Natural Variations in Key Maturity Genes Underpin Soybean Cultivars Adaptation Beyond 50° N in Northeast China
Abstract
1. Introduction
2. Results
2.1. Precise Classification of Test Soybean Cultivars into Relative Maturity Groups (RMGs)
2.2. Phenotypic Analysis in Multiple Environments
2.3. Photothermal and Temperature Sensitivity of Soybean in High-Latitude and High-Altitude Regions
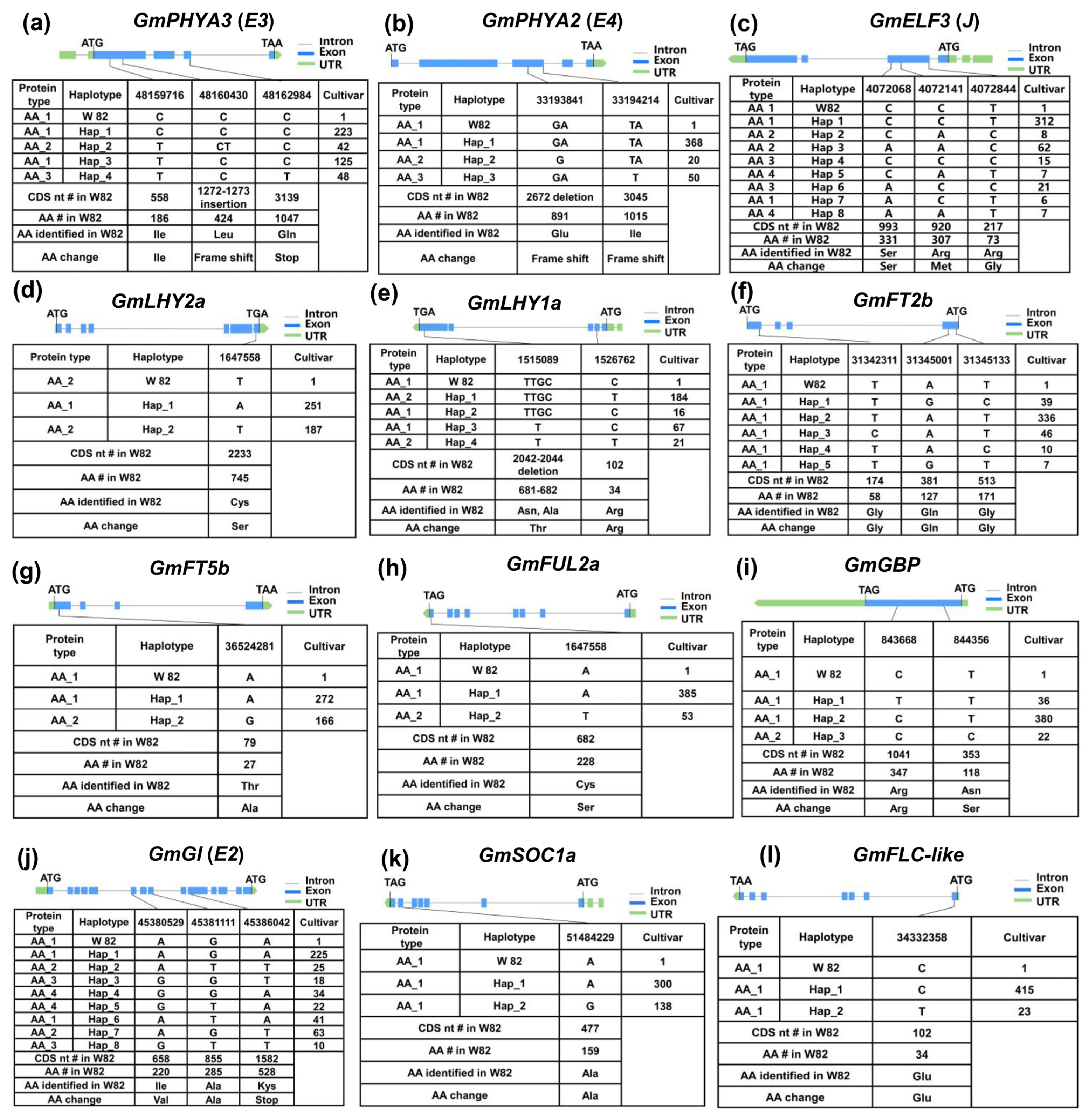
2.4. Genetic Variation Analysis and Haplotype Identification of Maturity Major-Effect Genes in Soybean
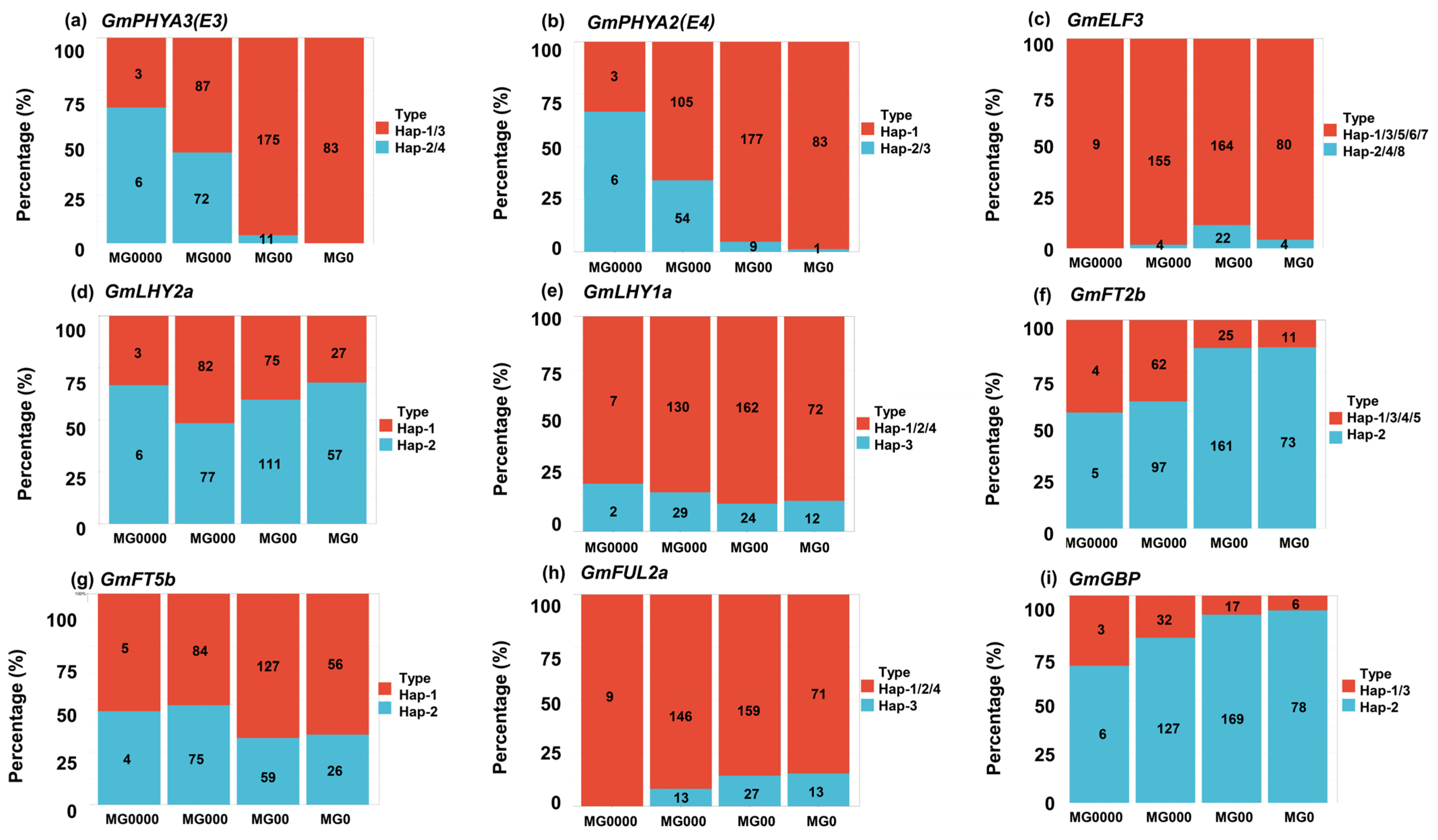
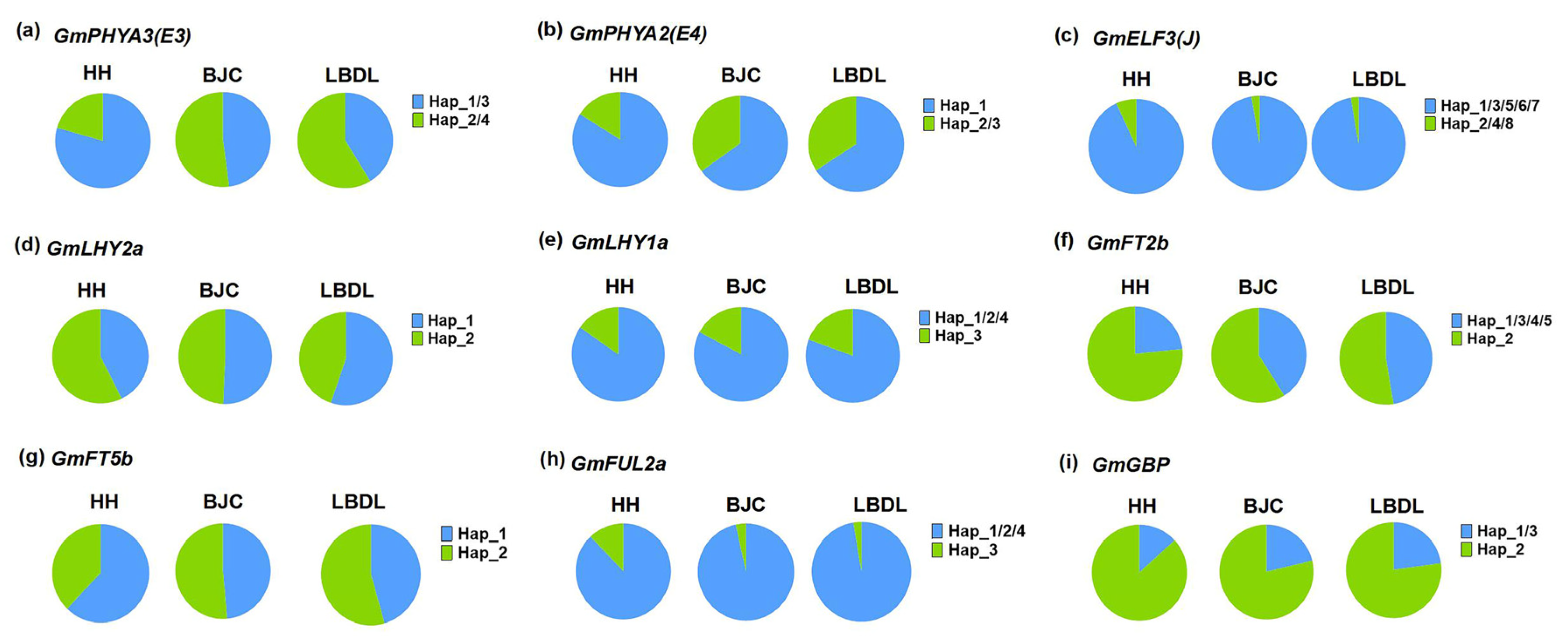
