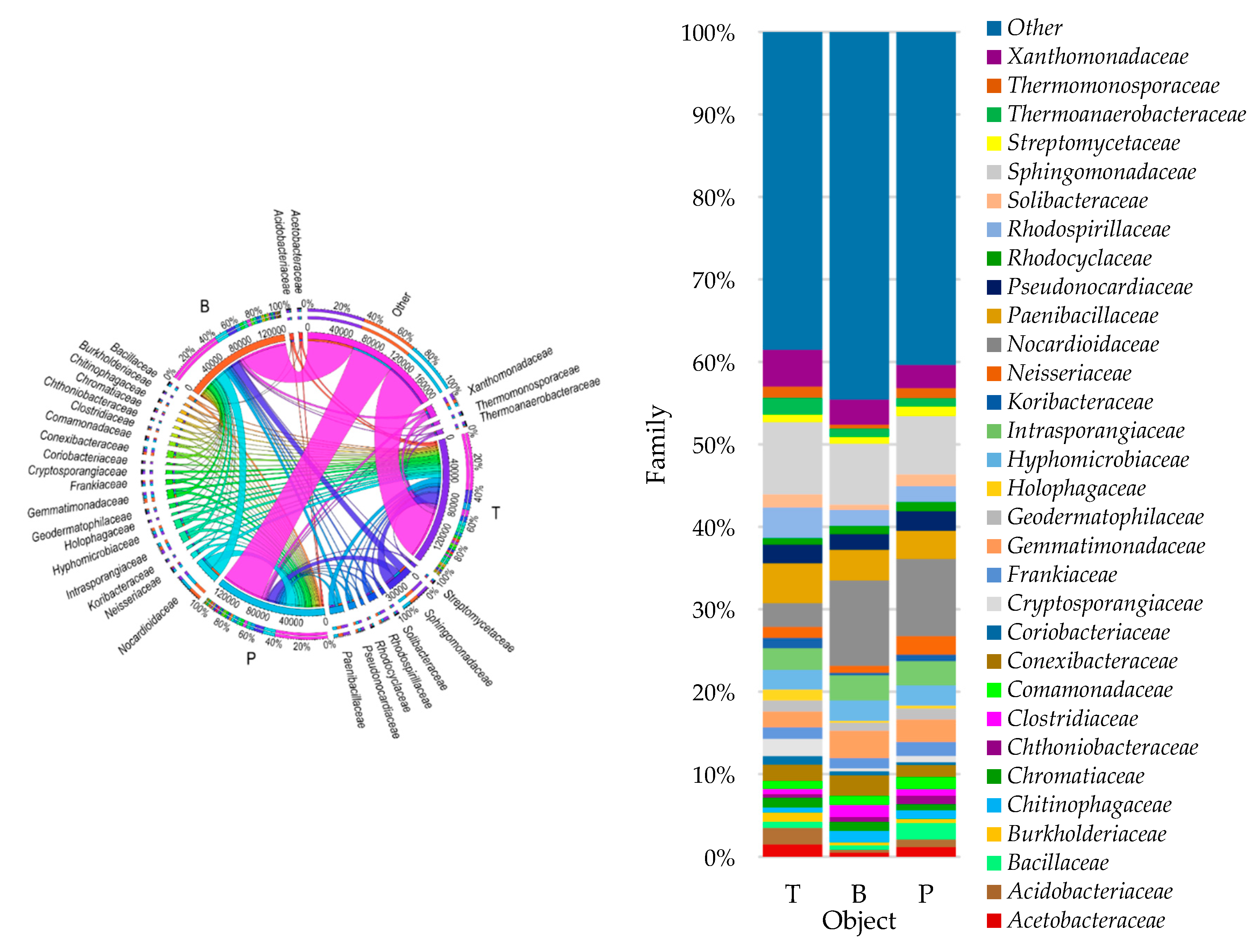
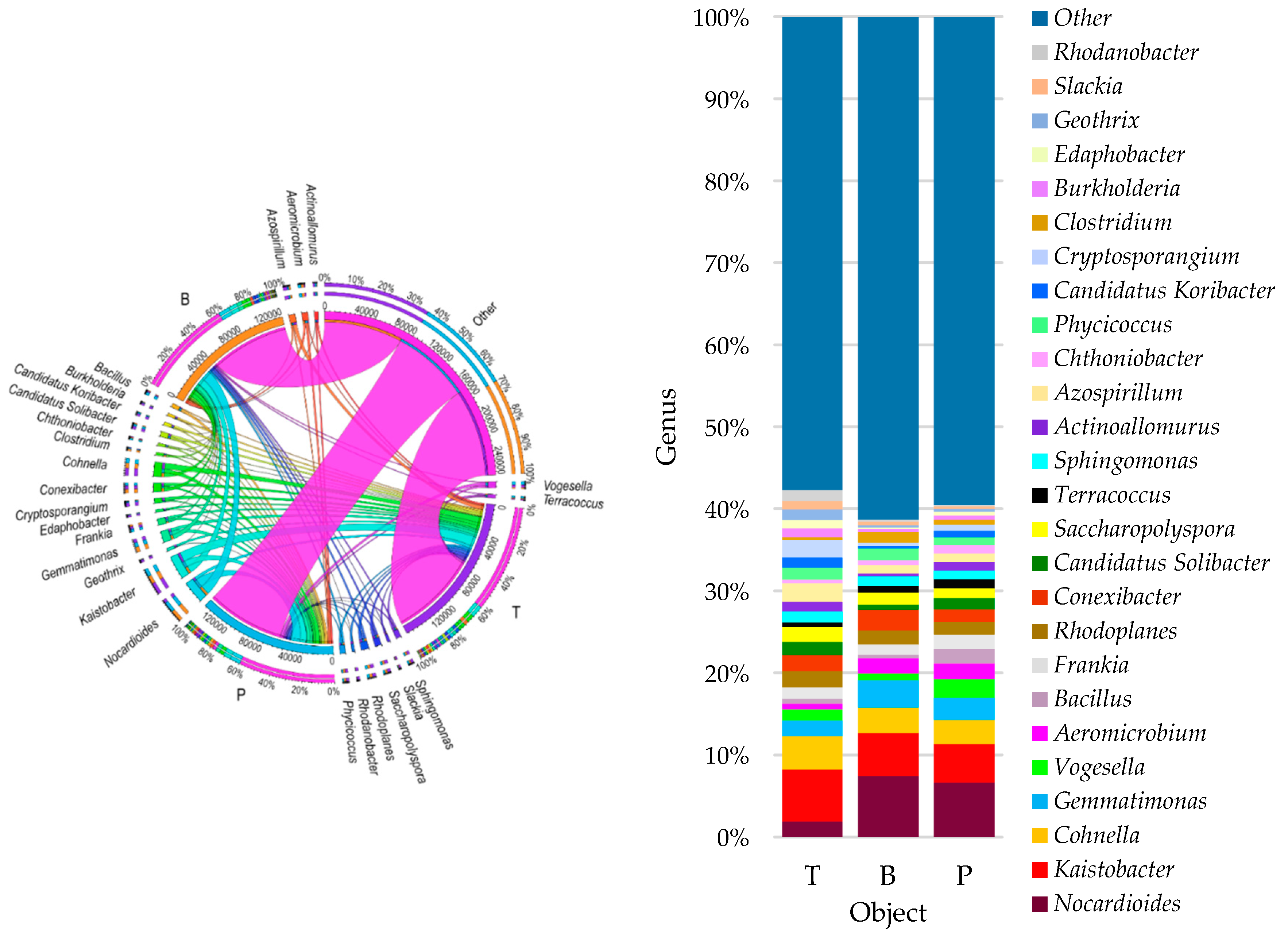
3.2. Counts and Diversity of Microorganisms
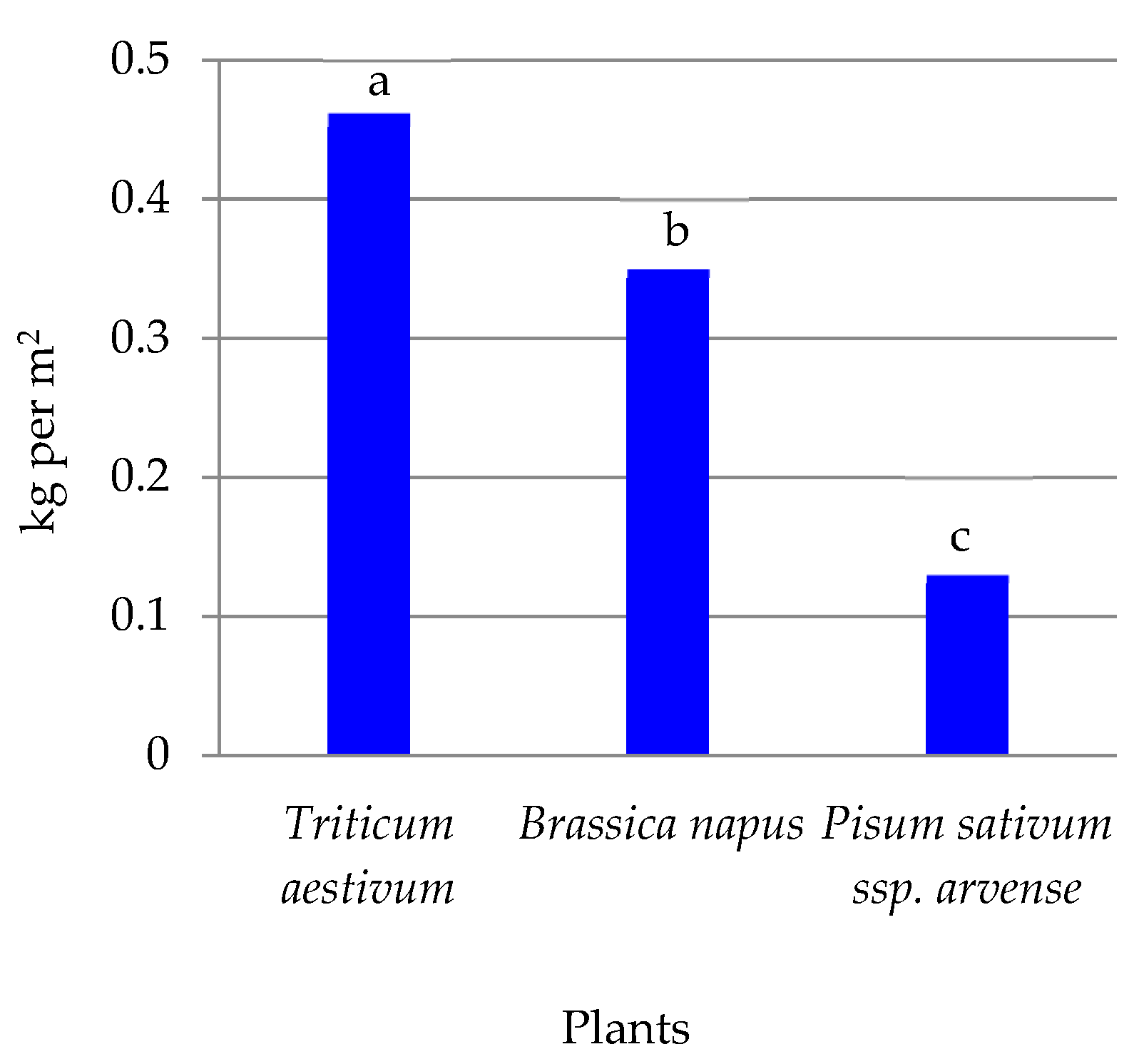
3.3. Enzymatic Activity of Soil
4. Discussion
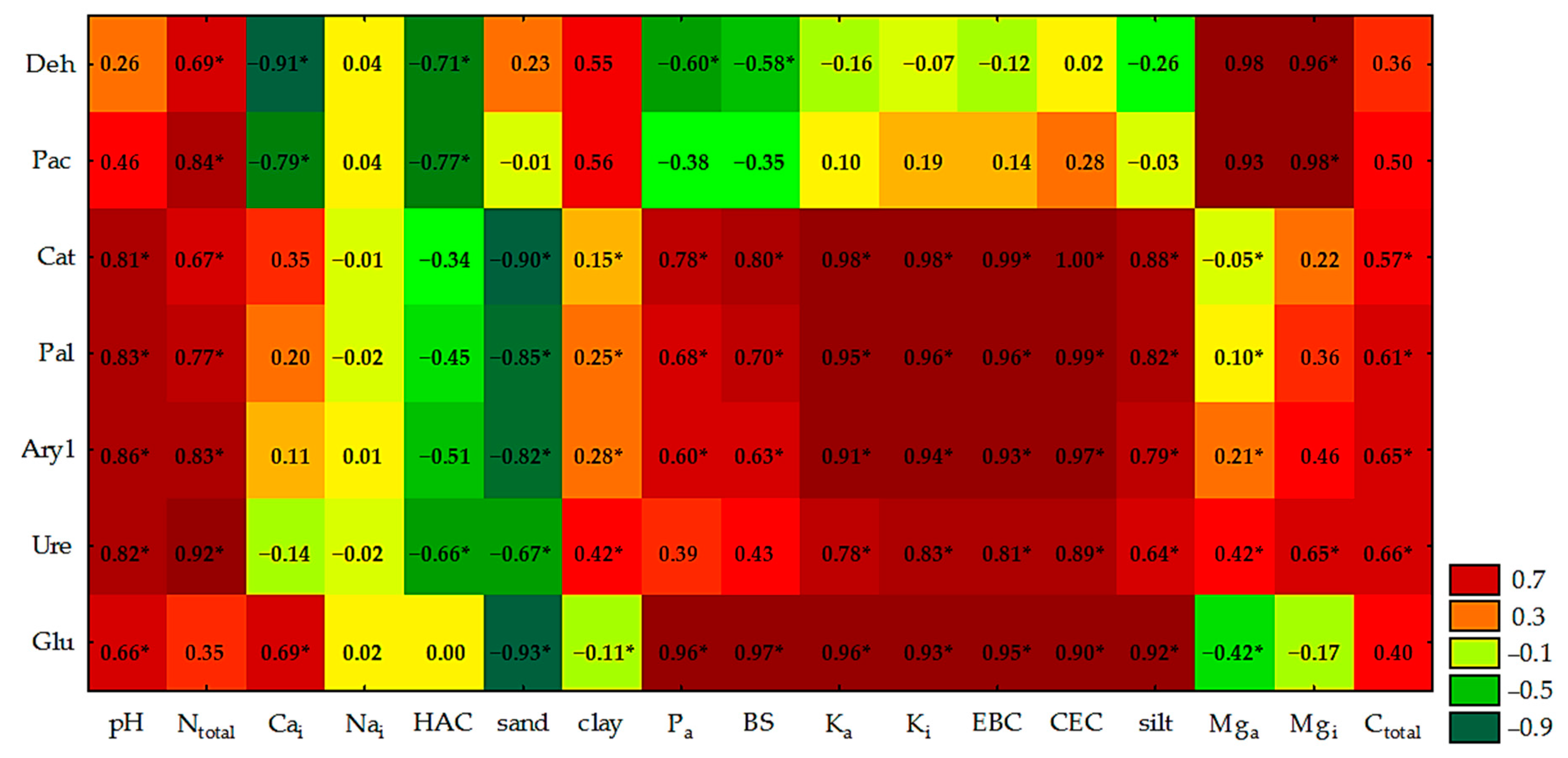
4.1. Physicochemical and Chemical Properties of Soil
4.2. Counts and Diversity of Bacteria
4.3. Enzymatic Activity of Soil
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Congreves, K.A.; Hayes, A.; Verhallen, E.A.; Van Eerd, L.L. Long-term impact of tillage and crop rotation on soil health at four temperate agroecosystems. Soil Tillage Res. 2015, 152, 17–28. [Google Scholar] [CrossRef]
- Lehman, M.R.; Cambardella, A.C.; Stott, E.D.; Acosta-Martinez, V.; Manter, K.D.; Buyer, S.J.; Maul, E.J.; Smith, L.J.; Collins, P.H.; Halvorson, J.J.; et al. Understanding and Enhancing Soil Biological Health: The Solution for Reversing Soil Degradation. Sustainability 2015, 7, 988. [Google Scholar] [CrossRef]
- Baveye, P.C.; Baveye, J.; Gowdy, J. Soil “ecosystem” services and natural capital: Critical appraisal of research on uncertain ground. Front. Environ. Sci. 2016, 4, 1–49. [Google Scholar] [CrossRef]
- Doran, J.W. Soil health and global sustainability: Translating science into practice. Agric. Ecosyst. Environ. 2002, 88, 119–127. [Google Scholar] [CrossRef]
- Cardoso, E.J.B.N.; Vasconcellos, R.L.F.; Bini, D.; Miyauchi, M.Y.H.; dos Santos, C.A.; Alves, P.R.L.; de Paula, A.M.; Nakatani, A.S.; Pereira, J.M.; Nogueira, M.A. Soil health: Looking for suitable indicators. What should be considered to assess the effects of use and management on soil health? Sci. Agric. 2013, 70, 274–289. [Google Scholar] [CrossRef]
- Legaz, B.V.; Maia De Souza, D.; Teixeira, R.F.M.; Antón, A.; Putman, B.; Sala, S. Soil quality, properties, and functions in life cycle assessment: An evaluation models. J. Clean. Prod. 2017, 140, 502–515. [Google Scholar] [CrossRef]
- Bünemann, E.K.; Bongiorno, G.; Bai, Z.; Creamer, R.E.; De Deyn, G.; de Goede, R.; Fleskens, L.; Geissen, V.; Kuyper, T.W.; Mäder, P.; et al. Soil quality—A critical review. Soil Biol. Biochem. 2018, 120, 105–125. [Google Scholar] [CrossRef]
- Veum, K.S.; Sudduth, K.A.; Kremer, R.J.; Kitchen, N.R. Sensor data fusion for soil health assessment. Geoderma 2017, 305, 53–61. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2003, 54, 655–670. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Kucharski, M.; Kucharski, J. Applicability of biochemical indices to quality assessment of soil pulluted with heavy metal. J. Elem. 2013, 18, 723–732. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Clement, C.; Barka, A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [PubMed]
