Altered Neuroinflammatory Transcriptomic Profile in the Hippocampal Dentate Gyrus Three Weeks After Lateral Fluid Percussion Injury in Rats
Abstract
1. Introduction
2. Results
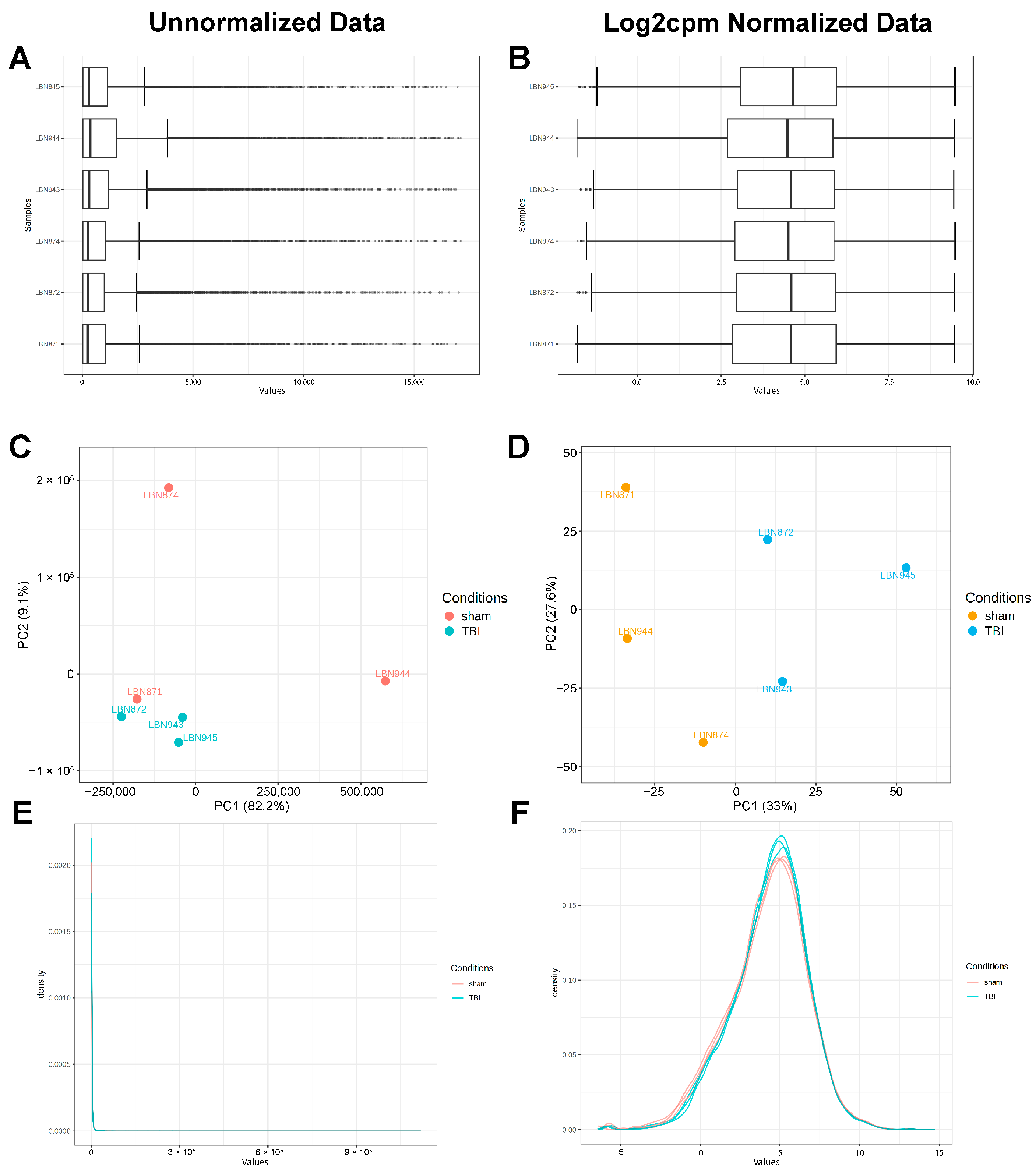
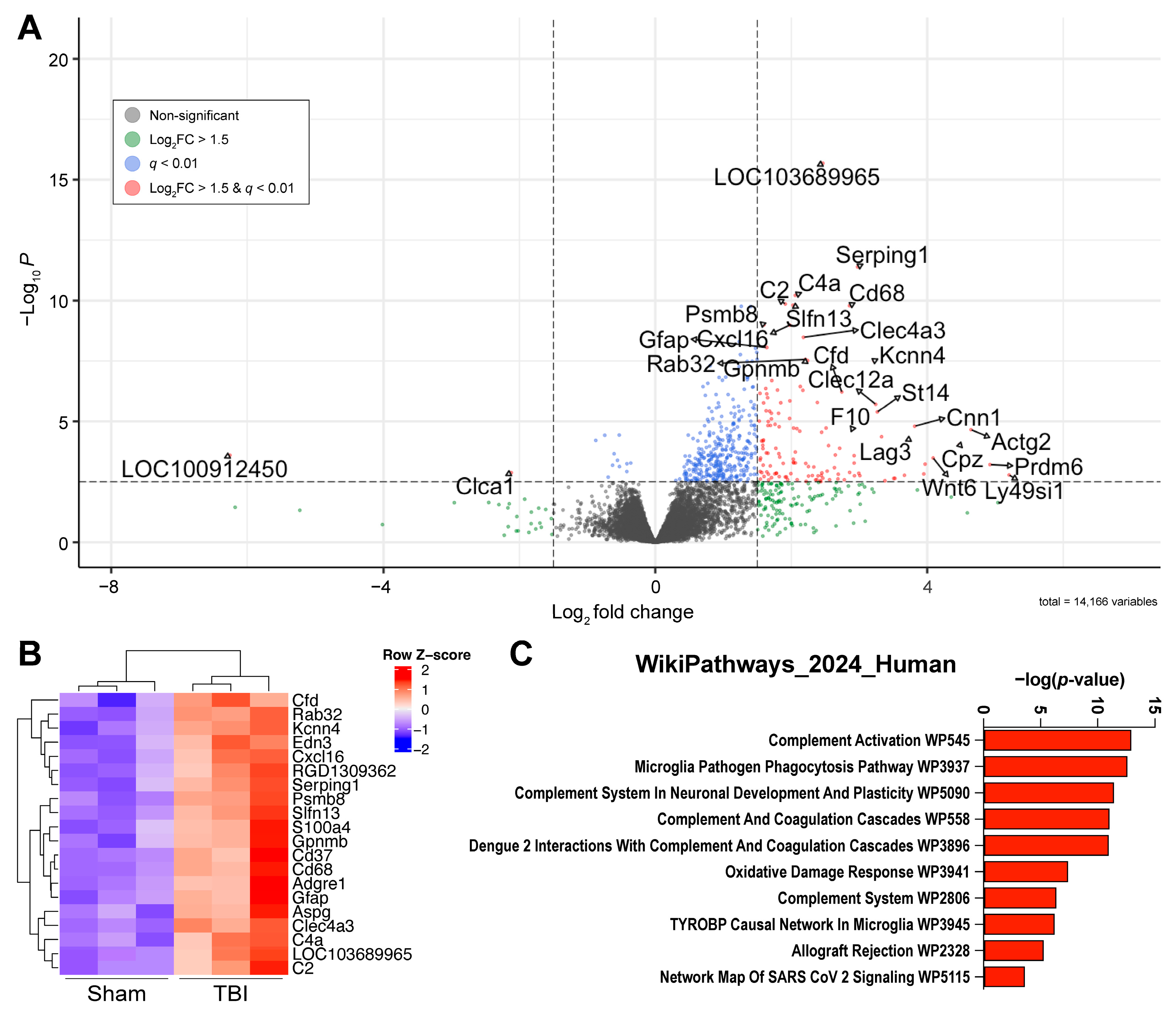
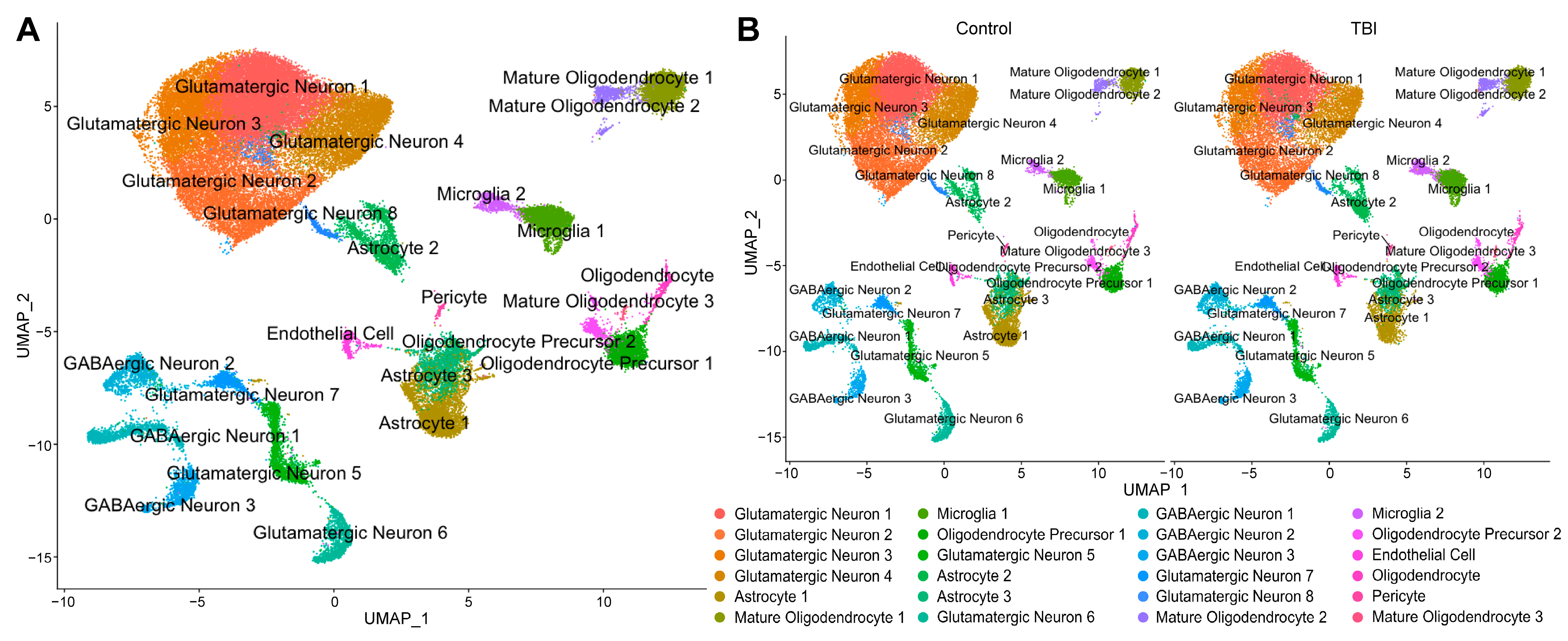
2.1. Bulk RNA Sequencing Reveals a Neuroinflammatory Response in the Dentate Gyrus Three Weeks After LFPI
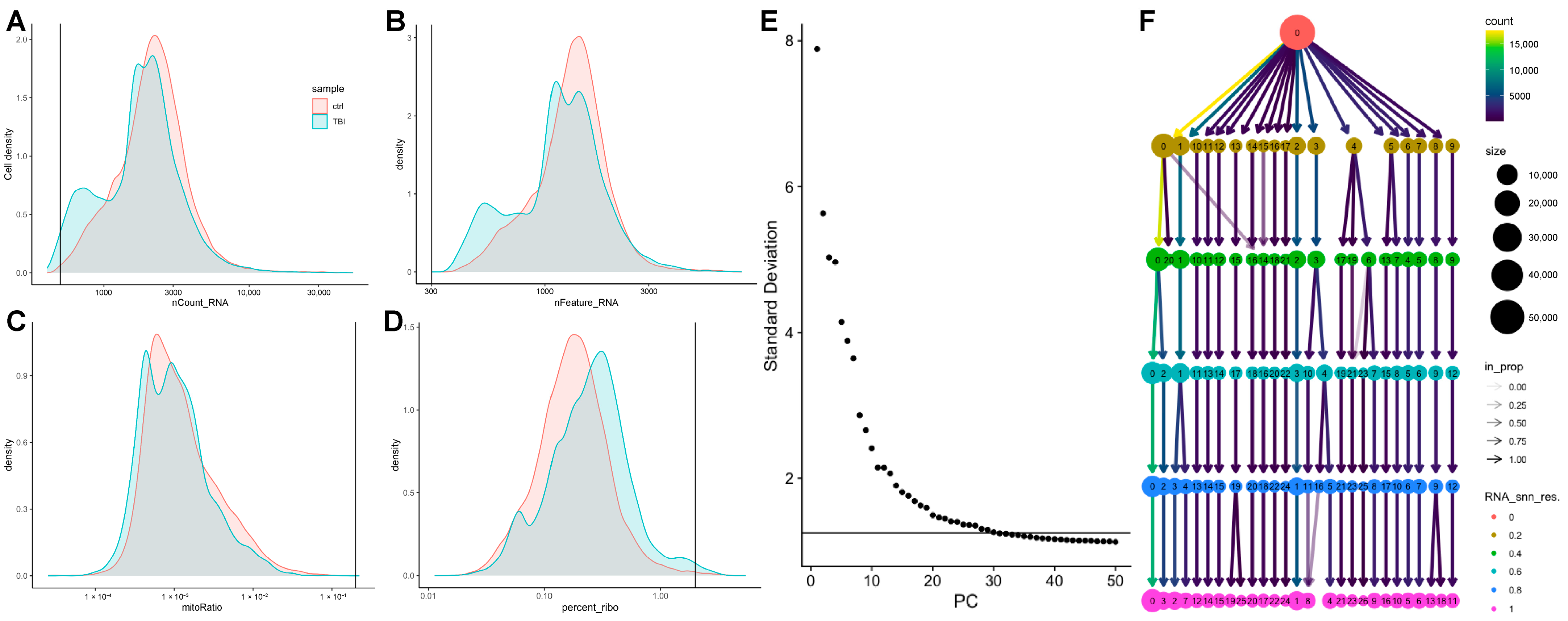
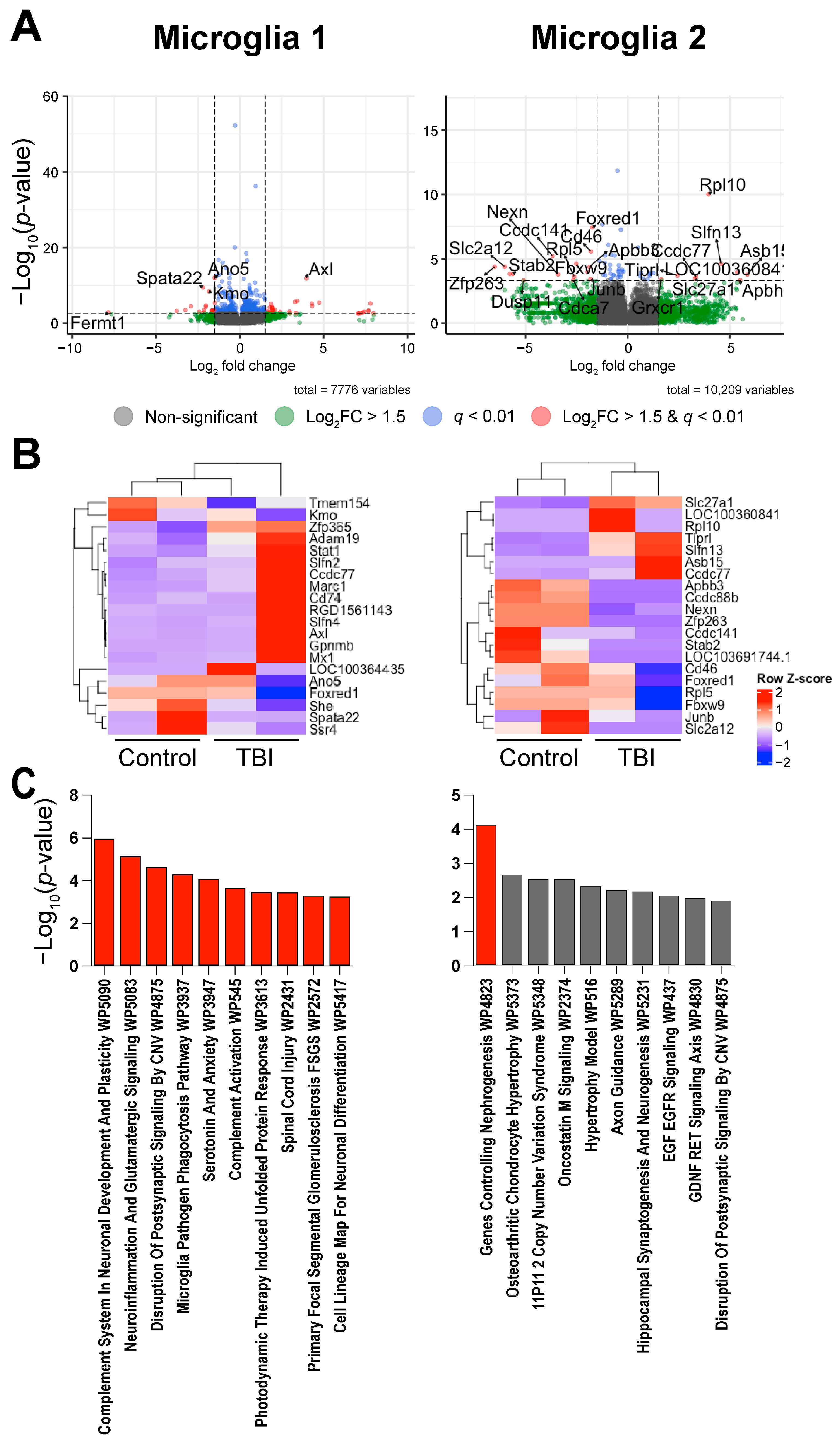
2.2. A Subset of Dentate Gyrus Microglia Express Genes Associated with an Activated Phenotype
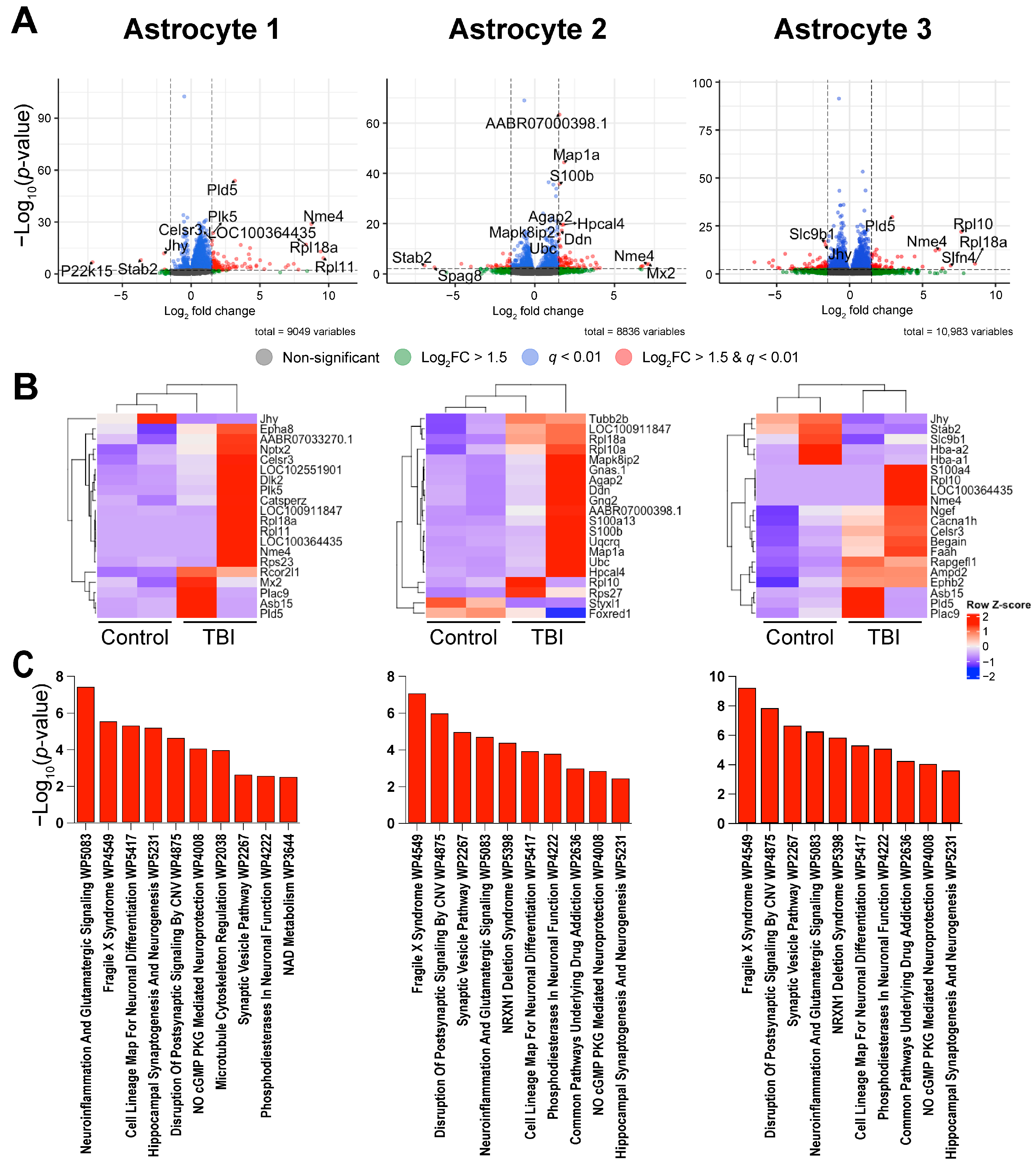
3. Discussion
4. Materials and Methods
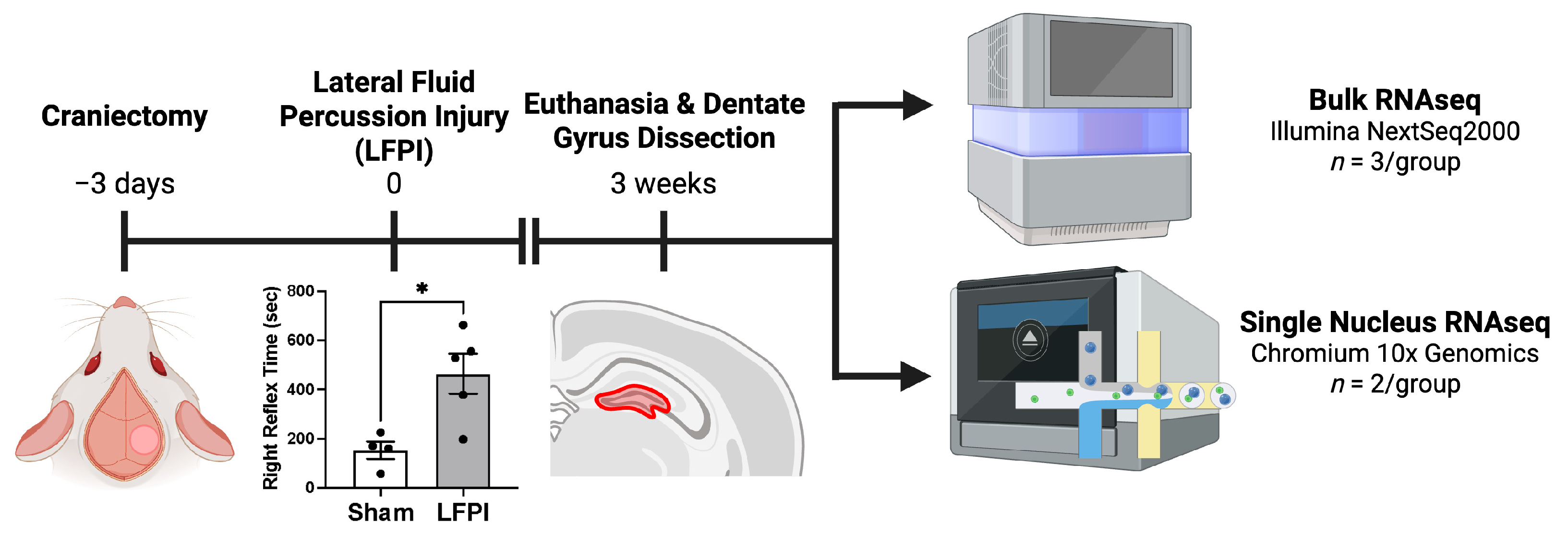
4.1. Animals and Experimental Design
4.2. Lateral Fluid Percussion Injury Model
4.3. Euthanasia and Tissue Acquisition
4.4. Bulk RNA-Seq Sample Preparation and Analysis
4.5. Single-Nucleus RNA-Seq Sample Preparation and Analysis
4.6. Pathway Analysis
Supplementary Materials
Author Contributions
Funding
Institution Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TBI | Traumatic Brain Injury |
| LFPI | Lateral Fluid Percussion Injury |
| RRT | Righting Reflex Time |
| UMAP | Uniform Manifold Approximation and Projection |
| DEG | Differentially Expressed Gene |
| ACT | Annotation of Cell Type |
| MAC | Membrane Attack Complex |
| fB | Factor B |
| sCR1 | Soluble CR1 |
| sCrry | Soluble Complement Inhibitor |
| PBS | Phosphate-Buffered Saline |
| logCPM | Log2 Counts Per Million |
| log2FC | Log2 Fold Change |
| UMI | Unique Molecular Identifier |
| PCA | Principal Component Analysis |
| CCA | Canonical Correlation Analysis |
| Stdev | Standard Deviation |
| RNA-seq | Ribonucleic Acid Sequencing |
Appendix A
| Cluster Name |
Nuclei Counts
(Control) | Nuclei Counts (TBI) | Control % of Total | TBI % of Total |
|---|---|---|---|---|
| Astrocyte 1 | 2307 | 1308 | 8.18% | 4.86% |
| Astrocyte 2 | 753 | 1144 | 2.67% | 4.25% |
| Astrocyte 3 | 743 | 696 | 2.63% | 2.59% |
| Endothelial Cell | 197 | 237 | 0.70% | 0.88% |
| GABAergic Neuron 1 | 623 | 702 | 2.21% | 2.61% |
| GABAergic Neuron 2 | 531 | 681 | 1.88% | 2.53% |
| GABAergic Neuron 3 | 460 | 699 | 1.63% | 2.60% |
| Glutamatergic Neuron 1 | 6610 | 5348 | 23.43% | 19.88% |
| Glutamatergic Neuron 2 | 3936 | 3096 | 13.95% | 11.51% |
| Glutamatergic Neuron 3 | 3082 | 2220 | 10.92% | 8.25% |
| Glutamatergic Neuron 4 | 2949 | 2327 | 10.45% | 8.65% |
| Glutamatergic Neuron 5 | 902 | 996 | 3.20% | 3.70% |
| Glutamatergic Neuron 6 | 672 | 662 | 2.38% | 2.46% |
| Glutamatergic Neuron 7 | 419 | 377 | 1.48% | 1.40% |
| Glutamatergic Neuron 8 | 342 | 442 | 1.21% | 1.64% |
| Mature Oligodendrocyte 1 | 848 | 1667 | 3.01% | 6.20% |
| Mature Oligodendrocyte 2 | 263 | 458 | 0.93% | 1.70% |
| Mature Oligodendrocyte 3 | 19 | 30 | 0.07% | 0.11% |
| Microglia 1 | 900 | 1611 | 3.19% | 5.99% |
| Microglia 2 | 188 | 447 | 0.67% | 1.66% |
| Oligodendrocyte | 155 | 194 | 0.55% | 0.72% |
| Oligodendrocyte Precursor 1 | 995 | 1128 | 3.53% | 4.19% |
| Oligodendrocyte Precursor 2 | 251 | 369 | 0.89% | 1.37% |
| Pericyte | 71 | 68 | 0.25% | 0.25% |
References
- Dewan, M.C.; Rattani, A.; Gupta, S.; Baticulon, R.E.; Hung, Y.C.; Punchak, M.; Agrawal, A.; Adeleye, A.O.; Shrime, M.G.; Rubiano, A.M.; et al. Estimating the global incidence of traumatic brain injury. J. Neurosurg. 2019, 130, 1080–1097. [Google Scholar] [CrossRef]
- Lai, J.Q.; Shi, Y.C.; Lin, S.; Chen, X.R. Metabolic disorders on cognitive dysfunction after traumatic brain injury. Trends Endocrinol. Metab. 2022, 33, 451–462. [Google Scholar] [CrossRef]
- Lambez, B.; Vakil, E. The effectiveness of memory remediation strategies after traumatic brain injury: Systematic review and meta-analysis. Ann. Phys. Rehabil. Med. 2021, 64, 101530. [Google Scholar] [CrossRef] [PubMed]
- Jamora, C.W.; Young, A.; Ruff, R.M. Comparison of subjective cognitive complaints with neuropsychological tests in individuals with mild vs more severe traumatic brain injuries. Brain Inj. 2012, 26, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Bay, E.; Kalpakjian, C.; Giordani, B. Determinants of subjective memory complaints in community-dwelling adults with mild-to-moderate traumatic brain injury. Brain Inj. 2012, 26, 941–949. [Google Scholar] [CrossRef] [PubMed]
- McGuire, J.L.; Correll, E.A.; Lowery, A.C.; Rhame, K.; Anwar, F.N.; McCullumsmith, R.E.; Ngwenya, L.B. Pioglitazone improves working memory performance when administered in chronic TBI. Neurobiol. Dis. 2019, 132, 104611. [Google Scholar] [CrossRef]
- Thompson, H.J.; LeBold, D.G.; Marklund, N.; Morales, D.M.; Hagner, A.P.; McIntosh, T.K. Cognitive evaluation of traumatically brain-injured rats using serial testing in the Morris water maze. Restor. Neurol. Neurosci. 2006, 24, 109–114. [Google Scholar] [CrossRef]
- Whiting, M.D.; Hamm, R.J. Traumatic Brain Injury Produces Delay-Dependent Memory Impairment in Rats. J. Neurotrauma 2006, 23, 1529–1534. [Google Scholar] [CrossRef]