2.5. Distribution Pattern of Different Haplotypes of Maturity Groups and Geographical Regions
3. Discussion
3.1. Characteristics of Adaptive Soybean Cultivars for High Latitudes and High Altitudes
3.2. Conservation and Variation Patterns of Major-Effect Maturity Genes in Soybean Varieties North of 50° N Latitude
3.3. Molecular Breeding Strategies for Improving Super-Early Soybean Cultivars in High-Latitude Regions
4. Materials and Methods
4.1. Plant Materials and Multiple-Site Field Experiments
4.2. Photothermal Sensitivity Calculation
4.3. DNA Extraction, Resequencing, SNP Calling, and Natural Variation Analysis
4.4. Haplotype Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hou, Z.; Fang, C.; Liu, B.; Yang, H.; Kong, F. Origin, Variation, and Selection of Natural Alleles Controlling Flowering and Adaptation in Wild and Cultivated Soybean. Mol. Breed. 2023, 43, 36. [Google Scholar] [PubMed]
- Kim, M.Y.; Van, K.; Kang, Y.J.; Kim, K.H.; Lee, S.H. Tracing Soybean Domestication History: From Mucleotide to Genome. Breed. Sci. 2012, 61, 445–452. [Google Scholar] [PubMed]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176. [Google Scholar]
- USDA Foreign Agricultural Service. China: Oilseeds and Products Update. 2024. Available online: https://www.fas.usda.gov/ (accessed on 28 June 2024).