- Zaborowska, M.; Woźny, G.; Wyszkowska, J.; Kucharski, J. Biostimulation of the activity of microorganisms and soil enzymes through fertilisation with composts. Soil Res. 2018, 56, 737–751. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J. Remediation of soil contaminated with diesel oil. J. Elem. 2018, 23, 767–788. [Google Scholar] [CrossRef]
- Zaborowska, M.; Kucharski, J.; Wyszkowska, J. Biochemical and microbiological activity of soil contaminated with o-cresol and biostimulated with Perna canaliculus mussel meal. Environ. Monit. Assess. 2018, 190, 602. [Google Scholar] [CrossRef]
- Sharifi, R.; Ryu, C.M. Sniffing bacterial volatile compounds for healthier plants. Curr. Opin. Plant Biol. 2018, 44, 88–97. [Google Scholar] [CrossRef]
- Bell, C.W.; Acosta-Martinez, V.; McIntyre, N.E.; Cox, S.; Tissue, D.T.; Zak, J.C. Linking microbial community structure and function to seasonal differences in soil moisture and temperature in a Chihuahuan Desert Grassland. Microb. Ecol. 2009, 58, 827–842. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J. Soil moisture as a factor affecting the microbiological and biochemical activity of soil. PlantSoil Environ. 2016, 62, 250–255. [Google Scholar] [CrossRef]
- Gordon, H.; Haygarth, P.M.; Bardgett, R.D. Drying and rewetting effects on soil microbial community composition and nutrient leaching. Soil Biol. Biochem. 2008, 40, 302–311. [Google Scholar] [CrossRef]
- Singurindy, O.; Molodovskaya, M.; Richards, B.K.; Steenhuis, T.S. Nitrous oxide emission at low temperatures from manure-amended soiils under corn (Zea mays L.). Agric. Ecosyst. Environ. 2009, 132, 74–81. [Google Scholar] [CrossRef]
- Craine, J.; Spurr, R.; McLauchlan, K.; Fierer, N. Lanscape-level variation in temperature sensitivity of soil organic carbon decomposition. Soil Biol. Biochem. 2010, 42, 373–375. [Google Scholar] [CrossRef]
- Silva, C.C.; Guido, M.L.; Ceballos, J.M.; Marsch, R.; Dendooven, L. Production of carbon dioxide and nitrous oxide in alkaline saline soil of Texcoco at different water contents amended with urea: A laboratory study. Soil Biol. Biochem. 2008, 40, 1813–1822. [Google Scholar] [CrossRef]
- Asuming-Brempong, S.; Ganter, S.; Adiku, S.G.K.; Archer, G.; Edusei, V.; Tiedje, J.M. Changes in the biodiversity of microbial populations in tropical soils under different fallow treatments. Siol Biol. Biochem. 2008, 40, 2811–2818. [Google Scholar] [CrossRef]
- Garcia-Ruiz, R.; Ochoa, V.; Hinojosa, M.B.; Carreira, J.A. Suitability of enzyme activities for the monitoring of soil quality improvement in organic agricultural systems. Soil Biol. Biochem. 2008, 40, 2137–2145. [Google Scholar] [CrossRef]
- Udawatta, R.P.; Kremer, R.J.; Garrett, H.E.; Anderson, S.H. Soil enzyme activities and physical propertie4s in a watershed managed under agroforestry and row-crop systems. Agric. Ecosyst. Environ. 2009, 131, 98–104. [Google Scholar] [CrossRef]
