Mismatch Repair Deficiency and the Role of Non-Canonical Functions in Cancer: Diagnosis and Therapeutic Implications
Abstract
1. Methodology
2. Introduction
3. MMR Pathway in Normal Human Cells
4. Emerging Molecular Mechanisms of MMR Deficiency
4.1. The Non-Canonical Roles of MMR Genes
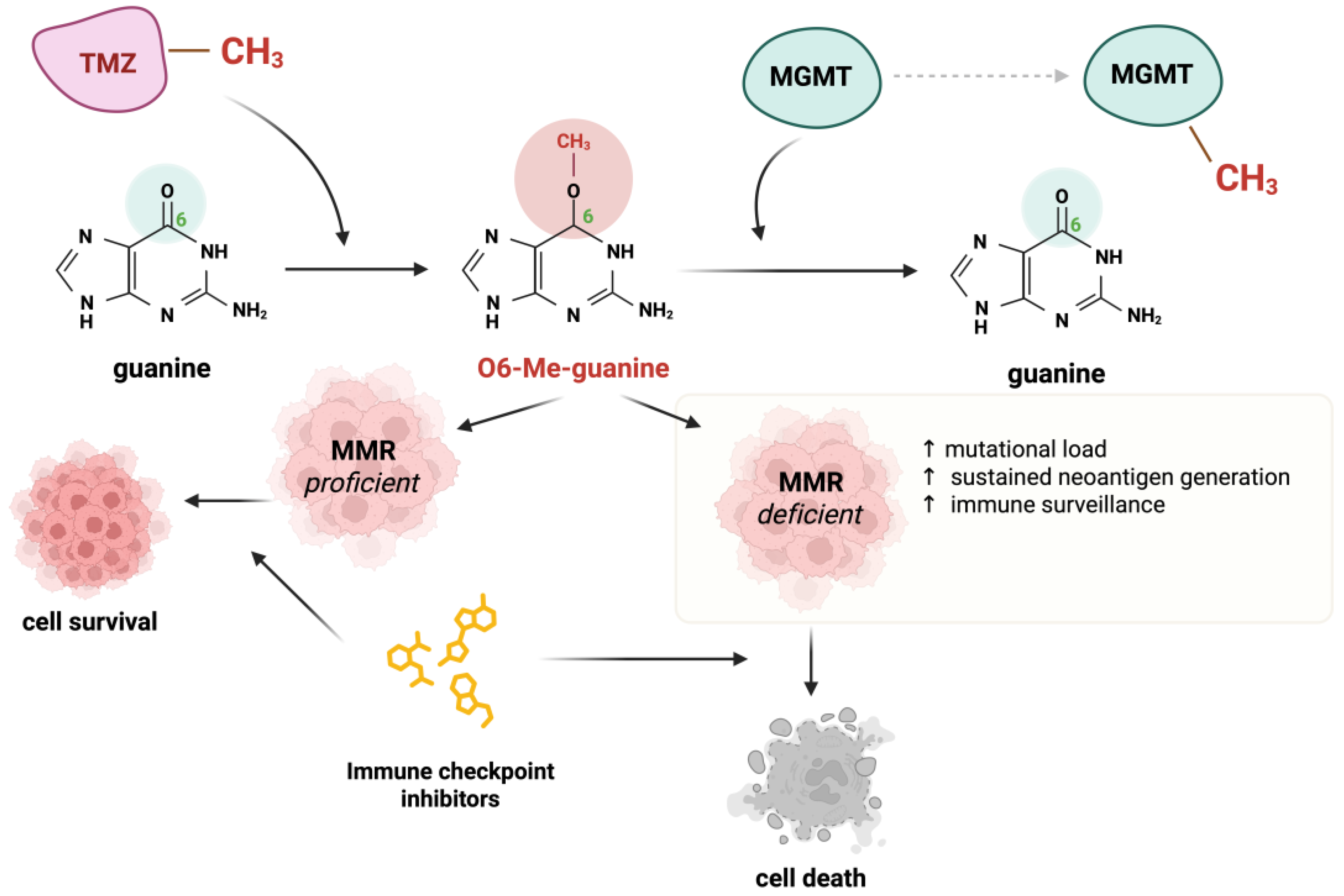
4.2. Alkylation Damage in Cells
4.3. MMR Heterogeneity
5. MMR Deficiency in Understudied Cancer Types
5.1. Constitutional MMR Deficiency Syndrome
5.2. MMR Deficiency in Rare Solid and Hematologic Tumors
6. Novel Diagnostic Approaches for MMR Deficiency
6.1. Liquid Biopsy and ctDNA
6.2. Functional Assays for MMR Activity
7. MMR Altering Therapies
8. MMR Deficiency in Early-Stage vs. Advanced Cancer
9. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADDM | active DNA demethylation; |
| AE | adverse event; |
| AID | activation-induced deaminase; |
| ATR | ataxia telangiectasia and Rad3–related kinase; |
| BER | base excision repair; |
| cHL | classical Hodgkin Lymphoma; |
| CMMRD | constitutional mismatch repair deficiency; |
| CR/PR | complete/partial response; |
| CRC | colorectal cancer; |
| ctDNA | circulating tumor DNA; |
| CTLA-4 | cytotoxic T-lymphocyte–associated antigen 4; |
| DCR | disease control rate (CR + PR + stable disease); |
| DDM | DNA demethylation; |
| DLBCL | diffuse large B-cell lymphoma; |
| dMMR | deficient mismatch repair (MMRdeficient); |
| dPCR | digital polymerase chain reaction; |
| EC | endometrial cancer; |
| EXO1 | exonuclease 1; |
| GE | gastroesophageal; |
| HeCOG | hellenic cooperative oncology group; |
| HR | hazard ratio; |
| ICI | immune checkpoint inhibitors; |
| IDL | insertion-deletion loops; |
| IHC | Immunohistochemistry; |
| IO | immunotherapy; |
| LS | lynch syndrome; |
| MAF | mutant allele frequencies; |
| mCRC | metastatic colorectal cancers; |
| MEF | mouse embryonic fibroblasts; |
| MGMT | methylguanine DNA methyltransferase; |
| MLH | MutL homolog; |
| MMR | mismatch repair; |
| MSH | MutS homolog; |
| MSI | microsatellite instability; |
| MSI-H | microsatellite instability high; |
| MSS | microsatellite stable; |
| MutLα | MLH1-PMS2; |
| MutSα | MSH2-MSH6; |
| NEC | Neuroendocrine Cancer; |
| NET | neuroendocrine tumors; |
| NGS | next-generation sequencing; |
| ORR | objective response rate; |
| OS | overall survival; |
| PCNA | proliferating cell nuclear antigen; |
| PCR | polymerase chain reaction; |
| PD-1/PD-L1 | programmed cell death-1/its ligand; |
| PFS | progression-free survival |
| PINK1 | PTEN-induced putative kinase 1; |
| PMBCL | primary mediastinal B-cell lymphoma; |
| pMMR | proficient MMR (MMR-intact); |
| POLβ | polymerase beta; |
| POLγ | polymerase gamma; |
| ROS | reactive oxygen species; |
| RPA | replication protein A; |
| SCE | sister chromatid exchange; |
| SHM | somatic hypermutation; |
| SS | synovial sarcomas; |
| TLS | post-translesion synthesis; |
| TMB | tumor mutational burden; |
| TMZ | Temozolomide; |
| UV | ultraviolet; |
| VUS | variants of unknown significance; |
References
- Olave, M.C.; Graham, R.P. Mismatch Repair Deficiency: The What, How and Why It Is Important. Genes Chromosomes Cancer 2022, 61, 314–321. [Google Scholar] [CrossRef]
- Rahimian, E.; Amini, A.; Alikarami, F.; Pezeshki, S.M.S.; Saki, N.; Safa, M. DNA Repair Pathways as Guardians of the Genome: Therapeutic Potential and Possible Prognostic Role in Hematologic Neoplasms. DNA Repair 2020, 96, 102951. [Google Scholar] [CrossRef]
