Nitrogen Fixing and Phosphate Mineralizing Bacterial Communities in Sweet Potato Rhizosphere Show a Genotype-Dependent Distribution
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Genotypes, Experimental Field Design and Conditions
2.2. TC-DNA Extraction and PCR Amplification
2.3. DGGE and Statistical Analyses
2.4. Determination of alp and nifH Genes Copy Numbers
3. Results
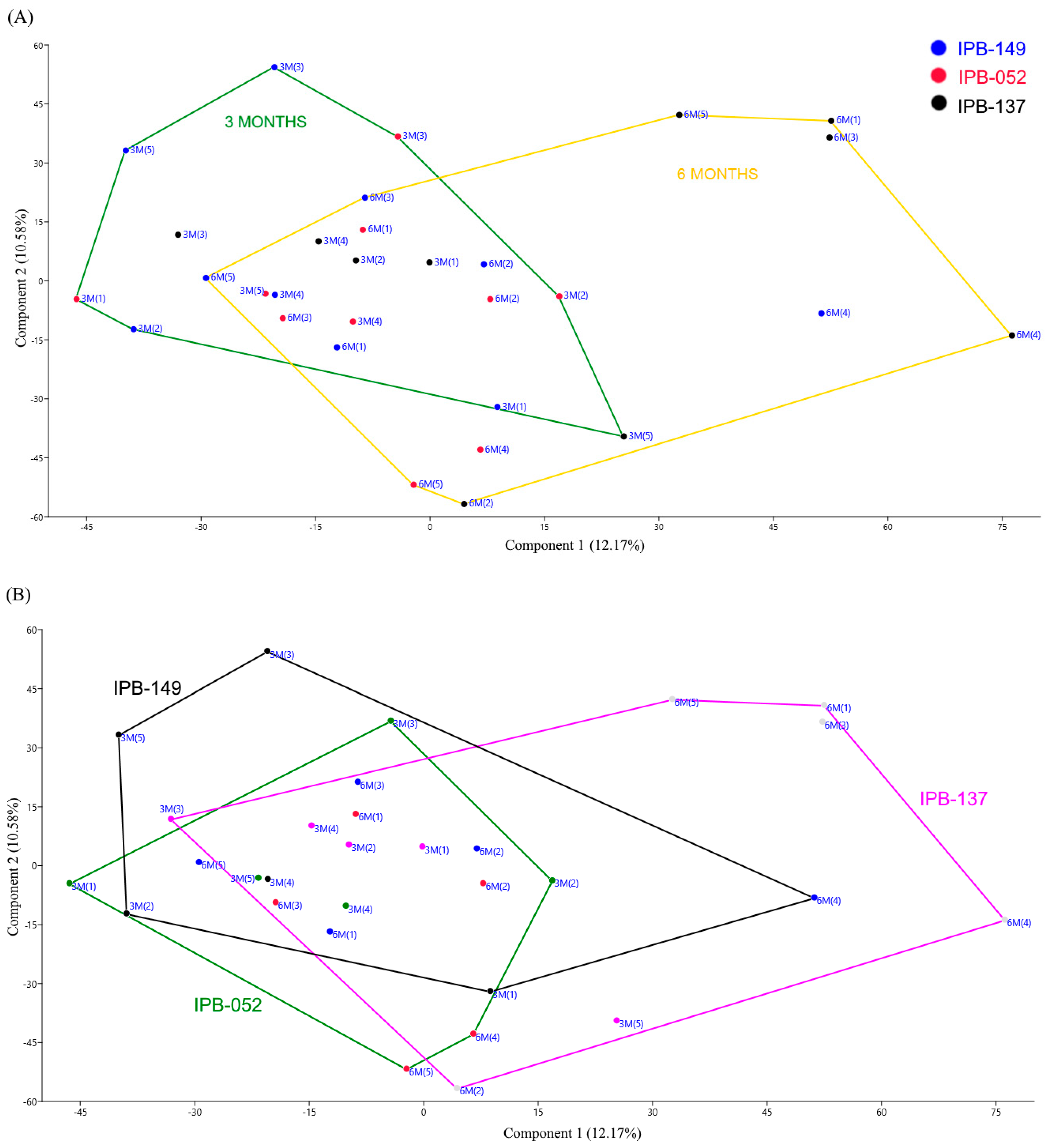
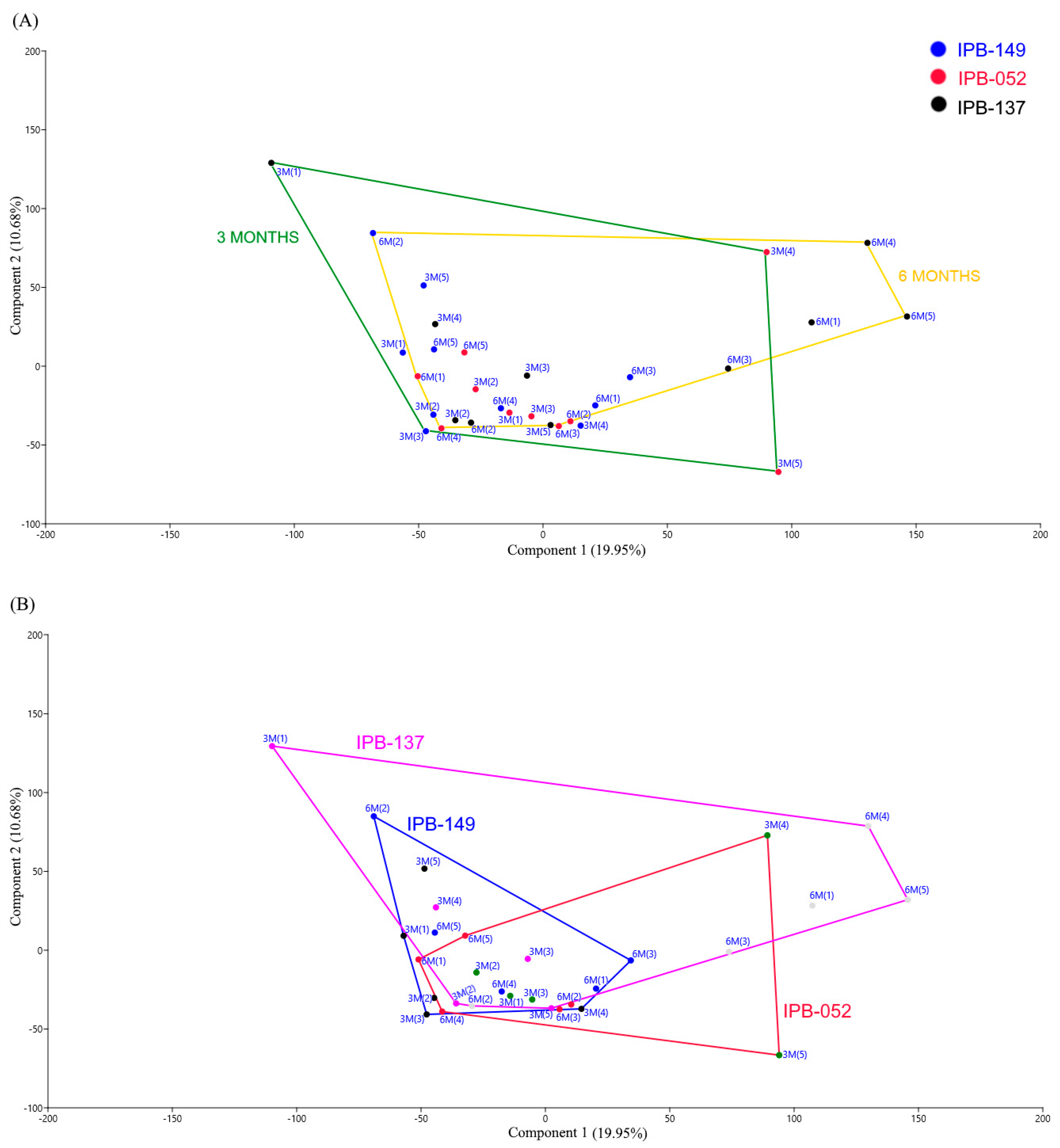
3.1. Structure of Phosphate Mineralizing and Nitrogen Fixing Bacterial Communities Analyzed by DGGE
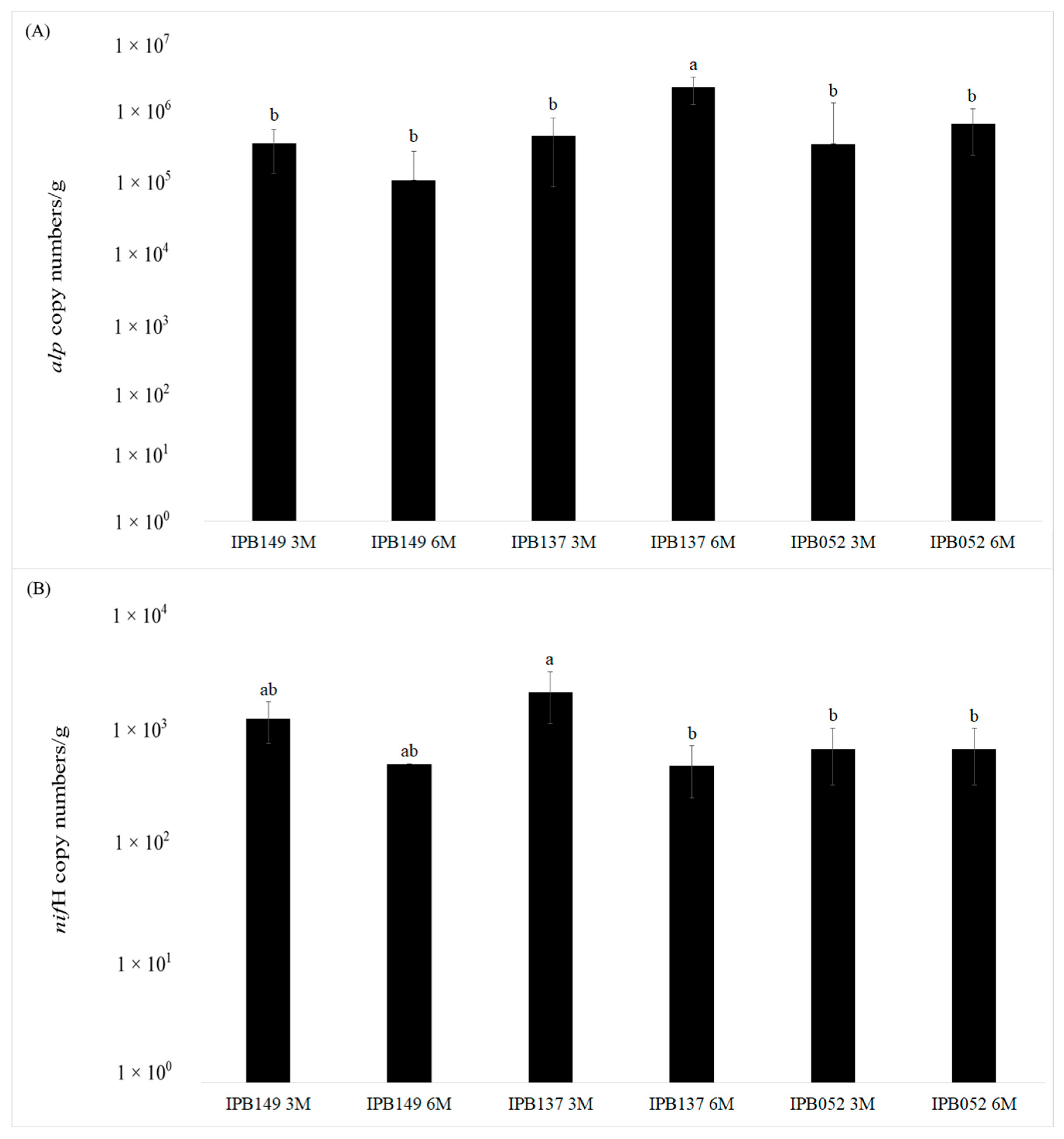
3.2. Quantitative Real-time PCR of alp and nifH Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Podile, A.R. Plant growth promoting rhizobacteria (PGPR): The bugs to debug the root zone. Crit. Ver. Microbiol. 2010, 36, 232–344. [Google Scholar] [CrossRef]
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, O.S.; Staton, M.E. Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019, 19, 201. [Google Scholar] [CrossRef] [PubMed]
- Babalola, O.O. Beneficial bacteria of agricultural importance. Biotechnol. Lett. 2010, 32, 1559–1570. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Lifshitz, R.; Zablotowicz, R.M. Free-living bacterial inocula for enhancing crop productity. Trends Biotechnol. 1989, 7, 39–43. [Google Scholar] [CrossRef]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability—A Review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Lugtenberg, B. Use of plant growth-promoting rhizobacteria to alleviate salinity stress in plants. In Use of Microbes for the Alleviation of Soil Stresses; Miransari, M., Ed.; Springer: New York, NY, USA, 2014; pp. 73–96. [Google Scholar]
- Glick, B.R. The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 1995, 41, 109–117. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Bulgarelli, D. The plant microbiome at work. Mol. Plant Microbe Interact. 2015, 28, 212–217. [Google Scholar] [CrossRef]
