Cellulase Production by Ultraviolet-Derived Mutant Trichoderma sp. Mut-4 Under Submerged Fermentation: Parameter Optimization and Large-Scale Application
Abstract
1. Introduction
2. Results and Discussion
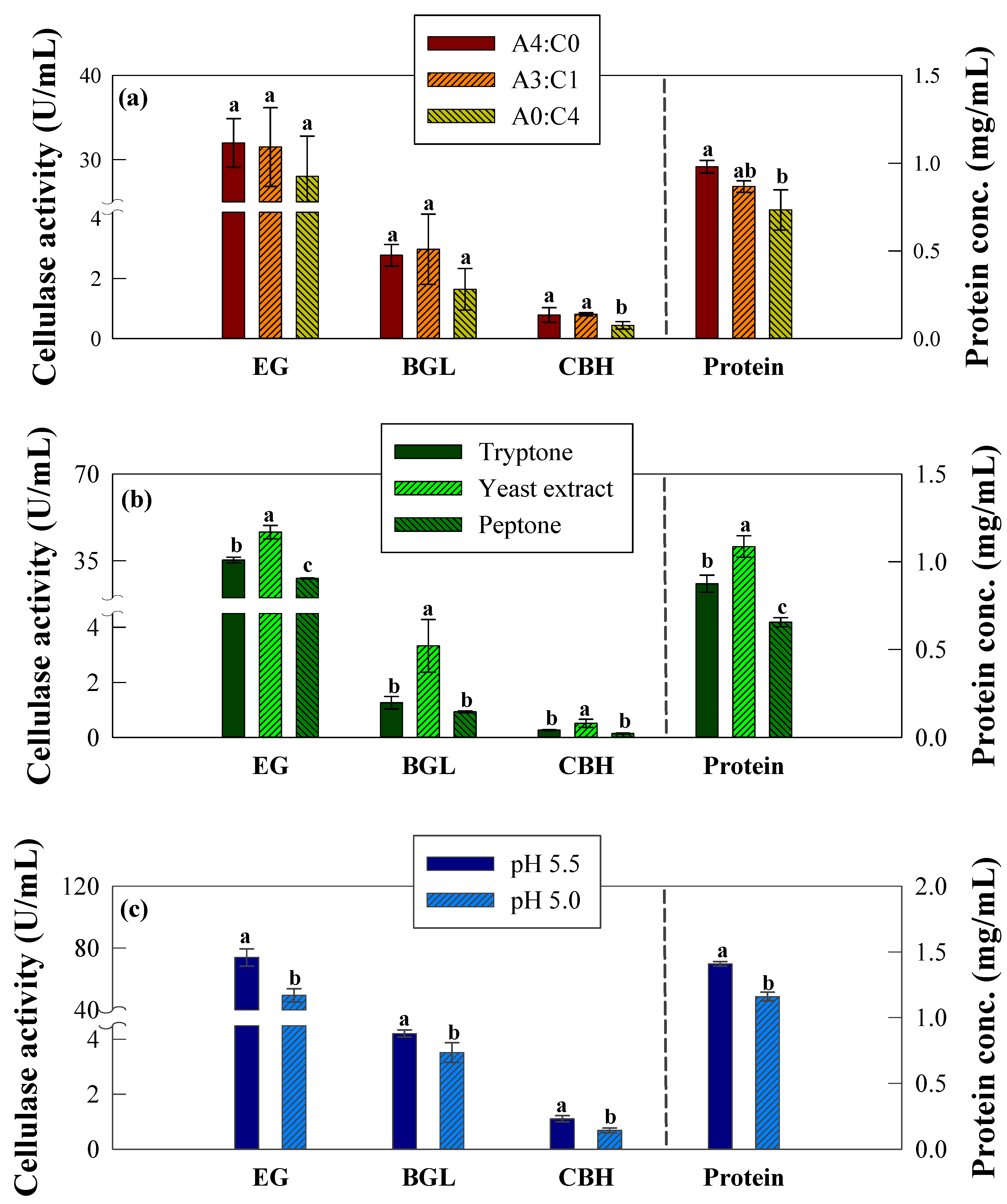
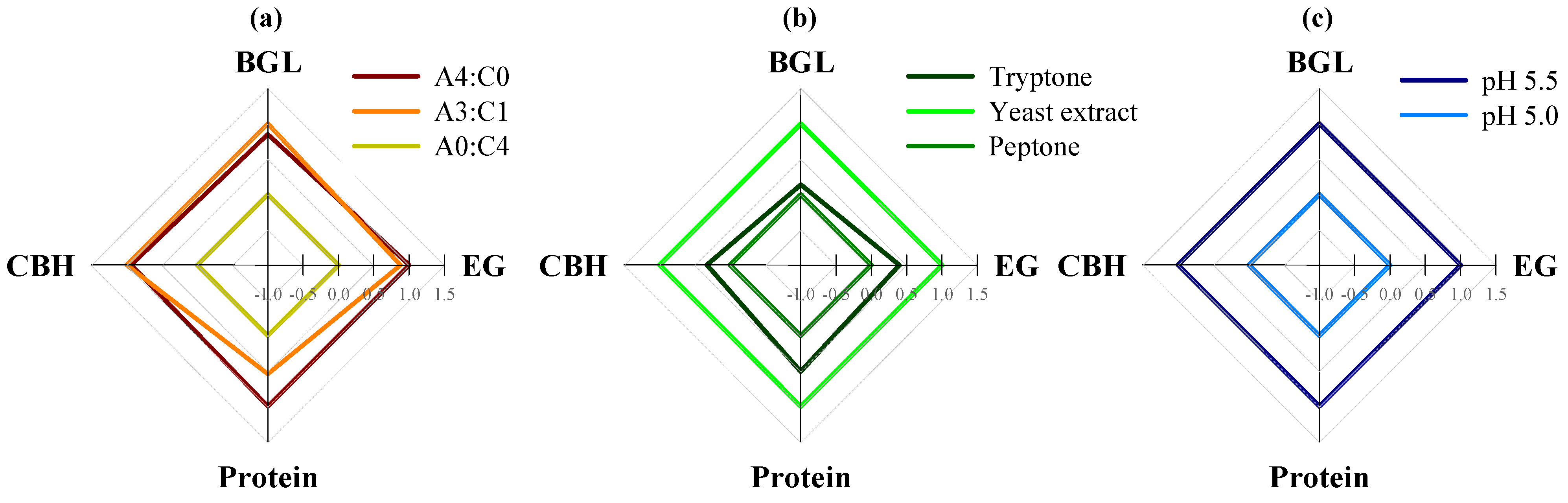
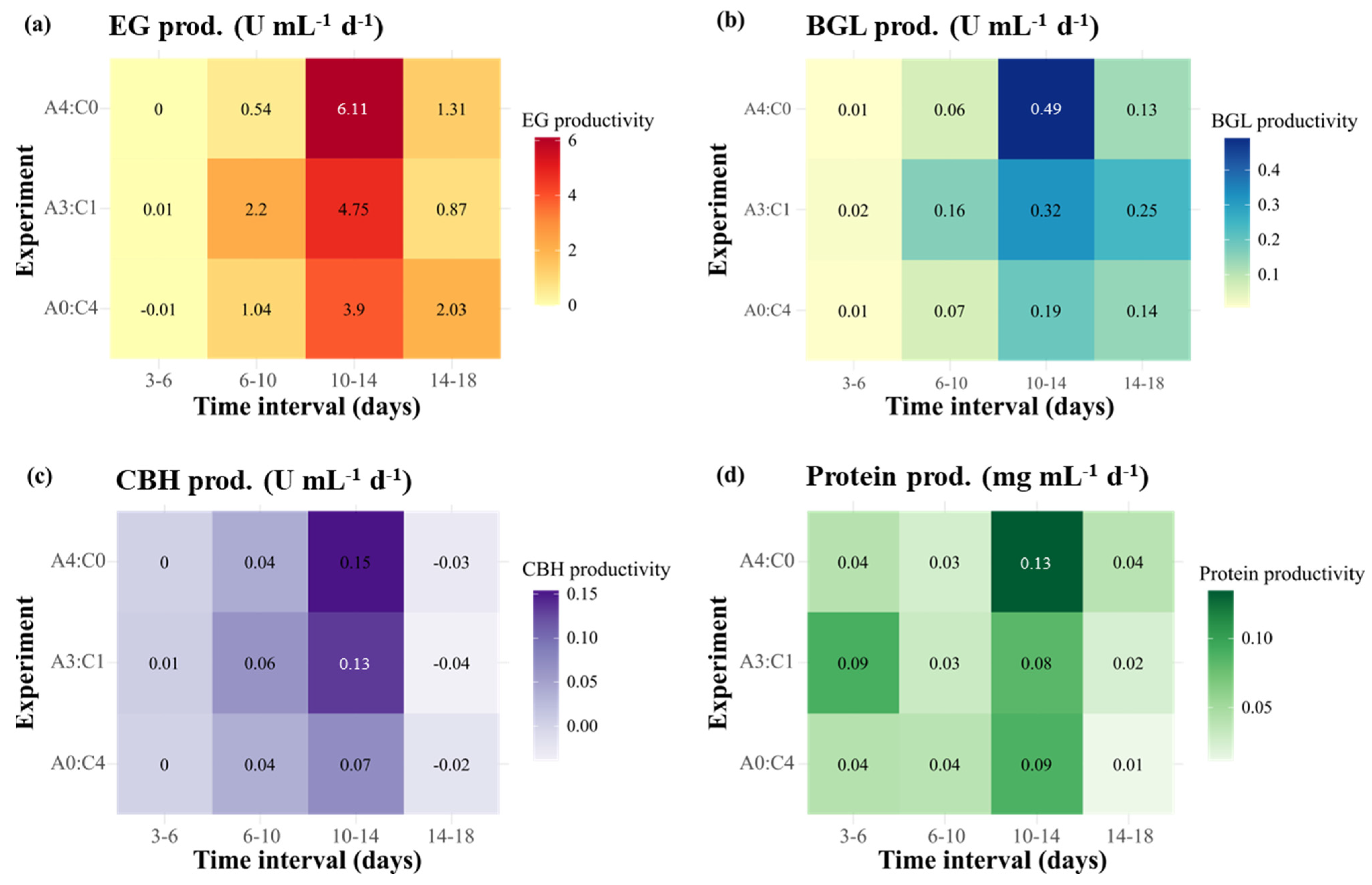
2.1. Effect of the Carbon Sources on Cellulase Activity
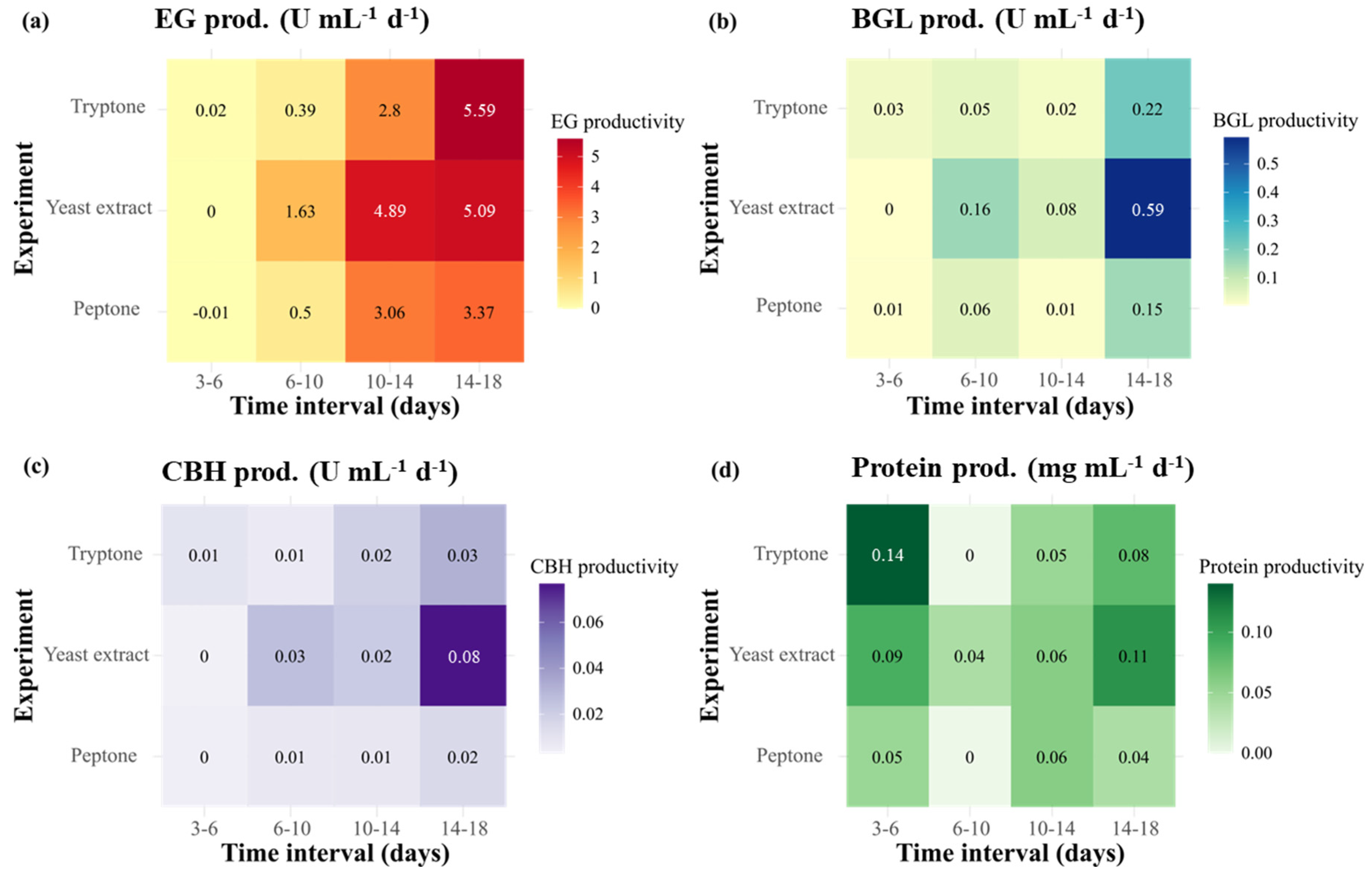
2.2. Effect of Nitrogen Sources on Cellulase Activity
2.3. Effect of Initial pH on Cellulase Activity
2.4. Effect of Optimal Conditions on Cellulase Production in a Bioreactor
3. Materials and Methods
3.1. Microorganism and Inoculum Preparation
3.2. Effect of the Blending Ratio of the Carbon Source on Cellulase Production
3.3. Effect of the Type of Nitrogen Source on Cellulase Production
3.4. Effect of Initial pH on Cellulase Production
3.5. Evaluation of Cellulase Production in SmF Bioreactors Under Various Conditions
3.6. Assay Method
3.6.1. Enzymatic Activity
3.6.2. Protein Concentration
3.6.3. Statistical Analysis
3.7. X-Ray Diffraction (XRD) Analysis and Segal Crystallinity Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Rodionova, M.V.; Bozieva, A.M.; Zharmukhamedov, S.K.; Leong, Y.K.; Lan, J.C.; Veziroglu, A.; Veziroglu, T.N.; Tomo, T.; Chang, J.; Allakhverdiev, S.I. A comprehensive review on lignocellulosic biomass biorefinery for sustainable biofuel production. Int. J. Hydrogen Energy 2022, 47, 1481–1498. [Google Scholar] [CrossRef]