- Mandal, A.; Patra, A.K.; Singh, D.; Swarup, A.; Masto, R.E. Effect of long-term application of manure and fertilizer on biological and biochemical activities in soil during corp development stages. Bioresour. Technol. 2007, 98, 3585–3592. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Simpson, M.J. Temperature and substrate controls on microbial phospholipid fatty acid composition during incubation of grassland soils contrasting in organic matter quality. Soil Biol. Biochem. 2009, 41, 804–812. [Google Scholar] [CrossRef]
- Liu, Z.; Fu, B.; Zheng, X.; Liu, G. Plant biomass, soil water content and soil N: P ratio regulating soil microbial functional diversity in a temperature steppe: A regional scale study. Soil Biol. Biochem. 2010, 42, 445–450. [Google Scholar] [CrossRef]
- Boros-Lajszner, E.; Wyszkowska, J.; Kucharski, J. Use of zeolite to neutralise nickel in a soil environment. Environ. Monit. Assess. 2018, 190, 54. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Boros-Lajszner, E.; Borowik, A.; Kucharski, J.; Baćmaga, M.; Tomkiel, M. Changes in the microbiological and biochemical properties of soil contaminated with zinc. J. Elem. 2017, 22, 437–451. [Google Scholar] [CrossRef]
- Kucharski, J.; Jastrzębska, E. Effects of heating oil on the count of microorganisms and physico-chemical properties of soil. Pol. J. Environ. Stud. 2005, 14, 195–204. [Google Scholar]
- Lipińska, A.; Wyszkowska, J.; Kucharski, J. Diversity of organotrophic bacteria, activity of dehydrogenases and urease as well as seed germination and root growth Lepidium sativum, Sorghum saccharatum and Sinapis alba under the influence of polycyclic aromatic hydrocarbons. Environ. Sci. Pollut. Res. 2015, 22, 18519–18530. [Google Scholar] [CrossRef] [PubMed]
- Wyszkowska, J.; Borowik, A.; Kucharski, J. The resistance of Lolium perenne L. × hybridum, Poa pratensis, Festuca rubra, F. arundinacea, Phleum pratense and Dactylis glomerata to soil pollution by diesel oil and petroleum. PlantSoil Environ. 2019, 65, 307–312. [Google Scholar] [CrossRef]
- Baćmaga, M.; Wyszkowska, J.; Kucharski, J. Biostimulation as a process aiding tebuconazole degradation in soil. J. Soil Sediment. 2019, 19, 3728–3741. [Google Scholar] [CrossRef]
- Kucharski, J.; Tomkiel, M.; Baćmaga, M.; Borowik, A.; Wyszkowska, J. Enzyme activity and microorganisms diversity in soil contaminated with the Boreal 58 WG herbicide. J. Environ. Sci. Health B. 2016, 51, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Dong, J.; Ren, L.; Zhu, Q.; Huang, W.; Liu, Y.; Lu, D. Biodegradation of chlorothalonil by Enterobacter cloacae TUAH-1. Int. Biodeter. Biodegr. 2017, 121, 122–130. [Google Scholar] [CrossRef]
- Hund-Rinke, K.; Simon, M. Bioavailability assessment of contaminants in soils via respiration and nitrification tests. Environ. Pollut. 2008, 153, 468–475. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, X.; Liu, J.; Chen, Q.; Gao, L. Microbial properties of rhizosphere soils as affected by rotation, grafting, and soil sterilization in intensive vegetable production systems. Sci. Hortic. 2009, 123, 139–147. [Google Scholar] [CrossRef]