- Souza, R.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef]
- Marques, J.M.; da Silva, T.F.; Vollú, R.E.; Blank, A.F.; Ding, G.-C.; Seldin, L.; Smalla, K. Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol. Ecol. 2014, 88, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Hennion, N.; Durandb, M.; Vrieta, C.; Doidya, J.; Maurousseta, L.; Lemoinea, R.; Pourtaua, N. Sugars en route to the roots. Transport; metabolism and storage within plant roots and towards microorganisms of the rhizosphere. Physiol. Plant. 2019, 165, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Q.; Zhang, D. Gene expression activity and pathway selection for sucrose metabolism in developing storage root of sweet potato. Plant Cell Physiol. 2003, 44, 630–636. [Google Scholar] [CrossRef]
- CIP. The International Potato Center. 2019. Available online: https://cipotato.org (accessed on 29 April 2019).
- El Sheikha, A.F.; Ray, R.C. Potential impacts of bioprocessing of sweet potato: Review. Crit. Rev. Food Sci. Nutr. 2017, 57, 455–471. [Google Scholar] [CrossRef]
- Albuquerque, J.R.T.; Ribeiro, R.M.P.; Pereira, L.A.F.; Barros Júnior, A.P.; da Silveira, L.M.; dos Santos, M.G.; de Souza, A.R.E.; Lins, H.A.; Bezerra Neto, F. Sweet potato cultivars grown and harvested at different times in semiarid Brazil. Afr. J. Agric. Res. 2016, 11, 4810–4818. [Google Scholar]
- Yasmin, F.; Othman, R.; Sijam, K.; Saad, M.S. Effect of PGPR inoculation on growth and yield of sweet potato. J. Biol. Sci. 2007, 7, 421–424. [Google Scholar]
- Sahoo, R.K.; Bhardwaj, D.; Tuteja, N. Biofertilizers: A Sustainable Eco-Friendly Agricultural Approach to Crop Improvement. In Plant Acclimation to Environmental Stress; Tuteja, N., Gill, S.S., Eds.; Springer: New York, NY, USA, 2013; pp. 403–432. [Google Scholar]
- Dawwam, G.E.; Elbeltagy, A.; Emara, H.M.; Abbas, I.H.; Hassan, M.M. Beneficial effect of plant growth promoting bacteria isolated from the roots of potato plant. Ann. Agric. Sci. 2013, 58, 195–201. [Google Scholar] [CrossRef]