- Kurian, J.K.; Nair, G.R.; Hussain, A.; Raghavan, G.V. Feedstocks, logistics and pre-treatment processes for sustainable lignocellulosic biorefineries: A comprehensive review. Renew. Sustain. Energy Rev. 2013, 25, 205–219. [Google Scholar] [CrossRef]
- Mangal, M.; Rao, C.V.; Banerjee, T. Bioplastic: An eco-friendly alternative to non-biodegradable plastic. Polym. Int. 2023, 72, 984–996. [Google Scholar] [CrossRef]
- Siqueira, J.G.W.; Rodrigues, C.; de Souza Vandenberghe, L.P.; Woiciechowski, A.L.; Soccol, C.R. Current advances in on-site cellulase production and application on lignocellulosic biomass conversion to biofuels: A review. Biomass Bioenergy 2020, 132, 105419. [Google Scholar] [CrossRef]
- Shah, M.; Rajhans, S.; Pandya, H.A.; Mankad, A.U. Bioplastic for future: A review then and now. World J. Adv. Res. Rev. 2021, 9, 56. [Google Scholar] [CrossRef]
- Yu, D.; Guo, J.; Meng, J.; Sun, T. Biofuel production by hydro-thermal liquefaction of municipal solid waste: Process characterization and optimization. Chemosphere 2023, 328, 138606. [Google Scholar] [CrossRef]