- Hicks, R.R.; Smith, D.H.; Lowenstein, D.H.; Saint Marie, R.; McIntosh, T.K. Mild experimental brain injury in the rat induces cognitive deficits associated with regional neuronal loss in the hippocampus. J. Neurotrauma 1993, 10, 405–414. [Google Scholar] [CrossRef]
- Dash, P.K.; Mach, S.A.; Moore, A.N. Enhanced neurogenesis in the rodent hippocampus following traumatic brain injury. J. Neurosci. Res. 2001, 63, 313–319. [Google Scholar] [CrossRef]
- Correll, E.A.; Ramser, B.J.; Knott, M.V.; McCullumsmith, R.E.; McGuire, J.L.; Ngwenya, L.B. Deficits in pattern separation and dentate gyrus proliferation after rodent lateral fluid percussion injury. IBRO Neurosci. Rep. 2021, 10, 31–41. [Google Scholar] [CrossRef]
- Ngwenya, L.B.; Danzer, S.C. Impact of Traumatic Brain Injury on Neurogenesis. Front. Neurosci. 2018, 12, 1014. [Google Scholar] [CrossRef]
- Borzello, M.; Ramirez, S.; Treves, A.; Lee, I.; Scharfman, H.; Stark, C.; Knierim, J.J.; Rangel, L.M. Assessments of dentate gyrus function: Discoveries and debates. Nat. Rev. Neurosci. 2023, 24, 502–517. [Google Scholar] [CrossRef]
- Neuberger, E.J.; Swietek, B.; Corrubia, L.; Prasanna, A.; Santhakumar, V. Enhanced Dentate Neurogenesis after Brain Injury Undermines Long-Term Neurogenic Potential and Promotes Seizure Susceptibility. Stem Cell Rep. 2017, 9, 972–984. [Google Scholar] [CrossRef]
- Bielefeld, P.; Martirosyan, A.; Martín-Suárez, S.; Apresyan, A.; Meerhoff, G.F.; Pestana, F.; Poovathingal, S.; Reijner, N.; Koning, W.; Clement, R.A.; et al. Traumatic brain injury promotes neurogenesis at the cost of astrogliogenesis in the adult hippocampus of male mice. Nat. Commun. 2024, 15, 5222. [Google Scholar] [CrossRef] [PubMed]
- Urrea, C.; Castellanos, D.A.; Sagen, J.; Tsoulfas, P.; Bramlett, H.M.; Dietrich, W.D. Widespread cellular proliferation and focal neurogenesis after traumatic brain injury in the rat. Restor. Neurol. Neurosci. 2007, 25, 65–76. [Google Scholar] [CrossRef]
- Diaz-Aparicio, I.; Paris, I.; Sierra-Torre, V.; Plaza-Zabala, A.; Rodríguez-Iglesias, N.; Márquez-Ropero, M.; Beccari, S.; Huguet, P.; Abiega, O.; Alberdi, E.; et al. Microglia Actively Remodel Adult Hippocampal Neurogenesis through the Phagocytosis Secretome. J. Neurosci. 2020, 40, 1453. [Google Scholar] [CrossRef] [PubMed]
- Sultan, S.; Li, L.; Moss, J.; Petrelli, F.; Cassé, F.; Gebara, E.; Lopatar, J.; Pfrieger, F.W.; Bezzi, P.; Bischofberger, J.; et al. Synaptic Integration of Adult-Born Hippocampal Neurons Is Locally Controlled by Astrocytes. Neuron 2015, 88, 957–972. [Google Scholar] [CrossRef]
- Kreisel, T.; Wolf, B.; Keshet, E.; Licht, T. Unique role for dentate gyrus microglia in neuroblast survival and in VEGF-induced activation. Glia 2019, 67, 594–618. [Google Scholar] [CrossRef] [PubMed]
- Dejanovic, B.; Wu, T.; Tsai, M.C.; Graykowski, D.; Gandham, V.D.; Rose, C.M.; Bakalarski, C.E.; Ngu, H.; Wang, Y.; Pandey, S.; et al. Complement C1q-dependent excitatory and inhibitory synapse elimination by astrocytes and microglia in Alzheimer’s disease mouse models. Nat. Aging 2022, 2, 837–850. [Google Scholar] [CrossRef]
- Caplan, H.W.; Cardenas, F.; Gudenkauf, F.; Zelnick, P.; Xue, H.; Cox, C.S.; Bedi, S.S. Spatiotemporal Distribution of Microglia After Traumatic Brain Injury in Male Mice. ASN Neuro 2020, 12, 1759091420911770. [Google Scholar] [CrossRef]
- Robinson, C.; Apgar, C.; Shapiro, L.A. Astrocyte Hypertrophy Contributes to Aberrant Neurogenesis after Traumatic Brain Injury. Neural Plast. 2016, 2016, 1347987. [Google Scholar] [CrossRef]
- Littlejohn, E.L.; DeSana, A.J.; Williams, H.C.; Chapman, R.T.; Joseph, B.; Juras, J.A.; Saatman, K.E. IGF1-Stimulated Posttraumatic Hippocampal Remodeling Is Not Dependent on mTOR. Front. Cell Dev. Biol. 2021, 9, 663456. [Google Scholar] [CrossRef]
- Simon, D.W.; McGeachy, M.J.; Bayır, H.; Clark, R.S.B.; Loane, D.J.; Kochanek, P.M. The far-reaching scope of neuroinflammation after traumatic brain injury. Nat. Rev. Neurol. 2017, 13, 171–191, Erratum in Nat. Rev. Neurol. 2017, 13, 572. [Google Scholar] [CrossRef]
- Boone, D.R.; Weisz, H.A.; Willey, H.E.; Torres, K.E.O.; Falduto, M.T.; Sinha, M.; Spratt, H.; Bolding, I.J.; Johnson, K.M.; Parsley, M.A.; et al. Traumatic brain injury induces long-lasting changes in immune and regenerative signaling. PLoS ONE 2019, 14, e0214741. [Google Scholar] [CrossRef]
- Catta-Preta, R.; Zdilar, I.; Jenner, B.; Doisy, E.T.; Tercovich, K.; Nord, A.S.; Gurkoff, G.G. Transcriptional Pathology Evolves over Time in Rat Hippocampus after Lateral Fluid Percussion Traumatic Brain Injury. Neurotrauma Rep. 2021, 2, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Zhuang, Y.; Ying, Z.; Agrawal, R.; Yang, X.; Gomez-Pinilla, F. Traumatic Brain Injury Induces Genome-Wide Transcriptomic, Methylomic, and Network Perturbations in Brain and Blood Predicting Neurological Disorders. EBioMedicine 2017, 16, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Ren, L.; Xu, H.; Zhao, L.; Wang, Z.H.; Hu, G.D.; Wei, Z.L. Single-Cell RNA Sequencing Reveals Cellular and Transcriptional Changes Associated With Traumatic Brain Injury. Front. Genet. 2022, 13, 861428. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, W.; Yan, K.; Zhao, H.; Zhang, Z.; Wang, Y.; Zhu, W.; Chen, S. RNA-seq reveals Nup62 as a potential regulator for cell division after traumatic brain injury in mice hippocampus. PeerJ 2023, 11, e14913. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.Z.; Xing, J.; Huang, Q.; Yang, X.T.; Zhao, C.Y.; Li, X.Y. Integration of single-cell and bulk RNA sequencing data reveals key cell types and regulators in traumatic brain injury. Math. Biosci. Eng. 2021, 18, 1201–1214. [Google Scholar] [CrossRef]