- Amemiya, K.; Hirotsu, Y.; Nagakubo, Y.; Watanabe, S.; Amemiya, S.; Mochizuki, H.; Oyama, T.; Kondo, T.; Omata, M. Simple IHC Reveals Complex MMR Alternations than PCR Assays: Validation by LCM and next-Generation Sequencing. Cancer Med. 2022, 11, 4479–4490. [Google Scholar] [CrossRef] [PubMed]
- Grillo, F.; Angerilli, V.; Parente, P.; Vanoli, A.; Luchini, C.; Sciallero, S.; Puccini, A.; Bergamo, F.; Lonardi, S.; Valeri, N.; et al. Prevalence and Type of MMR Expression Heterogeneity in Colorectal Adenocarcinoma: Therapeutic Implications and Reporting. Virchows Arch. 2024, 485, 131–135. [Google Scholar] [CrossRef]
- Buchler, T. Microsatellite Instability and Metastatic Colorectal Cancer—A Clinical Perspective. Front. Oncol. 2022, 12, 888181. [Google Scholar] [CrossRef]
- Moss, E.L.; Gorsia, D.N.; Collins, A.; Sandhu, P.; Foreman, N.; Gore, A.; Wood, J.; Kent, C.; Silcock, L.; Guttery, D.S. Utility of Circulating Tumor DNA for Detection and Monitoring of Endometrial Cancer Recurrence and Progression. Cancers 2020, 12, 2231. [Google Scholar] [CrossRef] [PubMed]
- Martín-Broto, J.; Moura, D.S.; van Tine, B.A. Facts and Hopes in Immunotherapy of Soft-Tissue Sarcomas. Clin. Cancer Res. 2020, 26, 5801–5808. [Google Scholar] [CrossRef]
- Germano, G.; Amirouchene-Angelozzi, N.; Rospo, G.; Bardelli, A. The Clinical Impact of the Genomic Landscape of Mismatch Repair–Deficient Cancers. Cancer Discov. 2018, 8, 1518–1528. [Google Scholar] [CrossRef]
- Martin, S.A.; Lord, C.J.; Ashworth, A. Therapeutic Targeting of the DNA Mismatch Repair Pathway. Clin. Cancer Res. 2010, 16, 5107–5113. [Google Scholar] [CrossRef] [PubMed]
- Guillotin, D.; Martin, S.A. Exploiting DNA Mismatch Repair Deficiency as a Therapeutic Strategy. Exp. Cell Res. 2014, 329, 110–115. [Google Scholar] [CrossRef]
- Brierley, D.J.; Martin, S.A. Oxidative Stress and the DNA Mismatch Repair Pathway. Antioxid. Redox Signal. 2013, 18, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.; McCabe, N.; Mullarkey, M.; Cummins, R.; Burgess, D.J.; Nakabeppu, Y.; Oka, S.; Kay, E.; Lord, C.J.; Ashworth, A. DNA Polymerases as Potential Therapeutic Targets for Cancers Deficient in the DNA Mismatch Repair Proteins MSH2 or MLH1. Cancer Cell 2010, 17, 235–248. [Google Scholar] [CrossRef]
- Martin, S.A.; Hewish, M.; Sims, D.; Lord, C.J.; Ashworth, A. Parallel High-Throughput RNA Interference Screens Identify PINK1 as a Potential Therapeutic Target for the Treatment of DNA Mismatch Repair-Deficient Cancers. Cancer Res. 2011, 71, 1836–1848. [Google Scholar] [CrossRef]
- Santos, F.; Peat, J.; Burgess, H.; Rada, C.; Reik, W.; Dean, W. Active Demethylation in Mouse Zygotes Involves Cytosine Deamination and Base Excision Repair. Epigenetics Chromatin 2013, 6, 29. [Google Scholar] [CrossRef]
- Franchini, D.M.; Chan, C.F.; Morgan, H.; Incorvaia, E.; Rangam, G.; Dean, W.; Santos, F.; Reik, W.; Petersen-Mahrt, S.K. Processive DNA Demethylation via DNA Deaminase-Induced Lesion Resolution. PLoS ONE 2014, 9, e97754. [Google Scholar] [CrossRef]
- Grin, I.; Ishchenko, A.A. An Interplay of the Base Excision Repair and Mismatch Repair Pathways in Active DNA Demethylation. Nucleic Acids Res. 2016, 44, 3713–3747. [Google Scholar] [CrossRef]
- Ijsselsteijn, R.; Jansen, J.G.; de Wind, N. DNA Mismatch Repair-Dependent DNA Damage Responses and Cancer. DNA Repair 2020, 93, 102923. [Google Scholar] [CrossRef]