- Kohler, J.; Caravaca, F.; Roldán, A. Effect of drought on the stability of rhizosphere soil aggregates of Lactuca sativa grown in a degreded soil inoculated with PGPR and AM fungi. Appl. Soil Ecol. 2009, 42, 160–165. [Google Scholar] [CrossRef]
- Hai, L.; Li, X.G.; Suo, D.R.; Guggenberger, G. Long-term fertilization and manuring effects on physically-separated soil organic matter pools under a wheat-wheat-maize cropping system in an arid region of China. Soil Biol. Biochem. 2010, 42, 253–259. [Google Scholar] [CrossRef]
- Carminati, A.; Schneider, C.L.; Moradi, A.B.; Zarebanadkouki, M.; Vetterlein, D.; Vogel, H.J.; Hildebrandt, A.; Weller, U.; Schüler, L.; Oswald, S.E. How the rhizosphere may favor water availability to roots. Vadose Zone J. 2011, 10, 988–998. [Google Scholar] [CrossRef]
- Shaikh, S.; Wani, S.; Sayyed, R. Impact of Interactions between Rhizosphere and Rhizobacteria: A Review. J. Bacteriol. Mycol. 2018, 5, 1058. [Google Scholar]
- Benard, P.; Zarebanadkouki, M.; Brax, M.; Kaltenbach, R.; Jerjen, I.; Marone, F.; Couradeau, E.; Felde, V.J.; Kaestner, A.; Carminati, A. Microhydrological niches in soils: How mucilage and EPS alter the biophysical properties of the rhizosphere and other biological hotspots. Vadose Zone J. 2019, 18, 1–10. [Google Scholar] [CrossRef]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Kang, B.G.; Kim, W.T.; Yun, H.S.; Chang, S.C. Use of plant growth-promoting rhizobacteria to control stress responses of plant roots. Plant Biotechnol. Rep. 2010, 4, 179–183. [Google Scholar] [CrossRef]
- Nannipieri, P.; Aschner, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G.; Valori, F. Microbial diversity and microbial activity in the rhizosphere. Ci Suelo (Argentina) 2007, 25, 89–97. [Google Scholar]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Hassan, M.K.; McInroy, J.A.; Kloepper, J.W. The Interactions of Rhizodeposits with Plant Growth-Promoting Rhizobacteria in the Rhizosphere: A Review. Agriculture 2019, 9, 142. [Google Scholar] [CrossRef]
- Mueller, C.W.; Carminati, A.; Kaiser, C.; Subke, J.A.; Gutjahr, C. Rhizosphere functioning and structural development as complex interplay between plants, microorganisms and soil minerals. Front. Environ. Sci. 2019, 7, 130. [Google Scholar] [CrossRef]
- Rausch, P.; Rühlemann, M.; Hermes, B.M.; Doms, S.; Dagan, T.; Dierking, K.; Domin, H.; Fraune, S.; von Frieling, J.; Hentschel, U.; et al. Comparative analysis of amplicon and metagenomic sequencing methods reveals key features in the evolution of animal metaorganisms. Microbiome 2019, 7, 133. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Wyszkowski, M. Resistance of aerobic microorganisms and soil enzyme response to soil contamination with Ekodiesel Ultra fuel. Environ. Sci. Pollut. Res. 2017, 24, 24346–24363. [Google Scholar] [CrossRef]