- Arneson, D.; Zhang, G.; Ying, Z.; Zhuang, Y.; Byun, H.R.; Ahn, I.S.; Gomez-Pinilla, F.; Yang, X. Single cell molecular alterations reveal target cells and pathways of concussive brain injury. Nat. Commun. 2018, 9, 3894. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, H.; Zhong, J.; Yang, J.; Darwazeh, R.; Tian, X.; Huang, Z.; Jiang, L.; Cheng, C.; Wu, Y.; et al. Significant changes in circular RNA in the mouse cerebral cortex around an injury site after traumatic brain injury. Exp. Neurol. 2019, 313, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Puhakka, N.; Bot, A.M.; Vuokila, N.; Debski, K.J.; Lukasiuk, K.; Pitkänen, A. Chronically dysregulated NOTCH1 interactome in the dentate gyrus after traumatic brain injury. PLoS ONE 2017, 12, e0172521. [Google Scholar] [CrossRef]
- Zhang, S.-J.; Steijaert, M.N.; Lau, D.; Schütz, G.; Delucinge-Vivier, C.; Descombes, P.; Bading, H. Decoding NMDA Receptor Signaling: Identification of Genomic Programs Specifying Neuronal Survival and Death. Neuron 2007, 53, 549–562. [Google Scholar] [CrossRef]
- Wahl, A.S.; Buchthal, B.; Rode, F.; Bomholt, S.F.; Freitag, H.E.; Hardingham, G.E.; Rønn, L.C.B.; Bading, H. Hypoxic/ischemic conditions induce expression of the putative pro-death gene Clca1 via activation of extrasynaptic N-methyl-d-aspartate receptors. Neuroscience 2009, 158, 344–352. [Google Scholar] [CrossRef]
- Stevens, B.; Allen, N.J.; Vazquez, L.E.; Howell, G.R.; Christopherson, K.S.; Nouri, N.; Micheva, K.D.; Mehalow, A.K.; Huberman, A.D.; Stafford, B.; et al. The classical complement cascade mediates CNS synapse elimination. Cell 2007, 131, 1164–1178. [Google Scholar] [CrossRef]
- Schafer, D.P.; Lehrman, E.K.; Kautzman, A.G.; Koyama, R.; Mardinly, A.R.; Yamasaki, R.; Ransohoff, R.M.; Greenberg, M.E.; Barres, B.A.; Stevens, B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 2012, 74, 691–705. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Li, Y.; Li, X.; Zeng, F.; Rao, Y.; He, Y.; Wang, Y.; Liu, M.; Li, D.; Xu, Z.; et al. Microglial debris is cleared by astrocytes via C4b-facilitated phagocytosis and degraded via RUBICON-dependent noncanonical autophagy in mice. Nat. Commun. 2022, 13, 6233. [Google Scholar] [CrossRef]
- Kim, S.K.; Nabekura, J.; Koizumi, S. Astrocyte-mediated synapse remodeling in the pathological brain. Glia 2017, 65, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Krukowski, K.; Nolan, A.; Becker, M.; Picard, K.; Vernoux, N.; Frias, E.S.; Feng, X.; Tremblay, M.-E.; Rosi, S. Novel microglia-mediated mechanisms underlying synaptic loss and cognitive impairment after traumatic brain injury. Brain Behav. Immun. 2021, 98, 122–135. [Google Scholar] [CrossRef]
- Gao, C.; Jiang, J.; Tan, Y.; Chen, S. Microglia in neurodegenerative diseases: Mechanism and potential therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 359. [Google Scholar] [CrossRef]
- Brandebura, A.N.; Paumier, A.; Onur, T.S.; Allen, N.J. Astrocyte contribution to dysfunction, risk and progression in neurodegenerative disorders. Nat. Rev. Neurosci. 2023, 24, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Quan, F.; Liang, X.; Cheng, M.; Yang, H.; Liu, K.; He, S.; Sun, S.; Deng, M.; He, Y.; Liu, W.; et al. Annotation of cell types (ACT): A convenient web server for cell type annotation. Genome Med. 2023, 15, 91. [Google Scholar] [CrossRef]
- Sontheimer, R.D.; Racila, E.; Racila, D.M. C1q: Its Functions within the Innate and Adaptive Immune Responses and its Role in Lupus Autoimmunity. J. Investig. Dermatol. 2005, 125, 14–23. [Google Scholar] [CrossRef]
- Cao, C.; Liu, T.; Peng, L.; Li, L.; Xu, Z.; Li, X.; Chen, G.; Li, H.; Bai, L. Targeting CD74 in microglia to modulate experimental cerebral ischemia and reperfusion injury: Insights from Single-Cell and bulk transcriptomics. Mol. Brain 2025, 18, 46. [Google Scholar] [CrossRef]
- Jahn, J.; Bollensdorf, A.; Kalischer, C.; Piecha, R.; Weiß-Müller, J.; Potru, P.S.; Ruß, T.; Spittau, B. Microglial CD74 Expression Is Regulated by TGFβ Signaling. Int. J. Mol. Sci. 2022, 23, 10247. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, C.; Chen, C.; Li, S.; Wang, Y.; Yang, T.; Stetler, R.A.; Bennett, M.V.L.; Dixon, C.E.; Chen, J.; et al. STAT1 Contributes to Microglial/Macrophage Inflammation and Neurological Dysfunction in a Mouse Model of Traumatic Brain Injury. J. Neurosci. 2022, 42, 7466. [Google Scholar] [CrossRef]