- Tsaalbi-Shtylik, A.; Ferrás, C.; Pauw, B.; Hendriks, G.; Temviriyanukul, P.; Carlée, L.; Calléja, F.; Van Hees, S.; Akagi, J.I.; Iwai, S.; et al. Excision of Translesion Synthesis Errors Orchestrates Responses to Helix-Distorting DNA Lesions. J. Cell Biol. 2015, 209, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Wada-Hiraike, O.; Yano, T.; Nei, T.; Matsumoto, Y.; Nagasaka, K.; Takizawa, S.; Oishi, H.; Arimoto, T.; Nakagawa, S.; Yasugi, T.; et al. The DNA Mismatch Repair Gene HMSH2 Is a Potent Coactivator of Oestrogen Receptor α. Br. J. Cancer 2005, 92, 2286–2291. [Google Scholar] [CrossRef] [PubMed]
- Sajjadi, E.; Venetis, K.; Piciotti, R.; Invernizzi, M.; Guerini-Rocco, E.; Haricharan, S.; Fusco, N. Mismatch Repair-Deficient Hormone Receptor-Positive Breast Cancers: Biology and Pathological Characterization. Cancer Cell Int. 2021, 21, 266. [Google Scholar] [CrossRef]
- DeWitt, J.T.; Raghunathan, M.; Haricharan, S. Nonrepair Functions of DNA Mismatch Repair Proteins: New Avenues for Precision Oncology. Trends Cancer 2024, 11, 49–61. [Google Scholar] [CrossRef]
- Drabløs, F.; Feyzi, E.; Aas, P.A.; Vaagbø, C.B.; Kavli, B.; Bratlie, M.S.; Peña-Diaz, J.; Otterlei, M.; Slupphaug, G.; Krokan, H.E. Alkylation Damage in DNA and RNA—Repair Mechanisms and Medical Significance. DNA Repair 2004, 3, 1389–1407. [Google Scholar] [CrossRef]
- Uechi, Y.; Fujikane, R.; Morita, S.; Tamaoki, S.; Hidaka, M. Bloom Syndrome DNA Helicase Mitigates Mismatch Repair-Dependent Apoptosis. Biochem. Biophys. Res. Commun. 2024, 723, 150214. [Google Scholar] [CrossRef]
- Robinson, H.M.R.; Black, E.J.; Brown, R.; Gillespie, D.A.F. DNA Mismatch Repair and Chk1-Dependent Centrosome Amplification in Response to DNA Alkylation Damage. Cell Cycle 2007, 6, 982–992. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dondi, G.; Coluccelli, S.; De Leo, A.; Ferrari, S.; Gruppioni, E.; Bovicelli, A.; Godino, L.; Coadă, C.A.; Morganti, A.G.; Giordano, A.; et al. An Analysis of Clinical, Surgical, Pathological and Molecular Characteristics of Endometrial Cancer According to Mismatch Repair Status. A Multidisciplinary Approach. Int. J. Mol. Sci. 2020, 21, 7188. [Google Scholar] [CrossRef] [PubMed]
- Riedinger, C.J.; Esnakula, A.; Haight, P.J.; Suarez, A.A.; Chen, W.; Gillespie, J.; Villacres, A.; Chassen, A.; Cohn, D.E.; Goodfellow, P.J.; et al. Characterization of Mismatch-Repair/Microsatellite Instability-Discordant Endometrial Cancers. Cancer 2024, 130, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Yu, T.; Li, L.; Zhang, Q.; Zhang, S.; Li, B.; Li, X.; Xiao, W.; Liu, G. Intraindividual Tumor Heterogeneity of Mismatch Repair Status in Metastatic Colorectal Cancer. Appl. Immunohistochem. Mol. Morphol. 2023, 31, 84–93. [Google Scholar] [CrossRef]
- Marinelli, D.; Sabatini, A.; Bengala, E.; Ciurluini, F.; Picone, V.; Santini, D.; Pietrantonio, F.; Rossini, D.; Cremolini, C. Systemic Treatment of Mismatch Repair Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer—Single versus Double Checkpoint Inhibition. ESMO Open 2024, 9, 103483. [Google Scholar] [CrossRef]
- André, T.; Shiu, K.-K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability–High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef]
- Franke, A.J.; Skelton, W.P.; Starr, J.S.; Parekh, H.; Lee, J.J.; Overman, M.J.; Allegra, C.; George, T.J. Immunotherapy for Colorectal Cancer: A Review of Current and Novel Therapeutic Approaches. J. Natl. Cancer Inst. 2019, 111, 1131–1141. [Google Scholar] [CrossRef]
- Strickler, J.H.; Willis, J.A. Treatment of Metastatic Colorectal Cancer: ASCO Guideline. Available online: www.asco.org/gastrointestinal-cancer-guidelines (accessed on 1 February 2025).