- De Leij, F.A.A.; Whipps, J.M.; Lynch, J.M. The use of colony development for the characterization of bacterial communities in soil and on roots. Microb. Ecol. 1994, 27, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Tomkiel, M.; Baćmaga, M.; Wyszkowska, J.; Kucharski, J.; Borowik, A. The effect of carfentrazone-ethyl on soil microorganisms and soil enzymes activity. Arch. Environ. Prot. 2015, 41, 3–10. [Google Scholar] [CrossRef][Green Version]
- De Santis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- ISO 10390. Soil Quality—Determination of pH; International Organization for Standardization: Geneva, Switzerland, 2005. [Google Scholar]
- Carter, M.R.; Gregorich, E.G. Soil Sampling and Methods of Analysis, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2008; p. 1224. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic matter. In Method of Soil Analysis: Chemical Methods; Sparks, D.L., Ed.; American Society of Agronomy: Madison, WI, USA, 1996; pp. 1201–1229. [Google Scholar]
- ISO 11261. Soil Quality—Determination of Total Nitrogen—Modified Kjeldahl Method; International Organization for Standardization: Geneva, Switzerland, 1995. [Google Scholar]
- Egner, H.; Riehm, H.; Domingo, W.R. Untersuchun-gen über die chemische Bodenanalyse als Grundlage für die Beurteilung des Nährsto_zustandes der Böden. II. Chemische Extractionsmethoden zur Phospor-und Kaliumbestimmung. Ann. R. Agric. Coll. Swed. 1960, 26, 199–215. [Google Scholar]
- Schlichting, E.; Blume, H.P.; Stahr, K. Bodenkundliches Praktikum. Pareys Studientexte; Blackwell Wissenschafts: Berlin, Germany, 1995; p. 81. [Google Scholar]
- ISO 11260 Preview. Soil Quality—Determination of E_ective Cation Exchange Capacity and Base Saturation Level Using Barium Chloride Solution; International Organization for Standardization: Geneva, Switzerland, 2018. [Google Scholar]
- Dell Inc. Dell Statistica (Data Analysis Software System), Version 13.1; Dell Inc.: Tulsa, OK, USA, 2016. [Google Scholar]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Krzywinski, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Garbeva, P.; van Veen, J.A.; van Elsas, J.D. Microbial diversity in soil: Selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 2004, 42, 243–270. [Google Scholar] [CrossRef]
- Köberl, M.; Müller, H.; Ramadan, E.M.; Berg, G. Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS ONE 2011, 6, e24452. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Maron, P.A.; Sarr, A.; Kaisermann, A.; Lévêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S.; et al. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 2018, 84, 1–13. [Google Scholar] [CrossRef]
- Doornbos, R.F.; van Loon, L.C.; Bakker, P.A.H. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 2012, 32, 227–243. [Google Scholar] [CrossRef]