- Nasution, R.A.; Tangapo, A.; Taufik, I.; Aditiawati, P. Comparison of plant growth promoting rhizobacteria (PGPR) diversity and dynamics during growth of Cilembu sweet potato (Ipomoea batatas L var. Rancing) in Cilembu and Jatinangor site, Indonesia. J. Pure Appl. Microbiol. 2017, 11, 837–845. [Google Scholar] [CrossRef]
- Radziah, O.; Zulkifli, H.S. Utilization of Rhizobacteria for Increased Growth of Sweet Potato. In Investing Innovation: Agriculture; Food and Forestry; Zaharah, A.R., Ed.; University Putra Malaysia Press: Serdang, Malaysia, 2003; pp. 255–258. [Google Scholar]
- Yasmin, F.; Othman, R.; Sijam, K.; Saad, M.S. Characterization of beneficial properties of plant growth-promoting rhizobacteria isolated from sweet potato rhizosphere. Afr. J. Microbiol. Res. 2009, 3, 815–821. [Google Scholar]
- Marques, J.M.; da Silva, T.F.; Vollú, R.E.; de Lacerda, J.R.M.; Blank, A.F.; Smalla, K.; Seldin, L. Bacterial endophytes of sweet potato tuberous roots affected by the plant genotype and growth stage. Appl. Soil Ecol. 2015, 96, 273–281. [Google Scholar] [CrossRef]
- Alves, R.P.; Blank, A.F.; Oliveira, A.M.S.; Santana, A.D.D.; Pinto, V.S.; Andrade, T.M. Morpho-agronomic characterization of sweet potato germplasm. Hort. Bras. 2017, 35, 525–541. [Google Scholar] [CrossRef][Green Version]
- Sakurai, M.; Wasaki, J.; Tomizawa, Y.; Shinano, T.; Osaki, M. Analysis of bacterial communities on alkaline phosphatase genes in soil supplied with organic matter. Soil Sci. Plant Nut. 2008, 54, 62–71. [Google Scholar] [CrossRef]
- Simonet, P.; Grosjean, M.C.; Misra, A.K.; Nazaret, S.; Cournoyer, B.; Normand, P. Frankia genus-specific characterization by polymerase chain reaction. Appl. Environ. Microbiol. 1991, 57, 3278–8326. [Google Scholar] [PubMed]
- Poly, F.; Monrozier, L.J.; Bally, R. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 2001, 152, 95–103. [Google Scholar] [CrossRef]