- Emiloju, O.E.; Zhu, M.; Xie, H.; Jin, Z.; Sinicrope, F.A.; Hubbard, J.M. Selecting Optimal First-Line Treatment for Microsatellite Stable and Non-Mutated RAS/BRAF Metastatic Colorectal Cancer. Curr. Treat. Options Oncol. 2023, 24, 1739–1757. [Google Scholar] [CrossRef]
- Baris, H.N.; Barnes-Kedar, I.; Toledano, H.; Halpern, M.; Hershkovitz, D.; Lossos, A.; Lerer, I.; Peretz, T.; Kariv, R.; Cohen, S.; et al. Constitutional Mismatch Repair Deficiency in Israel: High Proportion of Founder Mutations in MMR Genes and Consanguinity. Pediatr. Blood Cancer 2016, 63, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Buecher, B.; Le Mentec, M.; Doz, F.; Bourdeaut, F.; Gauthier-Villars, M.; Stoppa-Lyonnet, D.; Colas, C. Syndrome CMMRD (Déficience Constitutionnelle Des Gènes MMR): Bases Génétiques et Aspects Cliniques. Bull. Cancer 2019, 106, 162–172. [Google Scholar] [CrossRef]
- Wimmer, K.; Rosenbaum, T.; Messiaen, L. Connections between Constitutional Mismatch Repair Deficiency Syndrome and Neurofibromatosis Type 1. Clin. Genet. 2017, 91, 507–519. [Google Scholar] [CrossRef]
- Siju, J.; Sahu, A.; Bhattacharya, K.; Prasad, M.; Sarin, R.; Gupta, T. Demystifying the Mystery of Genes: A Case Report on Constitutional Mismatch Repair Deficiency. Indian J. Radiol. Imaging 2024, 34, 562–565. [Google Scholar] [CrossRef]
- Carsote, M.; Turturea, I.F.; Turturea, M.R.; Valea, A.; Nistor, C.; Gheorghisan-Galateanu, A.-A. Pathogenic Insights into DNA Mismatch Repair (MMR) Genes–Proteins and Microsatellite Instability: Focus on Adrenocortical Carcinoma and Beyond. Diagnostics 2023, 13, 1867. [Google Scholar] [CrossRef]
- Multone, E.; La Rosa, S.; Sempoux, C.; Uccella, S. PD-L1 Expression, Tumor-Infiltrating Lymphocytes, and Mismatch Repair Proteins Status in Digestive Neuroendocrine Neoplasms: Exploring Their Potential Role as Theragnostic and Prognostic Biomarkers. Virchows Arch. 2024, 485, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Ban, X.; Mo, S.; Lu, Z.; Jia, C.; Shao, H.; Chang, X.; Mao, X.; Zhang, Y.; Pang, J.; Zhang, Y.; et al. Expression and Methylation Status of MMR and MGMT in Well-Differentiated Pancreatic Neuroendocrine Tumors and Potential Clinical Applications. Endocrine 2022, 77, 538–545. [Google Scholar] [CrossRef]
- Tanaka, K.; Suzuki, K.; Miyashita, K.; Wakasa, K.; Kawano, M.; Nakatsu, Y.; Tsumura, H.; Yoshida, M.A.; Oda, S. Activation of Recombinational Repair in Ewing Sarcoma Cells Carrying EWS-FLI1 Fusion Gene by Chromosome Translocation. Sci. Rep. 2022, 12, 14764. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Abro, B.; Kaushal, M.; Chen, L.; Chen, T.; Gondim, M.; Yan, W.; Neidich, J.; Dehner, L.P.; Pfeifer, J.D. Tumor Mutation Burden and Checkpoint Immunotherapy Markers in Primary and Metastatic Synovial Sarcoma. Hum. Pathol. 2020, 100, 15–23. [Google Scholar] [CrossRef]
- Tawbi, H.A.; Burgess, M.; Bolejack, V.; Van Tine, B.A.; Schuetze, S.M.; Hu, J.; D’Angelo, S.; Attia, S.; Riedel, R.F.; Priebat, D.A.; et al. Pembrolizumab in Advanced Soft-Tissue Sarcoma and Bone Sarcoma (SARC028): A Multicentre, Two-Cohort, Single-Arm, Open-Label, Phase 2 Trial. Lancet Oncol. 2017, 18, 1493–1501, Erratum in Lancet Oncol. 2017, 18, e711. Erratum in Lancet Oncol. 2018, 19, e8. [Google Scholar] [CrossRef]
- Wienand, K.; Chapuy, B.; Stewart, C.; Dunford, A.J.; Wu, D.; Kim, J.; Kamburov, A.; Wood, T.R.; Cader, F.Z.; Ducar, M.D.; et al. Genomic Analyses of Flow-Sorted Hodgkin Reed-Sternberg Cells Reveal Complementary Mechanisms of Immune Evasion. Blood Adv. 2019, 3, 4065–4080. [Google Scholar] [CrossRef]
- Tian, T.; Li, J.; Xue, T.; Yu, B.; Li, X.; Zhou, X. Microsatellite Instability and Its Associations with the Clinicopathologic Characteristics of Diffuse Large B-Cell Lymphoma. Cancer Med. 2020, 9, 2330–2442. [Google Scholar] [CrossRef] [PubMed]