- Verma, J.P.; Yadav, J.; Tiwari, K.N.; Jaiswal, D.K. Evaluation of plant growth promoting activities of microbial strains and their effect on growth and yield of chickpea (Cicer arietinum L.) in India. Soil Biol. Biochem. 2014, 70, 33–37. [Google Scholar] [CrossRef]
- Kumar, A.; Maurya, B.R.; Raghuwanshi, R. Isolation and Characterization of PGPR and their effect on growth, yield and Nutrient content in wheat (Triticum aestivum L.). Biocatal. Agril. Biotechnol. 2014, 3, 121–128. [Google Scholar] [CrossRef]
- Kumar, A.; Bahadur, I.; Maurya, B.R.; Raghuwanshi, R.; Meena, V.S.; Singh, D.K.; Dixit, J. Does a plant growth promoting rhizobacteria enhance agricultural sustainability? J. Pure Appl. Microbio. 2015, 9, 715–724. [Google Scholar]
- Verma, J.P.; Yadav, J.; Tiwari, K.N.; Kumar, A. Effect of indigenous Mesorhizobium spp. And plant growth promoting rhizobacteria on yields and nutrients uptake of chickpea (Cicer arietinum L.) under sustainable agriculture. Ecol. Eng. 2013, 51, 282–286. [Google Scholar] [CrossRef]
- Jalali, G.; Lakzian, A.; Astaraei, A.; Haddad-Mashadrizeh, A.; Azadvar, M.; Esfandiarpour, E. Effects of land use on the structural diversity of soil bacterial communities in Southeastern Iran. Biosci. Biotech. Res. Asia. 2016, 13, 1739–1747. [Google Scholar] [CrossRef]
- Soliman, T.; Yang, S.Y.; Yamazaki, T.; Jenke-Kodama, H. Profiling soil microbial communities with next-generation sequencing: The influence of DNA kit selection and technician technical expertise. PeerJ 2017, 5, e4178. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.; Wang, G.; Jin, J.; Sui, Y.; Liu, J.; Liu, X. Comparison of microbial community structures in four Black soils along a climatic gradient in northeast China. Can. J. Soil Sci. 2012, 92, 543–549. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, G.; Jin, J.; Liu, J.; Zhang, Q.; Liu, X. Bacterial communities in soybean rhizosphere in response to soil type, soybean genotype, and their growth stage. Soil Biol. Biochem. 2009, 41, 919–925. [Google Scholar] [CrossRef]
- Attwood, G.T.; Wakelin, S.A.; Leahy, S.C.; Rowe, S.; Clarke, S.; Chapman, D.F.; Muirhead, R.R.; Jacobs, J.M.E. Applications of the soil, plant and rumen microbiomes in pastoral agriculture. Front. Nutr. 2019, 6, 107. [Google Scholar] [CrossRef]
- Bakker, M.G.; Chaparro, J.M.; Manter, D.K.; Vivanco, J.M. Impacts of bulk soil microbial community structure on rhizosphere microbiomes of Zea mays. Plant Soil. 2015, 392, 115. [Google Scholar] [CrossRef]
- Pascault, N.; Ranjard, L.; Kaisermann, A.; Bachar, D.; Christen, R.; Terrat, S.; Mathieu, O.; Lévêque, J.; Mougel, C.; Henault, C.; et al. Stimulation of different functional groups of bacteria by various plant residues as a driver of soil priming effect. Ecosystems 2013, 16, 810–822. [Google Scholar] [CrossRef]