- Monteiro, J.M.; Vollú, R.E.; Coelho, M.R.R.; Fonseca, A.; Gomes Neto, S.C.; Seldin, L. Bacterial communities within the rhizosphere and roots of vetiver (Chrysopogon zizanioides L. Roberty) sampled at different growth stages. Eur. J. Soil Biol. 2011, 47, 236–242. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M.H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar]
- Kropf, S.; Heuer, H.; Grüning, M.; Smalla, K. Significance test for comparing microbial community fingerprints using pairwise similarity measures. J. Microbiol. Meth. 2004, 57, 187–195. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Lacerda, J.R.M.; da Silva, T.F.; Vollú, R.E.; Marques, J.M.; Seldin, L. Generally recognized as safe (GRAS) Lactococcus lactis strains associated with Lippia sidoides Cham. are able to solubilize/mineralize phosphate. SpringerPlus 2016, 5, 828. [Google Scholar] [CrossRef]
- Taketani, R.G.; dos Santos, H.F.; van Elsas, J.D.; Rosado, A.S. Characterisation of the effect of a simulated hydrocarbon spill on diazotrophs in mangrove sediment mesocosm. Antonie Van Leeuwenhoek 2009, 96, 343–354. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abdel-Razzak, H.S.; Moussa, A.G.; Abd El-Fattah, M.A.; El-Morabet, G.A. Response of sweet potato to integrated effect of chemical and natural phosphorus fertilizer and their levels in combination with mycorrhizal inoculation. J. Biol. Sci. 2013, 13, 112–122. [Google Scholar] [CrossRef]
- Mukhongo, R.W.; Tumuhairwe, J.B.; Ebanyat, P.; AbdelGadir, A.H.; Thuita, M.; Masso, C. Combined application of biofertilizers and inorganic nutrients improves sweet potato yields. Front. Plant Sci. 2017, 8, 219. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.P.; Santos, J.F.; Cavalcante, L.F.; Pereira, W.E.; Santos, M.D.; Oliveira, A.N.; Silva, N.V. Yield of sweet potato fertilized with cattle manure and biofertilizer. Hort. Bras. 2010, 28, 277–281. [Google Scholar] [CrossRef][Green Version]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2013, 2, 587. [Google Scholar] [CrossRef]