- Jeong, A.R.; Ball, E.D.; Goodman, A.M. Predicting Responses to Checkpoint Inhibitors in Lymphoma: Are We Up to the Standards of Solid Tumors? Clin. Med. Insights Oncol. 2020, 14, 1179554920976366. [Google Scholar] [CrossRef]
- Miyashita, K.; Fujii, K.; Taguchi, K.; Shimokawa, M.; Yoshida, M.A.; Abe, Y.; Okamura, J.; Oda, S.; Uike, N. A Specific Mode of Microsatellite Instability Is a Crucial Biomarker in Adult T-Cell Leukaemia/Lymphoma Patients. J. Cancer Res. Clin. Oncol. 2017, 143, 399–408. [Google Scholar] [CrossRef]
- Gilson, P.; Merlin, J.L.; Harlé, A. Detection of Microsatellite Instability: State of the Art and Future Applications in Circulating Tumour Dna (Ctdna). Cancers 2021, 13, 1491. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Garcia, E.; Zhao, E.; Bratman, S.V.; Siu, L.L. Monitoring and Adapting Cancer Treatment Using Circulating Tumor DNA Kinetics: Current Research, Opportunities, and Challenges. Sci. Adv. 2022, 8, eabi8618. [Google Scholar] [CrossRef]
- Silveira, A.B.; Bidard, F.C.; Kasperek, A.; Melaabi, S.; Tanguy, M.L.; Rodrigues, M.; Bataillon, G.; Cabel, L.; Buecher, B.; Pierga, J.Y.; et al. High-Accuracy Determination of Microsatellite Instability Compatible with Liquid Biopsies. Clin. Chem. 2020, 66, 606–613. [Google Scholar] [CrossRef]
- Georgiadis, A.; Durham, J.N.; Keefer, L.A.; Bartlett, B.R.; Zielonka, M.; Murphy, D.; White, J.R.; Lu, S.; Verner, E.L.; Ruan, F.; et al. Noninvasive Detection of Microsatellite Instabilit and High Tumor Mutation Burden in Cancer Patients Treated with PD-1 Blockade. Clin. Cancer Res. 2019, 25, 7024–7034. [Google Scholar] [CrossRef]
- Tiwari, A.K.; Roy, H.K.; Lynch, H.T. Lynch Syndrome in the 21st Century: Clinical Perspectives. QJM 2016, 109, 151–158. [Google Scholar] [CrossRef]
- Martín-López, J.V.; Fishel, R. The Mechanism of Mismatch Repair and the Functional Analysis of Mismatch Repair Defects in Lynch Syndrome. Fam. Cancer 2013, 12, 159–168. [Google Scholar] [CrossRef]
- Boeri, M.; Signoroni, S.; Ciniselli, C.M.; Gariboldi, M.; Zanutto, S.; Rausa, E.; Segale, M.; Zanghì, A.; Ricci, M.T.; Verderio, P.; et al. Detection of (Pre)Cancerous Colorectal Lesions in Lynch Syndrome Patients by Microsatellite Instability Liquid Biopsy. Cancer Gene Ther. 2024, 31, 842–850. [Google Scholar] [CrossRef]
- Jones, J.J.; Jones, K.L.; Wong, S.Q.; Whittle, J.; Goode, D.; Nguyen, H.; Iaria, J.; Stylli, S.; Towner, J.; Pieters, T.; et al. Plasma CtDNA Enables Early Detection of Temozolomide Resistance Mutations in Glioma. Neurooncol. Adv. 2024, 6, vdae041. [Google Scholar] [CrossRef]
- Kang, S.Y.; Kim, D.G.; Ahn, S.; Ha, S.Y.; Jang, K.T.; Kim, K.M. Comparative Analysis of Microsatellite Instability by Next-Generation Sequencing, MSI PCR and MMR Immunohistochemistry in 1942 Solid Cancers. Pathol. Res. Pract. 2022, 233, 153874. [Google Scholar] [CrossRef]
- Gilson, P.; Levy, J.; Rouyer, M.; Demange, J.; Husson, M.; Bonnet, C.; Salleron, J.; Leroux, A.; Merlin, J.L.; Harlé, A. Evaluation of 3 Molecular-Based Assays for Microsatellite Instability Detection in Formalin-Fixed Tissues of Patients with Endometrial and Colorectal Cancers. Sci. Rep. 2020, 10, 16386. [Google Scholar] [CrossRef] [PubMed]
- Smithgall, M.C.; Remotti, H.; Hsiao, S.J.; Mansukhani, M.; Liu-Jarin, X.; Fernandes, H. Investigation of Discrepant Mismatch Repair Immunohistochemistry and Microsatellite Instability Polymerase Chain Reaction Test Results for Gynecologic Cancers Using Next-Generation Sequencing. Hum. Pathol. 2022, 119, 41–50. [Google Scholar] [CrossRef]
- Meyers, M.; Hwang, A.; Wagner, M.W.; Boothman, D.A. Role of DNA Mismatch Repair in Apoptotic Responses to Therapeutic Agents. Environ. Mol. Mutagen. 2004, 44, 249–264. [Google Scholar] [CrossRef]