- Yap, E.-L.; Greenberg, M.E. Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 2018, 100, 330–348. [Google Scholar] [CrossRef] [PubMed]
- D’Sa, K.; Guelfi, S.; Vandrovcova, J.; Reynolds, R.H.; Zhang, D.; Hardy, J.; Botía, J.A.; Weale, M.E.; Taliun, S.A.G.; Small, K.S.; et al. Analysis of subcellular RNA fractions demonstrates significant genetic regulation of gene expression in human brain post-transcriptionally. Sci. Rep. 2023, 13, 13874. [Google Scholar] [CrossRef] [PubMed]
- Grindberg, R.V.; Yee-Greenbaum, J.L.; McConnell, M.J.; Novotny, M.; O’Shaughnessy, A.L.; Lambert, G.M.; Araúzo-Bravo, M.J.; Lee, J.; Fishman, M.; Robbins, G.E.; et al. RNA-sequencing from single nuclei. Proc. Natl. Acad. Sci. USA 2013, 110, 19802–19807. [Google Scholar] [CrossRef]
- Hammad, A.; Westacott, L.; Zaben, M. The role of the complement system in traumatic brain injury: A review. J. Neuroinflamm. 2018, 15, 24, Correction in J. Neuroinflamm. 2018, 15, 59. [Google Scholar] [CrossRef]
- Stahel, P.F.; Barnum, S.R. Bacterial meningitis: Complement gene expression in the central nervous system. Immunopharmacology 1997, 38, 65–72. [Google Scholar] [CrossRef]
- Alawieh, A.; Elvington, A.; Tomlinson, S. Complement in the Homeostatic and Ischemic Brain. Front. Immunol. 2015, 6, 417. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.; Strohmeyer, R.; Kovelowski, C.J.; Li, R. Microglia and inflammatory mechanisms in the clearance of amyloid beta peptide. Glia 2002, 40, 260–269. [Google Scholar] [CrossRef]
- Stephan, A.H.; Barres, B.A.; Stevens, B. The complement system: An unexpected role in synaptic pruning during development and disease. Annu. Rev. Neurosci. 2012, 35, 369–389. [Google Scholar] [CrossRef]
- Rahpeymai, Y.; Hietala, M.A.; Wilhelmsson, U.; Fotheringham, A.; Davies, I.; Nilsson, A.K.; Zwirner, J.; Wetsel, R.A.; Gerard, C.; Pekny, M.; et al. Complement: A novel factor in basal and ischemia-induced neurogenesis. EMBO J. 2006, 25, 1364–1374. [Google Scholar] [CrossRef] [PubMed]
- Shinjyo, N.; Ståhlberg, A.; Dragunow, M.; Pekny, M.; Pekna, M. Complement-derived anaphylatoxin C3a regulates in vitro differentiation and migration of neural progenitor cells. Stem Cells 2009, 27, 2824–2832. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Fukuhara, T.; Britschgi, M.; He, Y.; Narasimhan, R.; Villeda, S.; Molina, H.; Huber, B.T.; Holers, M.; Wyss-Coray, T. Complement receptor 2 is expressed in neural progenitor cells and regulates adult hippocampal neurogenesis. J. Neurosci. 2011, 31, 3981–3989. [Google Scholar] [CrossRef]
- Bellander, B.M.; Singhrao, S.K.; Ohlsson, M.; Mattsson, P.; Svensson, M. Complement activation in the human brain after traumatic head injury. J. Neurotrauma 2001, 18, 1295–1311. [Google Scholar] [CrossRef]
- Kossmann, T.; Stahel, P.F.; Morganti-Kossmann, M.C.; Jones, J.L.; Barnum, S.R. Elevated levels of the complement components C3 and factor B in ventricular cerebrospinal fluid of patients with traumatic brain injury. J. Neuroimmunol. 1997, 73, 63–69. [Google Scholar] [CrossRef]
- Kaczorowski, S.L.; Schiding, J.K.; Toth, C.A.; Kochanek, P.M. Effect of soluble complement receptor-1 on neutrophil accumulation after traumatic brain injury in rats. J. Cereb. Blood Flow Metab. 1995, 15, 860–864. [Google Scholar] [CrossRef]
- Rancan, M.; Morganti-Kossmann, M.C.; Barnum, S.R.; Saft, S.; Schmidt, O.I.; Ertel, W.; Stahel, P.F. Central nervous system-targeted complement inhibition mediates neuroprotection after closed head injury in transgenic mice. J. Cereb. Blood Flow Metab. 2003, 23, 1070–1074. [Google Scholar] [CrossRef]
- Leinhase, I.; Schmidt, O.I.; Thurman, J.M.; Hossini, A.M.; Rozanski, M.; Taha, M.E.; Scheffler, A.; John, T.; Smith, W.R.; Holers, V.M.; et al. Pharmacological complement inhibition at the C3 convertase level promotes neuronal survival, neuroprotective intracerebral gene expression, and neurological outcome after traumatic brain injury. Exp. Neurol. 2006, 199, 454–464. [Google Scholar] [CrossRef]
- Todd, B.P.; Chimenti, M.S.; Luo, Z.; Ferguson, P.J.; Bassuk, A.G.; Newell, E.A. Traumatic brain injury results in unique microglial and astrocyte transcriptomes enriched for type I interferon response. J. Neuroinflamm. 2021, 18, 151. [Google Scholar] [CrossRef]
- Izzy, S.; Liu, Q.; Fang, Z.; Lule, S.; Wu, L.; Chung, J.Y.; Sarro-Schwartz, A.; Brown-Whalen, A.; Perner, C.; Hickman, S.E.; et al. Time-Dependent Changes in Microglia Transcriptional Networks Following Traumatic Brain Injury. Front. Cell Neurosci. 2019, 13, 307. [Google Scholar] [CrossRef]
- Zhong, J.; Jiang, L.; Cheng, C.; Huang, Z.; Zhang, H.; Liu, H.; He, J.; Cao, F.; Peng, J.; Jiang, Y.; et al. Altered expression of long non-coding RNA and mRNA in mouse cortex after traumatic brain injury. Brain Res. 2016, 1646, 589–600. [Google Scholar] [CrossRef]