- Rodionova, M.V.; Poudyal, R.S.; Tiwari, I.; Voloshin, R.A.; Zharmukhamedov, S.K.; Nam, H.G.; Zayadan, B.K.; Bruce, B.D.; Hou, H.J.; Allakhverdiev, S.I. Biofuel production: Challenges and opportunities. Int. J. Hydrogen Energy 2017, 42, 8450–8461. [Google Scholar] [CrossRef]
- McNamara, J.T.; Morgan, J.L.; Zimmer, J. A molecular description of cellulose biosynthesis. Annu. Rev. Biochem. 2015, 84, 895–921. [Google Scholar] [CrossRef]
- Isikgor, F.H.; Becer, C.R. Lignocellulosic biomass: A sustainable platform for the production of bio-based chemicals and polymers. Polym. Chem. 2015, 6, 4497–4559. [Google Scholar] [CrossRef]
- Brown, J.; Lindstrom, J.K.; Ghosh, A.; Rollag, S.A.; Brown, R.C. Production of sugars from lignocellulosic biomass via biochemical and thermochemical routes. Front. Energy Res. 2024, 12, 1347373. [Google Scholar] [CrossRef]
- Ding, K.; Liu, D.; Chen, X.; Zhang, H.; Shi, S.; Guo, X.; Zhou, L.; Han, L.; Xiao, W. Scalable lignocellulosic biorefineries: Technoeconomic review for efficient fermentable sugars production. Renew. Sustain. Energy Rev. 2024, 202, 114692. [Google Scholar] [CrossRef]
- Saritha, M.; Arora, A. Lata Biological pretreatment of lignocellulosic substrates for enhanced delignification and enzymatic digestibility. Indian J. Microbiol. 2012, 52, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Sant’Ana Júnior, D.B.; Kelbert, M.; Hermes de Araújo, P.H.; de Andrade, C.J. Physical pretreatments of lignocellulosic biomass for fermentable sugar production. Sustain. Chem. 2025, 6, 13. [Google Scholar] [CrossRef]