- Di Pietro, M.; Marra, G.; Cejka, P.; Stojic, L.; Menigatti, M.; Cattaruzza, M.S.; Jiricny, J. Mismatch Repair-Dependent Transcriptome Changes in Human Cells Treated with the Methylating Agent N-Methyl-N′-Nitro-N-Nitrosoguanidine. Cancer Res. 2003, 63, 8158–8166. [Google Scholar]
- Bouvet, D.; Bodo, S.; Munier, A.; Guillerm, E.; Bertrand, R.; Colas, C.; Duval, A.; Coulet, F.; Muleris, M. Methylation Tolerance-Based Functional Assay to Assess Variants of Unknown Significance in the MLH1 and MSH2 Genes and Identify Patients With Lynch Syndrome. Gastroenterology 2019, 157, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Li, G.-M. DNA Mismatch Repair in Cancer Immunotherapy. NAR Cancer 2023, 5, zcad031. [Google Scholar] [CrossRef] [PubMed]
- Lenz, H.J.; Van Cutsem, E.; Limon, M.L.; Wong, K.Y.M.; Hendlisz, A.; Aglietta, M.; García-Alfonso, P.; Neyns, B.; Luppi, G.; Cardin, D.B.; et al. First-Line Nivolumab Plus Low-Dose Ipilimumab for Microsatellite Instability-High/ Mismatch Repair-Deficient Metastatic Colorectal Cancer: The Phase II CheckMate 142 Study. J. Clin. Oncol. 2022, 40, 161–170. [Google Scholar] [CrossRef]
- Qu, F.; Wu, S.; Yu, W. Progress of Immune Checkpoint Inhibitors Therapy for PMMR/MSS Metastatic Colorectal Cancer. Onco Targets Ther. 2024, 17, 1223–1253. [Google Scholar] [CrossRef]
- Matthaios, D.; Balgkouranidou, I.; Neanidis, K.; Sofis, A.; Pikouli, A.; Romanidis, K.; Pappa, A.; Karamouzis, M.; Zygogianni, A.; Charalampidis, C.; et al. Revisiting Temozolomide’s Role in Solid Tumors: Old Is Gold? J. Cancer 2024, 15, 3254–3271. [Google Scholar] [CrossRef]
- Zhang, J.; FG Stevens, M.; D Bradshaw, T. Temozolomide: Mechanisms of Action, Repair and Resistance. Curr. Mol. Pharmacol. 2011, 5, 102–114. [Google Scholar] [CrossRef]
- Germano, G.; Lamba, S.; Rospo, G.; Barault, L.; Magri, A.; Maione, F.; Russo, M.; Crisafulli, G.; Bartolini, A.; Lerda, G.; et al. Inactivation of DNA Repair Triggers Neoantigen Generation and Impairs Tumour Growth. Nature 2017, 552, 116–120. [Google Scholar] [CrossRef]
- Crisafulli, G.; Sartore-Bianchi, A.; Lazzari, L.; Pietrantonio, F.; Amatu, A.; Macagno, M.; Barault, L.; Cassingena, A.; Bartolini, A.; Luraghi, P.; et al. Temozolomide Treatment Alters Mismatch Repair and Boosts Mutational Burden in Tumor and Blood of Colorectal Cancer Patients. Cancer Discov. 2022, 12, 1656–1675. [Google Scholar] [CrossRef]
- Sawada, M.; Hida, T.; Kamiya, T.; Minowa, T.; Kato, J.; Okura, M.; Idogawa, M.; Tokino, T.; Uhara, H. Effects of Temozolomide on Tumor Mutation Burden and Microsatellite Instability in Melanoma Cells. J. Dermatol. 2024, 51, 409–418. [Google Scholar] [CrossRef]
- Teng, J.Y.; Yang, D.P.; Tang, C.; Fang, H.S.; Sun, H.Y.; Xiang, Y.N.; Li, X.M.; Yang, F.; Xia, R.X.; Fan, F.; et al. Targeting DNA Polymerase β Elicits Synthetic Lethality with Mismatch Repair Deficiency in Acute Lymphoblastic Leukemia. Leukemia 2023, 37, 1204–1215. [Google Scholar] [CrossRef]
- Saldanha, J.; Rageul, J.; Patel, J.A.; Kim, H. The Adaptive Mechanisms and Checkpoint Responses to a Stressed DNA Replication Fork. Int. J. Mol. Sci. 2023, 24, 10488. [Google Scholar] [CrossRef]
- Wang, M.; Ran, X.; Leung, W.; Kawale, A.; Saxena, S.; Ouyang, J.; Patel, P.S.; Dong, Y.; Yin, T.; Shu, J.; et al. ATR Inhibition Induces Synthetic Lethality in Mismatch Repair-Deficient Cells and Augments Immunotherapy. Genes Dev. 2023, 37, 929–943. [Google Scholar] [CrossRef]
- Liu, C.; Wang, X.; Qin, W.; Tu, J.; Li, C.; Zhao, W.; Ma, L.; Liu, B.; Qiu, H.; Yuan, X. Combining Radiation and the ATR Inhibitor Berzosertib Activates STING Signaling and Enhances Immunotherapy via Inhibiting SHP1 Function in Colorectal Cancer. Cancer Commun. 2023, 43, 435–454. [Google Scholar] [CrossRef]