- Toutonji, A.; Mandava, M.; Guglietta, S.; Tomlinson, S. Chronic complement dysregulation drives neuroinflammation after traumatic brain injury: A transcriptomic study. Acta Neuropathol. Commun. 2021, 9, 126. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Engel, J.; Teichmann, S.A.; Lönnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kang, H.; Jo, A.; Yoo, S.A.; Lee, H.O. Perspectives on single-nucleus RNA sequencing in different cell types and tissues. J. Pathol. Transl. Med. 2023, 57, 52–59. [Google Scholar] [CrossRef]
- Makinde, H.M.; Just, T.B.; Gadhvi, G.T.; Winter, D.R.; Schwulst, S.J. Microglia Adopt Longitudinal Transcriptional Changes After Traumatic Brain Injury. J. Surg. Res. 2020, 246, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Malinow, R.; Schulman, H.; Tsien, R.W. Inhibition of postsynaptic PKC or CaMKII blocks induction but not expression of LTP. Science 1989, 245, 862–866. [Google Scholar] [CrossRef]
- Fink, C.C.; Bayer, K.U.; Myers, J.W.; Ferrell, J.E., Jr.; Schulman, H.; Meyer, T. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron 2003, 39, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.J.; Paylor, R.; Wehner, J.M.; Tonegawa, S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science 1992, 257, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Puram, S.V.; Kim, A.H.; Ikeuchi, Y.; Wilson-Grady, J.T.; Merdes, A.; Gygi, S.P.; Bonni, A. A CaMKIIβ signaling pathway at the centrosome regulates dendrite patterning in the brain. Nat. Neurosci. 2011, 14, 973–983. [Google Scholar] [CrossRef]
- Borghi, R.; Trivisano, M.; Specchio, N.; Tartaglia, M.; Compagnucci, C. Understanding the pathogenetic mechanisms underlying altered neuronal function associated with CAMK2B mutations. Neurosci. Biobehav. Rev. 2023, 152, 105299. [Google Scholar] [CrossRef]
- Paban, V.; Ogier, M.; Chambon, C.; Fernandez, N.; Davidsson, J.; Risling, M.; Alescio-Lautier, B. Molecular gene expression following blunt and rotational models of traumatic brain injury parallel injuries associated with stroke and depression. J. Transl. Sci. 2016, 2. [Google Scholar] [CrossRef]
- Halpain, S.; Dehmelt, L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006, 7, 224. [Google Scholar] [CrossRef]
- Giza, J.; Urbanski, M.J.; Prestori, F.; Bandyopadhyay, B.; Yam, A.; Friedrich, V.; Kelley, K.; D’Angelo, E.; Goldfarb, M. Behavioral and cerebellar transmission deficits in mice lacking the autism-linked gene islet brain-2. J. Neurosci. 2010, 30, 14805–14816. [Google Scholar] [CrossRef]
- Villablanca, C.; Vidal, R.; Gonzalez-Billault, C. Are cytoskeleton changes observed in astrocytes functionally linked to aging? Brain Res. Bull. 2023, 196, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Richetin, K.; Steullet, P.; Pachoud, M.; Perbet, R.; Parietti, E.; Maheswaran, M.; Eddarkaoui, S.; Bégard, S.; Pythoud, C.; Rey, M.; et al. Tau accumulation in astrocytes of the dentate gyrus induces neuronal dysfunction and memory deficits in Alzheimer’s disease. Nat. Neurosci. 2020, 23, 1567–1579. [Google Scholar] [CrossRef]
- Milenkovic, I.; Petrov, T.; Kovacs, G.G. Patterns of Hippocampal Tau Pathology Differentiate Neurodegenerative Dementias. Dement. Geriatr. Cogn. Disord. 2014, 38, 375–388. [Google Scholar] [CrossRef]
- Edwards, G., 3rd; Zhao, J.; Dash, P.K.; Soto, C.; Moreno-Gonzalez, I. Traumatic Brain Injury Induces Tau Aggregation and Spreading. J. Neurotrauma 2020, 37, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Dejanovic, B.; Huntley, M.A.; De Mazière, A.; Meilandt, W.J.; Wu, T.; Srinivasan, K.; Jiang, Z.; Gandham, V.; Friedman, B.A.; Ngu, H.; et al. Changes in the Synaptic Proteome in Tauopathy and Rescue of Tau-Induced Synapse Loss by C1q Antibodies. Neuron 2018, 100, 1322–1336.e7. [Google Scholar] [CrossRef]
- Milligan, G.W. An examination of the effect of six types of error perturbation on fifteen clustering algorithms. Psychometrika 1980, 45, 325–342. [Google Scholar] [CrossRef]
- Thrupp, N.; Sala Frigerio, C.; Wolfs, L.; Skene, N.G.; Fattorelli, N.; Poovathingal, S.; Fourne, Y.; Matthews, P.M.; Theys, T.; Mancuso, R.; et al. Single-Nucleus RNA-Seq Is Not Suitable for Detection of Microglial Activation Genes in Humans. Cell Rep. 2020, 32, 108189. [Google Scholar] [CrossRef] [PubMed]
- Cembrowski, M.S.; Wang, L.; Sugino, K.; Shields, B.C.; Spruston, N. Hipposeq: A comprehensive RNA-seq database of gene expression in hippocampal principal neurons. eLife 2016, 5, e14997. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Guide for the Care and Use of Laboratory Animals: Eighth Edition; The National Academies Press: Washington, DC, USA, 2011; p. 246.
- Best, F.V.; Hartings, J.A.; Ngwenya, L.B. Repetitive cortical spreading depolarizations are prolonged early after experimental traumatic brain injury. Exp. Neurol. 2025, 385, 115120. [Google Scholar] [CrossRef]
- Hagihara, H.; Toyama, K.; Yamasaki, N.; Miyakawa, T. Dissection of hippocampal dentate gyrus from adult mouse. J. Vis. Exp. 2009, 1543. [Google Scholar] [CrossRef]