- Zoghlami, A.; Paës, G. Lignocellulosic biomass: Understanding recalcitrance and predicting hydrolysis. Front. Chem. 2019, 7, 874. [Google Scholar] [CrossRef]
- Sun, W.; Li, X.; Zhao, J.; Qin, Y. Pretreatment strategies to enhance enzymatic hydrolysis and cellulosic ethanol production for biorefinery of corn stover. Int. J. Mol. Sci. 2022, 23, 13163. [Google Scholar] [CrossRef]
- Taherzadeh, M.J.; Karimi, K. Enzymatic-based hydrolysis processes for ethanol from lignocellulosic materials: A review. BioResources 2007, 2, 707–738. [Google Scholar] [CrossRef]
- Wang, H.; Squina, F.; Segato, F.; Mort, A.; Lee, D.; Pappan, K.; Prade, R. High-temperature enzymatic breakdown of cellulose. Appl. Environ. Microbiol. 2011, 77, 5199–5206. [Google Scholar] [CrossRef]
- Zhang, Z.; Xing, J.; Li, X.; Lu, X.; Liu, G.; Qu, Y.; Zhao, J. Review of research progress on the production of cellulase from filamentous fungi. Int. J. Biol. Macromol. 2024, 277, 134539. [Google Scholar] [CrossRef]
- de França Passos, D.; Pereira, N., Jr.; de Castro, A.M. A comparative review of recent advances in cellulases production by Aspergillus, Penicillium and Trichoderma strains and their use for lignocellulose deconstruction. Curr. Opin. Green Sustain. Chem. 2018, 14, 60–66. [Google Scholar] [CrossRef]