- World Grain. China Soybean Imports Top 100 Million Tonnes. 2023. Available online: https://www.world-grain.com/ (accessed on 11 May 2024).
- Liu, L.; Li, J.; Shu, Y.; Chen, X.; Tang, G. Current Situation of Soybean Production and Consumption in China and Strategies to Improve Self-sufficiency Rate. Chin. J. Oil Crop Sci. 2022, 44, 242–248. (In Chinese) [Google Scholar]
- Jia, H.; Jiang, B.; Wu, C.; Lu, W.; Hou, W.; Sun, S.; Yan, H.; Han, T. Maturity Group Classification and Maturity Locus Genotyping of Early-maturing Soybean Varieties from High-latitude Cold Regions. PLoS ONE 2014, 9, e94139. [Google Scholar]
- He, Y.; Liu, D.; Yao, Y.; Huang, Q.; Li, J.; Chen, Y.; Shi, S.; Wan, L.; Yu, S.; Wang, D. Spatializing Growth Suitability for Spring Soybean Cultivation in Northeast China. J. Appl. Meteorol. Clim. 2013, 52, 773–783. [Google Scholar]
- Yao, X.; Pachner, M.; Rittler, L.; Hahn, V.; Leiser, W.; Riedel, C.; Rezi, R.; Bétrix, C.A.; Nawracała, J.; Temchenko, I.; et al. Genetic Adaptation of Phenological Stages in Chinese and European Elite Soybeans (Glycine max [L.] Merr.) Across Latitudes in Central Europe. Plant Breed. 2024, 143, 695–705. [Google Scholar]
- Zhu, X.; Willmar, L.; Volker, H.; Tobias, W. The Genetic Architecture of Soybean Photothermal Adaptation to High Latitudes. J. Exp. Bot. 2023, 74, 2987–3002. [Google Scholar]
- Li, C.; Li, Y.H.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A Domestication-Associated Gene GmPRR3b Regulates the Circadian Clock and Flowering Time in Soybean. Mol. Plant. 2020, 13, 745–759. [Google Scholar]
- Lin, X.; Liu, B.; Weller, J.L.; Abe, J.; Kong, F. Molecular Mechanisms for the Photoperiodic Regulation of Flowering in Soybean. J. Integr. Plant Biol. 2021, 63, 981–994. [Google Scholar]
- Dong, L.; Cheng, Q.; Fang, C.; Kong, L.; Yang, H.; Hou, Z.; Li, Y.; Nan, H.; Zhang, Y.; Chen, Q. Parallel Selection of Distinct Tof5 Alleles Drove the Adaptation of Cultivated and Wild Soybean to High Latitudes. Mol. Plant. 2022, 15, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Li, S.; Wang, L.; Su, T.; Zhang, C.; Bi, Y.; Lai, Y.; Kong, L.; Wang, F.; Pei, X. The Genetic Basis of High-latitude Adaptation in Wild Soybean. Curr. Biol. 2023, 33, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 Wild and Cultivated Accessions Identifies Genes Related to Domestication and Improvement in Soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [PubMed]
- Chen, L.; Cai, Y.; Qu, M.; Wang, L.; Sun, H.; Jiang, B.; Wu, T.; Liu, L.; Sun, S.; Wu, C.; et al. Soybean Adaption to High-latitude Regions is Associated with Natural Variations of GmFT2b, an Ortholog of FLOWERING LOCUS T. Plant Cell Environ. 2020, 43, 934–944. [Google Scholar]
- Lin, X.; Dong, L.; Tang, Y.; Li, H.; Cheng, Q.; Li, H.; Zhang, T.; Ma, L.; Xiang, H.; Chen, L.; et al. Novel and Multifaceted Regulations of Photoperiodic Flowering by Phytochrome A in Soybean. Proc. Natl. Acad. Sci. USA 2022, 119, e2208708119. [Google Scholar]