- Liu, P.; Ewald, J.; Pang, Z.; Legrand, E.; Jeon, Y.S.; Sangiovanni, J.; Hacariz, O.; Zhou, G.; Head, J.A.; Basu, N.; et al. ExpressAnalyst: A unified platform for RNA-sequencing analysis in non-model species. Nat. Commun. 2023, 14, 2995. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R Package Version 1.22.0 2024. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Harvard Chan Bioinformatics Core. Introduction to Single-Cell RNA-seq. Available online: https://github.com/hbctraining (accessed on 1 July 2024).
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2024, 42, 293–304. [Google Scholar] [CrossRef]
- Zappia, L.; Oshlack, A. Clustering trees: A visualization for evaluating clusterings at multiple resolutions. GigaScience 2018, 7, giy083. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Sock, E.; Wegner, M. Transcriptional control of myelination and remyelination. Glia 2019, 67, 2153–2165. [Google Scholar] [CrossRef] [PubMed]
- Pilarczyk, M.; Fazel-Najafabadi, M.; Kouril, M.; Shamsaei, B.; Vasiliauskas, J.; Niu, W.; Mahi, N.; Zhang, L.; Clark, N.A.; Ren, Y.; et al. Connecting omics signatures and revealing biological mechanisms with iLINCS. Nat. Commun. 2022, 13, 4678. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Balcı, H.; Hanspers, K.; Coort, S.L.; Martens, M.; Slenter, D.N.; Ehrhart, F.; Digles, D.; Waagmeester, A.; Wassink, I.; et al. WikiPathways 2024: Next generation pathway database. Nucleic Acids Res. 2024, 52, D679–D689. [Google Scholar] [CrossRef]
| Term | DEGs in Dataset/Genes in Pathway | p-Value | Adjusted p-Value (q-Value) | Odds Ratio | Combined Score | DEGs in Pathway |
|---|---|---|---|---|---|---|
| Complement Activation WP545 | 8/23 | 9.66 × 10−14 | 2.05 × 10−11 | 120.54 | 3612.29 | Cfd; C1qb; C1qa; C4a; C1s; C1r; C2; C1qc |
| Microglia Pathogen Phagocytosis Pathway WP3937 | 9/40 | 2.23 × 10−13 | 2.36 × 10−11 | 66.32 | 1931.98 | C1qb; C1qq; Tyrobp; Ncf1; Arpc1b; Cyba; Ptpn6; Vav1; C1qc |
| Complement System In Neuronal Development And Plasticity WP5090 | 11/106 | 3.30 × 10−12 | 2.33 × 10−10 | 26.98 | 713.36 | C1qb; Cfd; C4a; C1qa; Tgfb; C1s; C1r; Axl; Serping1; C2; C1qc |
| Complement And Coagulation Cascades WP558 | 9/58 | 8.09 × 10−12 | 4.03 × 10−10 | 41.92 | 1070.62 | Cfd; C1qb; C1qa; C1s; F10; C1r; Serping1; C2; C1qc |
| Dengue 2 Interactions With Complement And Coagulation Cascades WP3896 | 9/59 | 9.50 × 10−12 | 4.03 × 10−10 | 41.08 | 1042.51 | Cfd; C1qb; C1qa; C1s; F10; C1r; Serping1; C2; C1qc |
| Oxidative Damage Response WP3941 | 6/40 | 3.51 × 10−8 | 1.24 × 10−6 | 38.96 | 668.74 | C1qb; C1qa; C1s; C1r; C2; C1qc |
| Complement System WP2806 | 7/95 | 3.68 × 10−7 | 1.12 × 10−5 | 17.71 | 262.38 | Cfd; C4a; C1s; F10; Serping1; Icam1; C2 |
| TYROBP Causal Network In Microglia WP3945 | 6/62 | 5.17 × 10−7 | 1.37 × 10−5 | 23.63 | 342.04 | Tyrobp; Npc2; Cd37; Capg; Cxcl16; C1qc |
| Allograft Rejection WP2328 | 6/90 | 4.70 × 10−6 | 1.11 × 10−4 | 15.73 | 192.98 | C1qb; C4a; C1qa; Tgfb1; C2; C1qc |
| Network Map of SARS-CoV 2 Signaling WP5115 | 7/254 | 2.24 × 10−4 | 0.00474164 | 6.26 | 52.61 | Lgals3bp; Trpm2; C1s; C1r; Ctsz; Ptpn6; Cxcl16 |
| Gene Name | p-Value | q-Value | Avg Log2FC | Pct.1 (Control) | Pct.2 (TBI) | |
|---|---|---|---|---|---|---|
| Microglia 1 | Ano5 | 1.02 × 10−12 | 1.30 × 10−10 | −1.52 | 0.136 | 0.055 |
| Axl | 1.41 × 10−12 | 1.53 × 10−10 | 3.97 | 0.006 | 0.065 | |
| Spata22 | 3.94 × 10−10 | 2.73 × 10−08 | −2.21 | 0.059 | 0.013 | |
| Kmo | 4.18 × 10−9 | 2.28 × 10−7 | −1.85 | 0.059 | 0.016 | |
| Slfn4 | 1.45 × 10−6 | 3.40 × 10−5 | 3.41 | 0.014 | 0.054 | |
| Mx1 | 2.38 × 10−6 | 4.97 × 10−5 | 3.28 | 0.004 | 0.034 | |
| Marc1 | 3.75 × 10−6 | 7.23 × 10−5 | 4.72 | 0.001 | 0.025 | |
| Tmem154 | 4.44 × 10−6 | 8.46 × 10−5 | −1.51 | 0.039 | 0.011 | |
| LOC100364435 | 6.30 × 10−6 | 1.13 × 10−4 | 7.77 | 0 | 0.019 | |
| Gpnmb | 6.69 × 10−6 | 1.19 × 10−4 | 4.27 | 0.002 | 0.026 | |
| Microglia 2 | Rpl10 | 9.75 × 10−11 | 6.39 × 10−8 | 3.95 | 0 | 0.018 |
| Foxred1 | 3.83 × 10−8 | 1.25 × 10−5 | −1.75 | 0.213 | 0.069 | |
| Cd46 | 2.73 × 10−6 | 4.47 × 10−4 | −1.80 | 0.186 | 0.047 | |
| Ccdc141 | 6.03 × 10−6 | 6.58 × 10−4 | −3.69 | 0.032 | 0.002 | |
| Ccdc88b | 2.44 × 10−5 | 2.02 × 10−3 | −2.53 | 0.09 | 0.013 | |
| Slfn13 | 2.47 × 10−5 | 2.02 × 10−3 | 4.57 | 0.005 | 0.101 | |
| Zfp263 | 4.17 × 10−5 | 2.73 × 10−3 | −6.52 | 0.037 | 0 | |
| Slc2a12 | 4.17 × 10−5 | 2.73 × 10−3 | −6.03 | 0.037 | 0 | |
| Apbb3 | 4.40 × 10−5 | 2.74 × 10−3 | −1.88 | 0.16 | 0.04 | |
| Fbxw9 | 1.06 × 10−4 | 4.78 × 10−3 | −1.98 | 0.106 | 0.025 |
| Term | DEGs in Dataset/Genes in Pathway | p-Value | Adjusted p-Value (q-Value) | Odds Ratio | Combined Score | DEGs in Pathway | |
|---|---|---|---|---|---|---|---|
| Microglia 1 | Complement System In Neuronal Development And Plasticity WP5090 | 7/106 | 1.03 × 10−6 | 2.07 × 10−4 | 15.05 | 207.58 | C1qb; C1qa; Axl; Mbp; Itgav; Mertk; C1qc |
| Neuroinflammation And Glutamatergic Signaling WP5083 | 7/140 | 6.59 × 10−6 | 6.66 × 10−4 | 11.19 | 133.45 | Camk2b; Gria2; Camk2D; Slc17a7; Disc1; Grm1; Gls | |
| Disruption Of Postsynaptic Signaling By CNV WP4875 | 4/33 | 2.15 × 10−5 | 1.45 × 10−3 | 28.55 | 306.79 | Camk2b; Camk2d; Nrxn3; Grm1 | |