- Bhati, N.; Shreya; Sharma, A.K. Cost-effective cellulase production, improvement strategies, and future challenges. J. Food Process Eng. 2021, 44, e13623. [Google Scholar] [CrossRef]
- Bhat, M. Cellulases and related enzymes in biotechnology. Biotechnol. Adv. 2000, 18, 355–383. [Google Scholar] [CrossRef]
- Myeong, S.; Lee, Y.; Yun, J. Optimization and bioreactor scale-up of cellulase production in Trichoderma sp. KMF006 for higher yield and performance. Int. J. Mol. Sci. 2025, 26, 3731. [Google Scholar] [CrossRef]
- Myeong, S.; Yun, J. Culture of Trichoderma sp. with biochar to produce high-activity cellulase in a laboratory. BioResources 2024, 19, 2029–2044. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Kumar, B.; Agrawal, K.; Verma, P. Current perspective on production and applications of microbial cellulases: A review. Bioresour. Bioprocess. 2021, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Singhania, R.R.; Sukumaran, R.K.; Patel, A.K.; Larroche, C.; Pandey, A. Advancement and comparative profiles in the production technologies using solid-state and submerged fermentation for microbial cellulases. Enzym. Microb. Technol. 2010, 46, 541–549. [Google Scholar] [CrossRef]