- Morano, F.; Raimondi, A.; Pagani, F.; Lonardi, S.; Salvatore, L.; Cremolini, C.; Murgioni, S.; Randon, G.; Palermo, F.; Antonuzzo, L.; et al. Temozolomide Followed by Combination With Low-Dose Ipilimumab and Nivolumab in Patients With Microsatellite-Stable, O 6-Methylguanine-DNA Methyltransferase-Silenced Metastatic Colorectal Cancer: The MAYA Trial. J. Clin. Oncol. 2022, 40, 1562–1573. [Google Scholar] [CrossRef]
- Fountzilas, E.; Kotoula, V.; Pentheroudakis, G.; Manousou, K.; Polychronidou, G.; Vrettou, E.; Poulios, C.; Papadopoulou, E.; Raptou, G.; Pectasides, E.; et al. Prognostic Implications of Mismatch Repair Deficiency in Patients with Nonmetastatic Colorectal and Endometrial Cancer. ESMO Open 2019, 4, e000474. [Google Scholar] [CrossRef] [PubMed]
- Chaowiwatkun, K.; Trongwongsa, T.; Rodpenpear, N.; Nutthachote, P. Comparison of Tissue Mismatch Repair Protein Deficiency between Early and Advanced-Stage Endometrial Cancer. Asian Pac. J. Cancer Prev. 2023, 24, 345. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yan, Z.; Hou, X.; Ren, K.; Hu, K.; Zhang, F. Mismatch Repair Status Is an Effective Prognostic Factor for Early-Stage Endometrial Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2023, 117, e553–e554. [Google Scholar] [CrossRef]
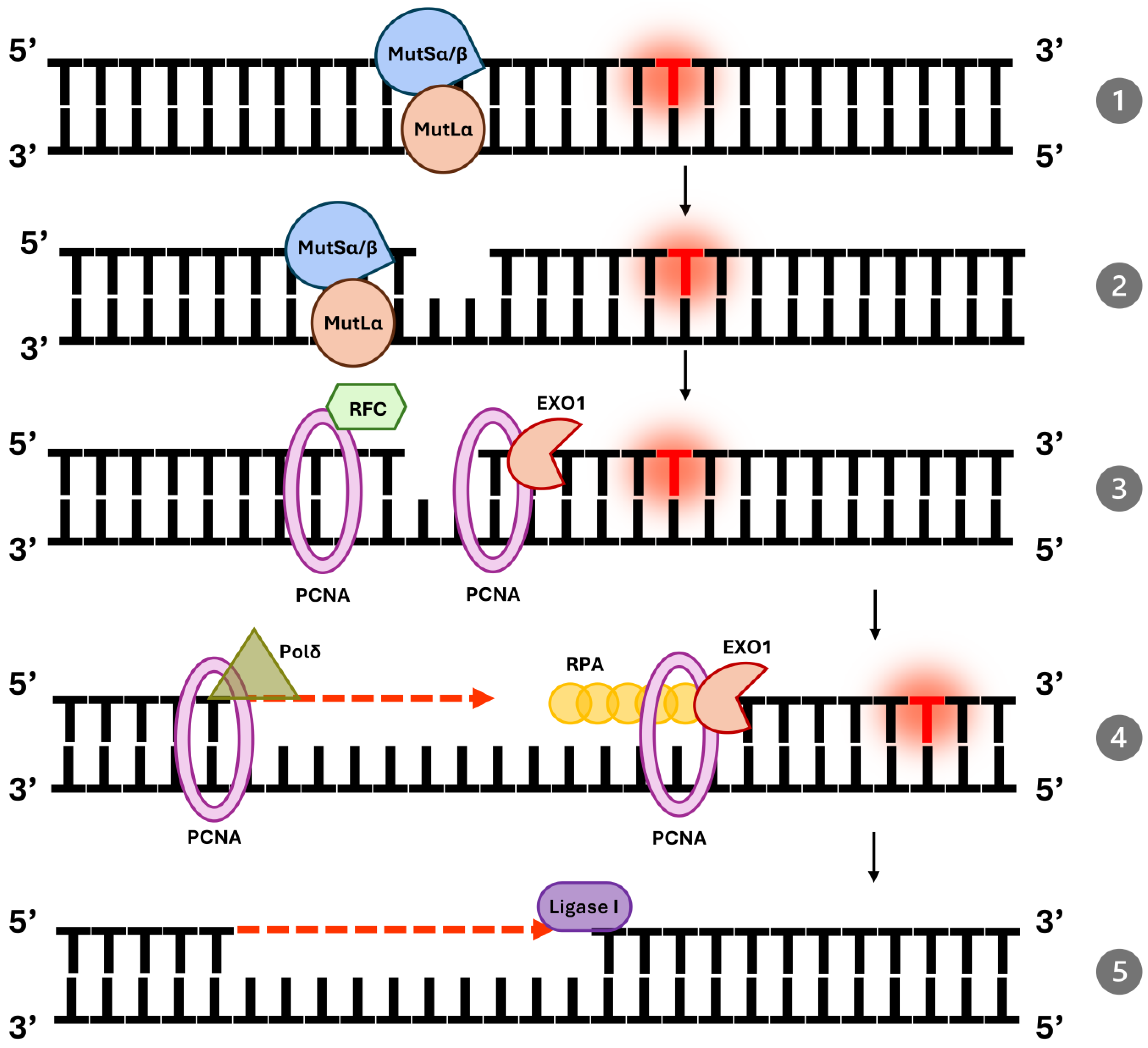

| Protein | Function | Complex |
|---|---|---|
| MSH2 | Scaffold protein in MutSα/β | MutSα/MutSβ |
| MSH6 | Recognizes base mismatches | MutSα |
| MSH3 | Recognizes insertion–deletion loops | MutSβ |
| MLH1 | Core component of MutL complexes | MutLα, MutLβ, MutLγ |
| PMS2 | Endonuclease activity | MutLα |
| EXO1 | Exonuclease for mismatch excision | N/A |
| PCNA | Coordinates DNA resynthesis | N/A |
| RPA | Protects single-stranded DNA | N/A |
| DNA Pol δ | DNA resynthesis | N/A |
| DNA Ligase I | DNA nicks ligation | N/A |
| Diagnostic Step | Methods | Possible Results | Clinical Consequences | Therapeutic Implications |
|---|---|---|---|---|
| 1. Initial MMR/MSI testing | IHC for MMR proteins (MLH1, MSH2, MSH6, PMS2); PCR-based MSI panels (e.g., pentaplex); targeted NGS panels that report MSI and TMB; ctDNA assays (liquid biopsy) | dMMR/MSI-H (loss of MMR protein expression or MSI detected) | High tumor mutational burden (often), increased neoantigen load and lymphocyte infiltration → higher probability of benefit from immune checkpoint inhibitors (ICI) | ICI are preferred/considered (e.g., pembrolizumab, nivolumab, dostarlimab) for eligible patients; consider clinical trials for combination strategies (dual checkpoint blockade, ATRi + ICI) |
| pMMR/MSS (intact MMR protein expression; MSS on MSI assay) | Lower immunogenicity on average; less likely to respond to single-agent ICI | Standard systemic therapy (chemotherapy ± targeted agents according to tumour type and molecular profile); consider experimental sensitization strategies (e.g., TMZ priming, ATRi + ICI) within trials | ||
| 2. Interpret discordant or borderline results | Re-test with complementary method (IHC ↔ PCR/NGS); evaluate sample quality; test metastatic site if clinically warranted | Heterogeneous/subclonal MMR or discordant results between assays | Potential for under- or over-estimation of dMMR; impacts treatment choice and trial eligibility | Confirmatory testing recommended before committing to long-term ICI; multidisciplinary tumour board discussion advised |
| 3. Special/adjunct testing | Germline testing for MMR genes (MLH1, MSH2, MSH6, PMS2, EPCAM) if suspicion of Lynch/CMMRD; MGMT methylation in TMZ contexts; functional assays or ctDNA serial monitoring | Lynch syndrome/CMMRD; MGMT methylation status; emerging MMR mutations in ctDNA (post-treatment) | Hereditary cancer risk identified → implications for surveillance and family testing; MGMT status may predict TMZ effects; ctDNA may detect emergent resistance | Offer genetic counseling and cascade testing for relatives; use MGMT and ctDNA to guide experimental approaches (e.g., TMZ priming → consider trial enrollment) |