- Tang, Y.; Lu, S.; Fang, C.; Liu, H.; Dong, L.; Li, H.; Su, T.; Li, S.; Wang, L.; Cheng, Q.; et al. Diverse Flowering Responses Subjecting to Ambient High Temperature in Soybean under Short-day Conditions. Plant Biotechnol. J. 2023, 21, 782–791. [Google Scholar]
- Liu, L.; Song, W.; Wang, L.; Sun, X.; Qi, Y.; Wu, T.; Sun, S.; Jiang, B.; Wu, C.; Hou, W.; et al. Allele Combinations of Maturity Genes E1-E4 Affect Adaptation of Soybean to Diverse Geographic Regions and Farming Systems in China. PLoS ONE. 2020, 15, e0235397. [Google Scholar] [CrossRef]
- Xia, Z.; Watanabe, S.; Yamada, T.; Tsubokura, Y.; Nakashima, H.; Zhai, H.; Anai, T.; Sato, S.; Yamazaki, T.; Lü, S.; et al. Positional Cloning and Characterization Reveal the Molecular Basis for Soybean Maturity Locus E1 that Regulates Photoperiodic Flowering. Proc. Natl. Aacd. Sci. USA 2012, 109, 2155–2164. [Google Scholar]
- Li, Y.; Hou, Z.; Li, W.; Li, H.; Lu, S.; Gan, Z.; Du, H.; Li, T.; Zhang, Y.; Kong, F.; et al. The Legume-specific Transcription Factor E1 Controls Leaf Morphology in Soybean. BMC Plant Biol. 2021, 21, 531. [Google Scholar]
- Fang, C.; Du, H.; Wang, L.; Liu, B.; Kong, F. Mechanisms Underlying Key Agronomic Traits and Implications for Molecular Breeding in Soybean. J. Genet. Genom. 2024, 51, 379–393. [Google Scholar] [CrossRef]
- Kong, F.; Liu, B.; Xia, Z.; Sato, S.; Kim, B.M.; Watanabe, S.; Yamada, T.; Tabata, S.; Kanazawa, A.; Harada, K. Two Coordinately Regulated Homologs of FLOWERING LOCUS T are Involved in the Control of Photoperiodic Flowering in Soybean. Plant Physiol. 2010, 154, 1220–1231. [Google Scholar] [PubMed]
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D.; et al. Stepwise Selection on Homologous PRR Genes Controlling Flowering and Maturity During Soybean Domestication. Nat. Genet. 2020, 52, 428–436. [Google Scholar] [PubMed]
- Kou, K.; Yang, H.; Li, H.; Fang, C.; Chen, L.; Yue, L.; Nan, H.; Kong, L.; Li, X.; Wang, F.; et al. A functionally Divergent SOC1 Homolog Improves Soybean Yield and Latitudinal Adaptation. Curr. Biol. 2022, 32, 1728–1742.e6. [Google Scholar]
- Jiang, B.; Nan, H.; Gao, Y.; Tang, L.; Yue, Y.; Lu, S.; Ma, L.; Cao, D.; Sun, S.; Wang, J.; et al. Allelic Combinations of Soybean Maturity Loci E1, E2, E3 and E4 Result in Diversity of Maturity and Adaptation to Different Latitudes. PLoS ONE 2014, 9, e106042. [Google Scholar]
- Jiang, B.; Zhang, S.; Song, W.; Mohammad, A.A.K.; Sun, S.; Zhang, C.; Wu, T.; Wu, C.; Han, T. Natural Variations of FT Family Genes in Soybean Varieties Covering a Wide Range of Maturity Groups. BMC Genom. 2019, 20, 230. [Google Scholar]
- Alliprandini, L.F.; Abatti, C.; Bertagnolli, P.F.; Cavassim, J.E.; Gabe, H.L.; Kurek, A.; Matsumoto, M.N.; de Oliveira, M.A.R.; Pitol, C.; Prado, L.C. Understanding Soybean Maturity Groups in Brazil: Environment, Cultivar Classification, and Stability. Crop Sci. 2009, 49, 801–808. [Google Scholar]