- Huang, X.F.; Chaparro, J.M.; Reardon, K.F.; Zhang, R.; Shen, Q.; Vivanco, J.M. Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany 2014, 92, 267–275. [Google Scholar] [CrossRef]
- Fernandes, C.C.; Kishi, L.T.; Lopes, E.M.; Omori, W.P.; Souza, J.A.M.; Alves, L.M.C.; Lemos, E.G. Bacterial communities in mining soils and surrounding areas under regeneration process in a former ore mine. Braz. J. Microbiol. 2018, 49, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Nelkner, J.; Henke, C.; Lin, T.W.; Pätzold, W.; Hassa, J.; Jaenicke, S.; Grosch, R.; Puhler, A.; Sczyrba, A.; Schlüter, A. Effect of Long-Term Farming Practices on Agricultural Soil Microbiome Members Represented by Metagenomically Assembled Genomes (MAGs) and Their Predicted Plant-Beneficial Genes. Genes 2019, 10, 424. [Google Scholar] [CrossRef]
- Fei, Y.; Huang, S.; Zhang, H.; Tong, Y.; Wen, D.; Xia, X.; Wang, H.; Luo, Y.; Barceló, D. Response of soil enzyme activities and bacterial communities to the accumulation of microplastics in an acid cropped soil. Sci. Total Environ. 2019, 135634. [Google Scholar] [CrossRef]
- Yeager, C.M.; Gallegos-Graves, L.V.; Dunbar, J.; Hesse, C.N.; Daligault, H.; Kuske, C.R. Polysaccharide Degradation Capability of Actinomycetales Soil Isolates from a Semiarid Grassland of the Colorado Plateau. Appl. Environ. Microbiol. 2017, 83, 6. [Google Scholar] [CrossRef]
- Jog, R.; Nareshkumar, G.; Rajkumar, S. Plant growth promoting potential and soil enzyme production of the most abundant Streptomycess pp. from wheat rhizosphere. J. Appl. Microbiol. 2012, 113, 1154–1164. [Google Scholar] [CrossRef]
- Lopes, A.A.C.; de Sousa, D.M.G.; Chaer, G.M.; Junior, F.B.R.; Goedert, W.J.; Mendes, I.C. Interpretation of microbial soil indicators as a function of crop yield and organic carbon. Soil Sci. Soc. Am. J. 2013, 77, 461–472. [Google Scholar] [CrossRef]
- Stott, D.E.; Cambardella, C.A.; Tomer, M.D.; Karlen, D.L.; Wolf, R. A soil quality assessment within the Iowa River South Fork watershed. Soil Sci. Soc. Am. J. 2011, 75, 2271–2282. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, M.; Kucharski, J. Implications of soil pollution with diesel oil and BP Petroleum with Active Technology for soil health. Int. J. Environ. Res. Public Health 2019, 16, 2474. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Chaudhuri, S.; Maiti, S.K. Soil dehydrogenase enzyme activity in natural and mine soil—A Review. Middle-EastJ. Sci. Res. 2013, 13, 898–906. [Google Scholar] [CrossRef]
- Merino, C.; Godoy, R.; Matus, F. Soil enzymes and biological activity at different levels of organic matter stability. J. Soil Sci. Plant Nutr. 2016, 16, 14–30. [Google Scholar] [CrossRef]
- Bautista-Cruz, A.; Ortiz-Hernández, Y.D. Hydrolytic soil enzymes and their response to fertilization: A short review. Commun. Sci. 2015, 6, 255–262. [Google Scholar] [CrossRef]