| Microglia Pathogen Phagocytosis Pathway WP3937 | 4/40 | 4.68 × 10−5 | 2.36 × 10−3 | 22.99 | 229.20 | C1qb; Vav3; C1qa; C1qc | |
| Serotonin And Anxiety WP3947 | 3/17 | 7.84 × 10−5 | 3.17 × 10−3 | 43.93 | 415.32 | Camk2b; Ppp3ca; Grm1 | |
| Complement Activation WP545 | 3/23 | 2.00 × 10−4 | 6.72 × 10−3 | 30.74 | 261.88 | C1qb; C1qa; C1qc | |
| Photodynamic Therapy-Induced Unfolded Protein Response WP3613 | 3/27 | 3.25 × 10−4 | 8.30 × 10−3 | 25.61 | 205.71 | Hspa5; Calr; Hsp90b1 | |
| Spinal Cord Injury WP2431 | 5/119 | 3.29 × 10−4 | 8.30 × 10−3 | 9.13 | 73.26 | C1qb; Fkbp1a; Ppp3ca; Ptpra; Mbp | |
| Primary Focal Segmental Glomerulosclerosis FSGS WP2572 | 4/72 | 4.66 × 10−4 | 1.05 × 10−2 | 12.15 | 93.21 | Camk2b; Trpc6; Ctsl; Itgav | |
| Cell Lineage Map For Neuronal Differentiation WP5417 | 5/132 | 5.29 × 10−4 | 1.07 × 10−2 | 8.19 | 61.82 | Map2; Mbp; Slc17a7; S100b; Gls | |
| Microglia 2 | Genes Controlling Nephrogenesis WP4823 | 4/44 | 6.85 × 10−5 | 1.10 × 10−2 | 20.69 | 198.37 | Robo2; Notch2; Gli3; Vegfa |
| Osteoarthritic Chondrocyte Hypertrophy WP5373 | 3/50 | 2.00 × 10−3 | 1.11 × 10−1 | 13.06 | 81.16 | Jund; Junb; Vegfa | |
| 11P11 2 Copy Number Variation Syndrome WP5348 | 3/56 | 2.77 × 10−3 | 1.11 × 10−1 | 11.58 | 68.18 | B4gat1; Nrxn1; Gli3 | |
| Oncostatin M Signaling WP2374 | 3/56 | 2.77 × 10−3 | 1.11 × 10−1 | 11.58 | 68.18 | Jund; Junb; Vegfa | |
| Hypertrophy Model WP516 | 2/20 | 4.43 × 10−3 | 1.42 × 10−1 | 22.54 | 122.14 | Jund; Vegfa | |
| Axon Guidance WP5289 | 3/72 | 5.64 × 10−3 | 1.45 × 10−1 | 8.89 | 46.03 | Robo2; Robo3; Lrrc4c | |
| Hippocampal Synaptogenesis And Neurogenesis WP5231 | 2/24 | 6.36 × 10−3 | 1.45 × 10−1 | 18.44 | 93.27 | Nrxn1; Ncam1 | |
| EGF EGFR Signaling WP437 | 4/159 | 8.36 × 10−3 | 1.67 × 10−1 | 5.31 | 25.40 | Ptprr; Jund; Inpp5d; Ncoa3 | |
| GDNF RET Signaling Axis WP4830 | 2/30 | 9.83 × 10−3 | 1.75 × 10−1 | 14.48 | 66.95 | Robo2; Gli3 | |
| Disruption of Postsynaptic Signaling by CNV WP4875 | 2/33 | 1.18 × 10−2 | 1.89 × 10−1 | 13.08 | 58.06 | Nrxn1; Dlgap1 |
| Gene Name | p-Value | q-Value | Avg Log2FC | Pct.1 (Control) | Pct.2 (TBI) | |
|---|---|---|---|---|---|---|
| Astrocyte 1 | Pld5 | 1.29 × 10−54 | 3.51 × 10−51 | 3.18 | 0.02 | 0.157 |
| Nme4 | 1.32 × 10−29 | 7.18 × 10−27 | 8.73 | 0 | 0.044 | |
| Plk5 | 2.35 × 10−24 | 5.81 × 10−22 | 1.60 | 0.043 | 0.148 | |
| Celsr3 | 6.06 × 10−20 | 5.89 × 10−18 | 1.52 | 0.025 | 0.098 | |
| LOC100364435 | 1.22 × 10−17 | 7.04 × 10−16 | 8.36 | 0 | 0.02 | |
| Dlk2 | 1.32 × 10−17 | 7.48 × 10−16 | 1.52 | 0.026 | 0.091 | |
| Rps23 | 2.04 × 10−17 | 1.12 × 10−15 | 3.61 | 0.004 | 0.087 | |
| Asb15 | 1.18 × 10−15 | 4.04 × 10−14 | 4.28 | 0.001 | 0.026 | |
| Rpl18a | 1.09 × 10−13 | 2.39 × 10−12 | 9.41 | 0 | 0.063 | |
| Epha8 | 2.69 × 10−13 | 5.5 × 10−12 | 1.64 | 0.027 | 0.084 | |
| Astrocyte 2 | AABR07000398.1 | 4.15 × 10−64 | 4.61 × 10−61 | 1.55 | 0.624 | 0.824 |
| Map1a | 4.42 × 10−45 | 3.28 × 10−42 | 1.83 | 0.26 | 0.551 | |
| S100b | 1.82 × 10−36 | 8.09 × 10−34 | 1.55 | 0.235 | 0.486 | |
| Hpcal4 | 2.49 × 10−20 | 4.26 × 10−18 | 1.81 | 0.142 | 0.316 | |
| Agap2 | 3.42 × 10−20 | 5.43 × 10−18 | 1.71 | 0.158 | 0.332 | |
| Ddn | 4.92 × 10−18 | 5.76 × 10−16 | 1.69 | 0.1 | 0.247 | |
| Mapk8ip2 | 2.06 × 10−16 | 1.64 × 10−14 | 1.51 | 0.116 | 0.265 | |
| Ubc | 1.36 × 10−14 | 8.40 × 10−13 | 1.55 | 0.1 | 0.23 | |
| Rpl10 | 5.95 × 10−12 | 2.50 × 10−10 | 1.58 | 0.005 | 0.028 | |
| Gnas.1 | 1.92 × 10−11 | 7.11 × 10−10 | 1.62 | 0.068 | 0.166 | |
| Astrocyte 3 | Pld5 | 2.25 × 10−30 | 7.50 × 10−28 | 2.93 | 0.051 | 0.256 |
| Rpl10 | 1.16 × 10−22 | 1.78 × 10−20 | 7.65 | 0 | 0.106 | |
| Slc9b1 | 2.17 × 10−17 | 1.38 × 10−15 | −1.71 | 0.244 | 0.085 | |
| Jhy | 1.16 × 10−15 | 5.95 × 10−14 | −1.63 | 0.237 | 0.083 | |
| Begain | 1.67 × 10−13 | 5.96 × 10−12 | 1.85 | 0.101 | 0.257 | |
| Nme4 | 1.91 × 10−13 | 6.76 × 10−12 | 6.14 | 0 | 0.049 | |
| LOC100364435 | 6.10 × 10−13 | 1.98 × 10−11 | 5.89 | 0 | 0.022 | |
| Rapgefl1 | 6.24 × 10−12 | 1.63 × 10−10 | 1.73 | 0.112 | 0.264 | |
| Hba-a2 | 3.53 × 10−11 | 7.71 × 10−10 | −4.42 | 0.077 | 0.006 | |
| Cacna1h | 5.23 × 10−11 | 1.10 × 10−9 | 1.50 | 0.102 | 0.239 |
| Term | DEGs in Dataset/Genes in Pathway | p-Value | Adjusted p-Value (q-Value) | Odds Ratio | Combined Score | DEGs in Pathway | |
|---|---|---|---|---|---|---|---|
| Astrocyte 1 | Neuroinflammation And Glutamatergic Signaling WP5083 | 9/140 | 3.45 × 10−8 | 5.08 × 10−6 | 14.93 | 256.43 | Camk2b; Camk2a; Slc1a2; Slc1a3; Nsmf; Slc17a7; Il6r; Camkk1; Grin1 |
| Fragile X Syndrome WP4549 | 7/122 | 2.65 × 10−6 | 1.94 × 10−4 | 12.95 | 166.31 | Camk2b; Map1b; Camk2a; Agap2; Dlgap3; Dnm; Grin1 | |
| Cell Lineage Map For Neuronal Differentiation WP5417 | 7/132 | 4.47 × 10−6 | 2.13 × 10−4 | 11.91 | 146.68 | Slc1a2; Rimbp2; Slc1a3; Slc17a7; Bsn; Cacng2; Grin1 | |
| Hippocampal Synaptogenesis And Neurogenesis WP5231 | 4/24 | 5.79 × 10−6 | 2.13 × 10−4 | 41.42 | 499.47 | Camk2b; Nrxn1; Camk2a; Camkk1 | |
| Disruption Of Postsynaptic Signaling By CNV WP4875 | 4/33 | 2.15 × 10−5 | 6.33 × 10−4 | 28.55 | 306.79 | Camk2b; Nrxn1; Camk2a; Grin1 | |