- Wu, T.; Lu, S.; Cai, Y.; Xu, X.; Zhang, L.; Chen, F.; Jiang, B.; Zhang, H.; Sun, S.; Zhai, H.; et al. Molecular Breeding for Improvement of Photothermal Adaptability in Soybean. Mol. Breed. 2023, 43, 60. [Google Scholar] [CrossRef]
- Liu, X.; Wu, J.A.; Ren, H.; Qi, Y.; Li, C.; Cao, J.; Zhang, X.; Zhang, Z.; Cai, Z.; Gai, J. Genetic Variation of World Soybean Maturity Date and Geographic Distribution of Maturity Groups. Breed. Sci. 2017, 67, 221–232. [Google Scholar]
- Jia, H.; Liang, X.; Lu, W.; Zhang, L.; Wu, T.; Song, W.; Sun, S.; Zhang, L.; Yan, H.; Guo, R.; et al. Fine Identification of Maturity Groups of Regional Trial Check Varieties in Northern Heilongjiang Province. Soybean Sci. Technol 2023, 1, 1–11. (In Chinese) [Google Scholar]
- Fu, M.; Wang, Y.; Ren, H.; Du, W.; Wang, D.; Bao, R.; Yang, X.; Tian, Z.; Fu, L.; Cheng, Y.; et al. Genetic Dynamics of Earlier Maturity Group Emergence in South-to-north Extension of Northeast China Soybeans. Theor. Appl. Genet. 2020, 133, 1839–1857. [Google Scholar]
- Tsubokura, Y.; Watanabe, S.; Xia, Z.; Kanamori, H.; Yamagata, H.; Kaga, A.; Katayose, Y.; Abe, J.; Ishimoto, M.; Harada, K. Natural Variation in the Genes Responsible for Maturity Loci E1, E2, E3 and E4 in Soybean. Ann. Bot. 2014, 113, 429–441. [Google Scholar] [PubMed]
- Liu, B.H.; Kanazawa, A.; Matsumura, H.; Takahashi, R.; Harada, K.; Abe, J. Genetic Redundancy in Soybean Photoresponses Associated with Duplication of the Phytochrome A Gene. Genetics 2008, 180, 995–1007. [Google Scholar] [PubMed]
- Watanabe, S.; Hideshima, R.; Xia, Z.; Tsubokura, Y.; Sato, S.; Nakamoto, Y.; Yamanaka, N.; Takahashi, R.; Ishimoto, M.; Anai, T.; et al. Map-based Cloning of Thegene Associated with the Soybean Maturity Locus E3. Genetics 2009, 182, 1251–1262. [Google Scholar] [PubMed]
- Dong, L.; Fang, C.; Cheng, Q.; Su, T.; Kou, K.; Kong, L.; Zhang, C.; Li, H.; Hou, Z.; Zhang, Y.; et al. Genetic Basis and Adaptation Trajectory of Soybean From its Temperate Origin to Tropics. Nat. Commun. 2021, 12, 5445. [Google Scholar]
- Lu, S.; Zhao, X.; Hu, Y.; Liu, S.; Nan, H.; Li, X.; Fang, C.; Cao, D.; Shi, X.; Kong, L.; et al. Natural Variation at the Soybean J Locus Improves Adaptation to the Tropics and Enhances Yield. Nat. Genet. 2017, 49, 773–779. [Google Scholar]
- Zhang, Y.; Zhao, L.; Li, H.; Gao, Y.; Li, Y.; Wu, X.; Teng, W.; Han, Y.; Zhao, X.; Li, W. GmGBP1, A Homolog of Human Ski Interacting Protein in Soybean, Regulates Flowering and Stress Tolerance in Arabidopsis. BMC Plant Biol. 2013, 13, 21. [Google Scholar]
- Zhao, L.; Li, M.; Xu, C.; Yang, X.; Li, D.; Zhao, X.; Wang, K.; Li, Y.; Zhang, X.; Liu, L.; et al. Natural Variation in GmGBP1 Promoter Affects Photoperiod Control of Flowering Time and Maturity in Soybean. Plant J. 2018, 96, 147–162. [Google Scholar]