- Foster, E.J.; Fogle, E.J.; Cotrufo, M.F. Sorption to biochar impacts β-glucosidase and phosphatase enzyme activities. Agriculture 2018, 8, 158. [Google Scholar] [CrossRef]

| Plants | Particle Diameter, mm | Granulometric Subgroups | |||
|---|---|---|---|---|---|
| < 0.002 | 0.002–0.020 | 0.020–0.050 | 0.050–2.0 | ||
| % | |||||
| Triticum aestivum | 3.9 a | 18.1 a | 17.7 a | 60.2 a | sandy loam |
| Brassica napus | 3.8 a | 16.9 b | 15.0 b | 64.4 a | sandy loam |
| Pisum sativum ssp. arvense | 4.0 a | 16.0 b | 14.0 b | 66.0 a | sandy loam |
| Plants | pHKCl | HAC | EBC | CEC | BS % |
|---|---|---|---|---|---|
| mmol (+) kg−1 d.m. of soil | |||||
| Triticum aestivum | 6.1 a | 19.4 b | 174.8 a | 194.2 a | 90.0 a |
| Brassica napus | 5.4 b | 21.2 a | 64.0 b | 85.2 b | 75.1 b |
| Pisum sativum ssp. arvense | 5.6 b | 19.2 b | 46.7 c | 65.9 c | 70.9 c |
| Plants | Content | Available Forms | Interchangeable Forms | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ntotal | Ctotal | P | K | Mg | K | Ca | Na | Mg | |||
| g kg−1 d.m. of Soil | mg kg−1 d.m. of Soil | ||||||||||
| Triticum aestivum | 1.0 a | 14.8 a | 69.1 a | 224.1 a | 53.0 b | 272.0 a | 500.0 b | 20.0 a | 83.3 b | ||
| Brassica napus | 0.8 c | 13.8 b | 59.4 b | 128.7 b | 45.0 c | 184.0 b | 533.3 a | 20.0 a | 59.5 c | ||
| Pisum sativum ssp. arvense | 0.9 b | 14.3 ab | 43.1 c | 107.9 c | 62.0 a | 176.0 b | 350.0 c | 20.0 a | 92.9 a | ||
| Plants | Deh µM TFF M O2 | Cat mM PNP | Pal | Pac | Aryl | Glu | Ure mM N-NH4 |
|---|---|---|---|---|---|---|---|
| mM PNP | |||||||
| Triticum aestivum | 5.67 b | 3.45 a | 2.41 a | 3.29 b | 0.51 a | 1.16 a | 2.07 a |
| Brassica napus | 5.18 c | 3.25 b | 0.39 C | 2.73 c | 0.19 c | 0.97 b | 0.94 C |
| Pisum sativum ssp. arvense | 6.16 a | 3.49 b | 0.69 b | 3.49 a | 0.27 b | 0.84 C | 1.47 b |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wyszkowska, J.; Borowik, A.; Olszewski, J.; Kucharski, J. Soil Bacterial Community and Soil Enzyme Activity Depending on the Cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense. Diversity 2019, 11, 246. https://doi.org/10.3390/d11120246
Wyszkowska J, Borowik A, Olszewski J, Kucharski J. Soil Bacterial Community and Soil Enzyme Activity Depending on the Cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense. Diversity. 2019; 11(12):246. https://doi.org/10.3390/d11120246
Chicago/Turabian StyleWyszkowska, Jadwiga, Agata Borowik, Jacek Olszewski, and Jan Kucharski. 2019. "Soil Bacterial Community and Soil Enzyme Activity Depending on the Cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense" Diversity 11, no. 12: 246. https://doi.org/10.3390/d11120246
APA StyleWyszkowska, J., Borowik, A., Olszewski, J., & Kucharski, J. (2019). Soil Bacterial Community and Soil Enzyme Activity Depending on the Cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense. Diversity, 11(12), 246. https://doi.org/10.3390/d11120246