| 4. Monitoring & follow-up | Serial imaging + ctDNA kinetics (when available) | Changes in ctDNA MSI/TMB signature or emergence of new MMR mutations | Early indication of therapy response or acquired resistance | Use ctDNA trends to time reassessment and consider switching/adding therapies or trial inclusion |
| Cancer Type | Trial & Agents (Phase) | NCT Number | Outcomes | Biomarker Criteria |
|---|---|---|---|---|
| Colorectal (mCRC)—Refractory (post-chemo) | CheckMate-142 (Phase II) Nivolumab ± Ipilimumab | NCT02060188 | ORR ~49% with combo; 1-yr OS ~85%; durable control in ~83% | dMMR (MSI-H) selected |
| Colorectal (mCRC)—First Line | KEYNOTE-177 (Phase III) Pembrolizumab vs. chemo | NCT02563002 | PFS 16.5 vs. 8.2 mo (HR = 0.60); ORR 44% vs. 33%; lower grade ≥ 3 AEs | dMMR (MSI-H) selected |
| Colorectal— Neoadjuvant (localized-colon) | NICHE (Phase II) Nivolumab + Ipilimumab | NCT03026140 | 100% pathologic response; 60% pCR in dMMR; some activity in pMMR | dMMR (MSI-H) tumors |
| Colorectal— Neoadjuvant (localized-rectal) | MSK Rectal trial (Phase II) Dostarlimab | NCT04165772 | 100% clinical CR in 14/14; no surgeries required at 6–12 mo follow-up | dMMR (MSI-H) tumors |
| Endometrial— Recurrent/Advanced | KEYNOTE-158 (Phase II) Pembrolizumab | NCT02628067 | ORR ~45% (16% CR); median DOR ~47 mo | dMMR (MSI-H) selected |
| Endometrial—First Line | RUBY (Phase III) Dostarlimab + CT vs. CT | NCT03981796 | PFS HR ~0.3 in dMMR; 2-yr OS 71% vs. 56%; OS benefit overall | Stratified by MMR status (largest benefit in dMMR) |
| Gastric/GE Junction—Advanced (post-chemo) | KEYNOTE-059 (Phase II) Pembrolizumab | NCT02335411 | ORR 57.1% in MSI-H vs. ~9% in MSS; median DoR 16.2 mo | MSI-H subgroup analysis |
| Gastric/GE—First Line/Subgroup | KEYNOTE-062 (Phase III) Pembrolizumab ± CT | NCT02494583 | ORR ~57% in MSI-H subgroup; trend toward improved OS/PFS | MSI-H subgroup analysis |
| Pan-Cancer (multiple types)—Refractory solid tumors | KEYNOTE-158 (Phase II) Pembrolizumab | NCT02628067 | ORR 30.8%; median PFS ~4.2 mo; median OS ~23.5 mo; DOR ~47.5 mo | dMMR or MSI-H |
| Multiple (MSS tumors)—ATR inhibitor + IO | Phase II Ceralasertib (ATRi) + Durvalumab (PD-L1) | NCT03770494 | ORR 22.6% in MSS mCRC (vs. ~0–5% with IO alone) | MSS (pMMR) tumors |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dąbrowska, A.; Mastalerz, J.; Łapińska, Z.; Deszcz, I.; Chwiłkowska, A.; Rembiałkowska, N. Mismatch Repair Deficiency and the Role of Non-Canonical Functions in Cancer: Diagnosis and Therapeutic Implications. Int. J. Mol. Sci. 2025, 26, 9312. https://doi.org/10.3390/ijms26199312
Dąbrowska A, Mastalerz J, Łapińska Z, Deszcz I, Chwiłkowska A, Rembiałkowska N. Mismatch Repair Deficiency and the Role of Non-Canonical Functions in Cancer: Diagnosis and Therapeutic Implications. International Journal of Molecular Sciences. 2025; 26(19):9312. https://doi.org/10.3390/ijms26199312
Chicago/Turabian StyleDąbrowska, Alicja, Jakub Mastalerz, Zofia Łapińska, Iwona Deszcz, Agnieszka Chwiłkowska, and Nina Rembiałkowska. 2025. "Mismatch Repair Deficiency and the Role of Non-Canonical Functions in Cancer: Diagnosis and Therapeutic Implications" International Journal of Molecular Sciences 26, no. 19: 9312. https://doi.org/10.3390/ijms26199312
APA StyleDąbrowska, A., Mastalerz, J., Łapińska, Z., Deszcz, I., Chwiłkowska, A., & Rembiałkowska, N. (2025). Mismatch Repair Deficiency and the Role of Non-Canonical Functions in Cancer: Diagnosis and Therapeutic Implications. International Journal of Molecular Sciences, 26(19), 9312. https://doi.org/10.3390/ijms26199312