- Fan, C.; Hu, R.; Zhang, X.; Wang, X.; Zhang, W.; Zhang, Q.; Ma, J.; Fu, Y. Conserved CO-FT Regulons Contribute to the Photoperiod Flowering Control in Soybean. BMC Plant Biol. 2014, 14, 9. [Google Scholar]
- Yuan, J.; Ott, T.; Hiltbrunner, A. Phytochromes and Flowering: Legumes Do It Another Way. Trends Plant Sci. 2023, 28, 379–381. [Google Scholar]
- Jaudal, M.; Wen, J.; Mysore, K.; Putterill, J. Medicago PHYA Promotes Flowering, Primary Stem Elongation and Expression of Flowering Time Genes in Long Days. BMC Plant Biol. 2020, 20, 329. [Google Scholar]
- Weller, J.; Ortega, R. Genetic Control of Flowering Time in Legumes. Front. Plant Sci. 2015, 6, 207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhai, H.; Wang, Y.; Tian, X.; Zhang, Y.; Wu, H.; Lü, S.; Yang, G.; Li, Y.; Wang, L.; et al. Functional Conservation and Diversification of the Soybean Maturity Gene E1 and Its Homologs in Legumes. Sci. Rep. 2016, 6, 29548. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Sun, C.; Li, J.; Gao, C. Targeted Genome-modification Tools and Their Advanced Applications in Crop Breeding. Nat. Rev. Genet. 2024, 25, 603–622. [Google Scholar] [CrossRef] [PubMed]
- Tuncel, A.; Pan, C.; Clem, J.; Liu, D.; Qi, Y. CRISPR-Cas Applications in Agriculture and Plant Research. Nat. Rev. Mol. Cell Biol. 2025. [Google Scholar] [CrossRef]
- Wang, L.; O’Conner, S.; Tanvir, R.; Zheng, W.; Cothron, S.; Towery, K.; Bi, H.; Ellison, E.; Yang, B.; Voytas, D.; et al. CRISPR/Cas9-based Editing of NF-YC4 Promoters Yields High-protein Rice and Soybean. New Phytol. 2025, 245, 2103–2116. [Google Scholar] [CrossRef]
- Liao, W.; Guo, R.; Li, J.; Liu, N.; Jiang, L.; Whelan, J.; Shou, H. CRISPR/Cas9-mediated Mutagenesis of SEED FATTY ACID REDUCER Genes Significantly Increased Seed oil Content in Soybean. Plant Cell Physiol. 2025, 66, 273–284. [Google Scholar]
- Fehr, W.R.; Caviness, C.E. Stages of Soybean Development. Agriculture and Home Economics Experiment Station; Iowa State University of Science and Technology: Ames, IA, USA, 1977. [Google Scholar]
- Bolger, A.M.; Lohse, B.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Li, H. A Statistical Framework for SNP Calling, Mutation Discovery, Association Mapping and Population Genetical Parameter Estimation from Sequencing Data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Cingolani, P.; Patel, V.M.; Coon, M.; Nguyen, T.; Land, S.J.; Ruden, D.M.; Lu, X. Using Drosophila Melanogaster as A Model for Genotoxic Chemical Mutational Studies with A New Program, SnpSift. Front. Genet. 2012, 3, 35. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar]
- Bates, B.D.; Maechler, M. lme4: Linear Mixed-Effects Models Using S4 Classes; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar] [CrossRef]
- Field, A. Discovering Statistics Using IBM SPSS Statistics; Sage Publications Ltd.: Thousand Oaks, CA, USA, 2013. [Google Scholar]