- Yoneyama, T.; Terakado-Tonooka, J.; Minamisawa, K. Exploration of bacterial N2-fixation systems in association with soil-grown sugarcane; sweet potato; and paddy rice: A review and synthesis. Soil Sci. Plant Nut. 2017, 63, 578–590. [Google Scholar] [CrossRef]
- Tarafdar, J.C.; Jungk, A. Phosphatase activity in the rhizosphere and its relation to the depletion of soil organic phosphorus. Biol. Fert. Soils 1987, 3, 199–204. [Google Scholar] [CrossRef]
- Kandeler, E.; Marschner, P.; Tscherko, D.; Gahoonia, T.S. Nielsen NE. Microbial community composition and functional diversity in the rhizosphere of maize. Plant Soil 2002, 238, 301–312. [Google Scholar] [CrossRef]
- Chen, X.; Kou, M.; Tang, Z.; Zhang, A.; Li, H.; Wei, M. Responses of root physiological characteristics and yield of sweet potato to humic acid urea fertilizer. PLoS ONE 2017, 12, e0189715. [Google Scholar] [CrossRef]
- Chung, H.; Park, M.; Madhaiyan, M.; Seshadri, S.; Song, J.; Cho, H.; Sa, T. Isolation and characterization of phosphate solubilizing bacteria from the rhizosphere of crop plants of Korea. Soil Biol. Biochem. 2005, 37, 1970–1974. [Google Scholar] [CrossRef]
- Hill, W.A.; Bacon-Hill, P.; Crossman, S.M.; Stevens, C. Characterization of N2-fixing bacteria associated with sweet potato roots. Can. J. Microbiol. 1983, 29, 860–862. [Google Scholar] [CrossRef]
- Reiter, B.; Bürgmann, H.; Burg, K.; Sessitsch, A. Endophytic nifH gene diversity in African sweet potato. Can. J. Microbiol. 2003, 49, 549–555. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Terakado-Tonooka, J.; Ohwaki, Y.; Yamakawa, H.; Tanaka, F.; Yoneyama, T.; Fujihara, S. Expressed nifH genes of endophytic bacteria detected in field-grown sweet potatoes (Ipomoea batatas L.). Microbes Environ. 2008, 23, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Weller, D.M.; Thomashow, L.S. Current challenges in introducing beneficial microorganisms into the rhizosphere. In Molecular Ecology of Rhizosphere Microorganisms: Biotechnology and Release of GMOs; O’Gara, F., Dowling, D.N., Boesten, B., Eds.; VCH: New York, NY, USA, 1994; pp. 1–18. [Google Scholar]