| NO cGMP PKG Mediated Neuroprotection WP4008 | 4/46 | 8.17 × 10−5 | 2.00 × 10−3 | 19.70 | 185.43 | Camk2b; Camk2a; Pde2a; Grin1 | |
| Microtubule Cytoskeleton Regulation WP2038 | 4/48 | 9.67 × 10−5 | 2.03 × 10−3 | 18.80 | 173.82 | Tiam1; Ntrk3; Map1b; Ephb2 | |
| Synaptic Vesicle Pathway WP2267 | 3/51 | 2.12 × 10−3 | 3.90 × 10−2 | 12.79 | 78.73 | Slc1a3; Slc17a7; Dnm1 | |
| Phosphodiesterases In Neuronal Function WP4222 | 3/54 | 2.50 × 10−3 | 4.08 × 10−2 | 12.04 | 72.12 | Camk2a; Pde2A; Grin1 | |
| NAD Metabolism WP3644 | 2/16 | 2.84 × 10−3 | 4.17 × 10−2 | 28.99 | 170.01 | Nt5e; Nmnat2 | |
| Astrocyte 2 | Disruption Of Postsynaptic Signaling By CNV WP4875 | 6/33 | 1.33 × 10−8 | 2.37 × 10−6 | 46.98 | 851.93 | Camk2b; Nlgn1; Dlg2; Nrxn1; Nrxn3; Dlgap1 |
| Neuroinflammation And Glutamatergic Signaling WP5083 | 8/140 | 5.10 × 10−7 | 4.54 × 10−5 | 13.02 | 188.67 | Camk2b; Grm5; Cfl1; Nsmf; Calm1; Adcy8; Glul; Gfap | |
| Cell Lineage Map For Neuronal Differentiation WP5417 | 7/132 | 4.47 × 10−6 | 2.65 × 10−4 | 11.91 | 146.68 | Nlgn1; Dlg2; Aqp4; Mbp; S100b; Glul; Gfap | |
| ADHD And Autism ASD Pathways WP5420 | 10/370 | 1.64 × 10−5 | 7.31 × 10−4 | 6.03 | 66.44 | Gabrb1; Grm5; Nlgn1; Dlg2; Syt1; Nrxn1; Nrxn3; Dlgap1; Calm1; Glul | |
| Common Pathways Underlying Drug Addiction WP2636 | 4/41 | 5.17 × 10−5 | 1.84 × 10−3 | 22.37 | 220.78 | Grm5; Adcy8; Calm1; Actb | |
| Calcium Regulation In Cardiac Cells WP536 | 6/151 | 1.11 × 10−4 | 3.31 × 10−3 | 8.70 | 79.15 | Camk2b; Ywhab; Calm1; Adcy8; Calm2; Kcnj3 | |
| Myometrial Relaxation And Contraction Pathways WP289 | 6/156 | 1.33 × 10−4 | 3.39 × 10−3 | 8.40 | 74.99 | Camk2b; Ywhab; Calm1; Adcy8; Calm2; Actb | |
| Hippocampal Synaptogenesis And Neurogenesis WP5231 | 3/24 | 2.27 × 10−4 | 5.06 × 10−3 | 29.28 | 245.59 | Camk2b; Nrxn1; Calm1 | |
| Glial Cell Differentiation WP2276 | 2/7 | 5.11 × 10−4 | 1.01 × 10−2 | 81.20 | 615.40 | Plb1; Mbp | |
| Sudden Infant Death Syndrome SIDS Susceptibility Pathways WP706 | 5/157 | 1.16 × 10−3 | 2.06 × 10−2 | 6.84 | 46.24 | Ywhab; Plp1; Aqp4; Aldoa; Sptbn1 | |
| Astrocyte 3 | Fragile X Syndrome WP4549 | 10/122 | 5.35 × 10−10 | 7.70 × 10−8 | 19.63 | 419.10 | Cyfip2; Gria1; Camk2b; Ppp3ca; Grin2a; Map1b; Camk2a; Agap2; Dnm1; Grin1 |
| Disruption Of Postsynaptic Signaling By CNV WP4875 | 6/33 | 1.33 × 10−8 | 9.60 × 10−7 | 46.98 | 851.93 | Camk2b; Ryr2; Grin2a; Nrxn1; Camk2a; Grin1 | |
| Synaptic Vesicle Pathway WP2267 | 6/51 | 2.02 × 10−7 | 9.67 × 10−6 | 28.16 | 434.20 | Snap25; Slc1a3; Atp1a2; Slc17a7; Cplx2; Dnm1 | |
| Neuroinflammation And Glutamatergic Signaling WP5083 | 8/140 | 5.10 × 10−7 | 1.84 × 10−5 | 13.02 | 188.67 | Gria1; Camk2b; Grin2a; Camk2a; Slc1a2; Slc1a3; Slc17a7; Grin1 | |
| NRXN1 Deletion Syndrome WP5398 | 4/17 | 1.33 × 10−6 | 3.84 × 10−5 | 63.74 | 862.34 | Gria1; Grin2a; Nrxn1; Grin1 | |
| Cell Lineage Map For Neuronal Differentiation WP5417 | 7/132 | 4.47 × 10−6 | 1.07 × 10−4 | 11.91 | 146.68 | Snap25; Slc1a2; Rimbp2; Slc1a3; Slc17a7; Bsn; Grin1 | |
| Phosphodiesterases In Neuronal Function WP4222 | 5/54 | 7.36 × 10−6 | 1.51 × 10−4 | 21.32 | 252.03 | Gria1; Pde11A; Grin2A; Camk2a; Grin1 | |
| Common Pathways Underlying Drug Addiction WP2636 | 4/41 | 5.17 × 10−5 | 9.30 × 10−4 | 22.37 | 220.78 | Gria1; Grin2a; Camk2a; Grin1 | |
| NO cGMP PKG Mediated Neuroprotection WP4008 | 4/46 | 8.17 × 10−5 | 1.31 × 10−3 | 19.70 | 185.43 | Camk2b; Grin2A; Camk2a; Grin1 | |
| Hippocampal Synaptogenesis And Neurogenesis WP5231 | 3/24 | 2.27 × 10−4 | 3.28 × 10−3 | 29.28 | 245.59 | CAamk2b; Nrxn1; Camk2a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
DeSana, A.J.; Alfawares, Y.; Khatri, R.; Hopkins, T.M.; Best, F.V.; McGuire, J.L.; Ngwenya, L.B. Altered Neuroinflammatory Transcriptomic Profile in the Hippocampal Dentate Gyrus Three Weeks After Lateral Fluid Percussion Injury in Rats. Int. J. Mol. Sci. 2025, 26, 9140. https://doi.org/10.3390/ijms26189140
DeSana AJ, Alfawares Y, Khatri R, Hopkins TM, Best FV, McGuire JL, Ngwenya LB. Altered Neuroinflammatory Transcriptomic Profile in the Hippocampal Dentate Gyrus Three Weeks After Lateral Fluid Percussion Injury in Rats. International Journal of Molecular Sciences. 2025; 26(18):9140. https://doi.org/10.3390/ijms26189140
Chicago/Turabian StyleDeSana, Anthony J., Yara Alfawares, Roshni Khatri, Tracy M. Hopkins, Faith V. Best, Jennifer L. McGuire, and Laura B. Ngwenya. 2025. "Altered Neuroinflammatory Transcriptomic Profile in the Hippocampal Dentate Gyrus Three Weeks After Lateral Fluid Percussion Injury in Rats" International Journal of Molecular Sciences 26, no. 18: 9140. https://doi.org/10.3390/ijms26189140
APA StyleDeSana, A. J., Alfawares, Y., Khatri, R., Hopkins, T. M., Best, F. V., McGuire, J. L., & Ngwenya, L. B. (2025). Altered Neuroinflammatory Transcriptomic Profile in the Hippocampal Dentate Gyrus Three Weeks After Lateral Fluid Percussion Injury in Rats. International Journal of Molecular Sciences, 26(18), 9140. https://doi.org/10.3390/ijms26189140