| A. Genes Functionally Conserved Across the Cultivars | ||
| Gene | Status | Type |
| E1La | Functional | Inhibitory |
| E1Lb | Functional | Inhibitory |
| GmFT1a | Functional | Inhibitory |
| GmFT1b | Functional | Inhibitory |
| GmFT2a | Functional | Promotive |
| GmFT3a | Functional | Promotive |
| GmFT5a | Functional | Promotive |
| GmFT4 | Functional | Inhibitory |
| GmLHY1b | Functional | Promotive |
| GmLHY2b | Functional | Promotive |
| GmLUX1 | Functional | Promotive |
| GmLUX2 | Functional | Promotive |
| GmPRR3a | Functional | Inhibitory |
| GmPRR3b | Functional | Inhibitory |
| GmFULa | Functional | Promotive |
| GmSOC1b | Functional | Promotive |
| QNE1 | Functional | Promotive |
| GmPIF4b | Functional | Promotive |
| GmRAV | Functional | Inhibitory |
| GmEID1 | Functional | Promotive |
| GmTFLc | Functional | Inhibitory |
| GmTFLd | Functional | Inhibitory |
| E1 | Mutation (e1-as) | Inhibitory |
| B. Genes Specifically Mutated in the Cultivars Adaptive for High Latitudes Beyond 50° N | ||
| Gene | Haplotype | Function |
| GmPHYA3(E3) | H2/H4 | Early maturity |
| GmPHYA2(E4) | H2/H3 | Early maturity |
| GmELF3 | H1/H3/H5/H6/H7 | Early maturity |
| GmLHY2a | H1 | Early maturity |
| GmLHY1a | H3 | Early maturity |
| GmFT2b | H1/H3/H4/H5 | Early maturity |
| GmFT5b | H2 | Early maturity |
| GmFUL2a | H2 | Early maturity |
| GmGBP | H1/H3 | Early maturity |
| E2 | H1-H8 | No significant |
| GmFLC-like | H1/H2 | No significant |
| GmSOC1a | H1/H2 | No significant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, H.; Sun, B.; Jiang, B.; Wang, P.; Naser, M.; Qian, S.; Wang, L.; Zhang, L.; Sinegovskii, M.; Sun, S.; et al. Natural Variations in Key Maturity Genes Underpin Soybean Cultivars Adaptation Beyond 50° N in Northeast China. Int. J. Mol. Sci. 2025, 26, 3362. https://doi.org/10.3390/ijms26073362
Jia H, Sun B, Jiang B, Wang P, Naser M, Qian S, Wang L, Zhang L, Sinegovskii M, Sun S, et al. Natural Variations in Key Maturity Genes Underpin Soybean Cultivars Adaptation Beyond 50° N in Northeast China. International Journal of Molecular Sciences. 2025; 26(7):3362. https://doi.org/10.3390/ijms26073362
Chicago/Turabian StyleJia, Hongchang, Baiquan Sun, Bingjun Jiang, Peiguo Wang, Mahmoud Naser, Shuqing Qian, Liwei Wang, Lixin Zhang, Mikhail Sinegovskii, Shi Sun, and et al. 2025. "Natural Variations in Key Maturity Genes Underpin Soybean Cultivars Adaptation Beyond 50° N in Northeast China" International Journal of Molecular Sciences 26, no. 7: 3362. https://doi.org/10.3390/ijms26073362
APA StyleJia, H., Sun, B., Jiang, B., Wang, P., Naser, M., Qian, S., Wang, L., Zhang, L., Sinegovskii, M., Sun, S., Lu, W., Sinegovskaya, V., Bai, J., & Han, T. (2025). Natural Variations in Key Maturity Genes Underpin Soybean Cultivars Adaptation Beyond 50° N in Northeast China. International Journal of Molecular Sciences, 26(7), 3362. https://doi.org/10.3390/ijms26073362