| Genes | Sampling Time (t1 × t2) | Dissimilarity (%) | Genotypes (t1) | Dissimilarity (%) | Genotypes (t2) | Dissimilarity (%) |
|---|---|---|---|---|---|---|
| alp | IPB-149 | 0.7 | IPB-149 × IPB-137 | 8.1* | IPB-149 × IPB-137 | 17.6* |
| IPB-137 | 0.4* | IPB-149 × IPB-052 | 0.2 | IPB-149 × IPB-052 | 3.1 | |
| IPB-052 | −0.3 | IPB-137 × IPB-052 | −1.2 | IPB-137 × IPB-052 | 4.1 | |
| nifH | IPB-149 | −1.9 | IPB-149 × IPB-137 | −1.8 | IPB-149 × IPB-137 | 9.0 |
| IPB-137 | 18.3 | IPB-149 × IPB-052 | 0.4 | IPB-149 × IPB-052 | 11.0* | |
| IPB-052 | 12.4 | IPB-137 × IPB-052 | 1.6 | IPB-137 × IPB-052 | 26.8* |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marques, J.M.; Mateus, J.R.; da Silva, T.F.; Couto, C.R.d.A.; Blank, A.F.; Seldin, L. Nitrogen Fixing and Phosphate Mineralizing Bacterial Communities in Sweet Potato Rhizosphere Show a Genotype-Dependent Distribution. Diversity 2019, 11, 231. https://doi.org/10.3390/d11120231
Marques JM, Mateus JR, da Silva TF, Couto CRdA, Blank AF, Seldin L. Nitrogen Fixing and Phosphate Mineralizing Bacterial Communities in Sweet Potato Rhizosphere Show a Genotype-Dependent Distribution. Diversity. 2019; 11(12):231. https://doi.org/10.3390/d11120231
Chicago/Turabian StyleMarques, Joana Montezano, Jackeline Rossetti Mateus, Thais Freitas da Silva, Camila Rattes de Almeida Couto, Arie Fitzgerald Blank, and Lucy Seldin. 2019. "Nitrogen Fixing and Phosphate Mineralizing Bacterial Communities in Sweet Potato Rhizosphere Show a Genotype-Dependent Distribution" Diversity 11, no. 12: 231. https://doi.org/10.3390/d11120231
APA StyleMarques, J. M., Mateus, J. R., da Silva, T. F., Couto, C. R. d. A., Blank, A. F., & Seldin, L. (2019). Nitrogen Fixing and Phosphate Mineralizing Bacterial Communities in Sweet Potato Rhizosphere Show a Genotype-Dependent Distribution. Diversity, 11(12), 231. https://doi.org/10.3390/d11120231