- Park, S.; Yoon, S.; Kim, Y.; Kim, Y.; Yun, J. Optimization of culture conditions for cellulase production in Acanthophysium sp. KMF001 using response surface methodology. BioResources 2021, 16, 3520–3542. [Google Scholar] [CrossRef]
- Ye, M.; Sun, L.; Yang, R.; Wang, Z.; Qi, K. The optimization of fermentation conditions for producing cellulase of Bacillus amyloliquefaciens and its application to goose feed. R. Soc. Open Sci. 2017, 4, 171012. [Google Scholar] [CrossRef]
- Gutiérrez-Correa, M.; Villena, G.K. Surface adhesion fermentation: A new fermentation category. Rev. Peru. Biol. 2003, 10, 113–124. [Google Scholar] [CrossRef]
- Lehmann, J.; Joseph, S. Biochar for Environmental Management: Science, Technology and Implementation; Taylor & Francis: Abingdon, UK, 2024. [Google Scholar]
- Cherry, J.R.; Fidantsef, A.L. Directed evolution of industrial enzymes: An update. Curr. Opin. Biotechnol. 2003, 14, 438–443. [Google Scholar] [CrossRef]
- Kim, S.; Ha, I.; Lee, Y.; Lee, J.; Yun, J. Development of a high-performance Trichoderma mutant for enhanced cellulase production through UV-induced random mutagenesis. J. Fungi 2025, 11, 439. [Google Scholar] [CrossRef]
- Payne, C.M.; Knott, B.C.; Mayes, H.B.; Hansson, H.; Himmel, M.E.; Sandgren, M.; Stahlberg, J.; Beckham, G.T. Fungal cellulases. Chem. Rev. 2015, 115, 1308–1448. [Google Scholar] [CrossRef]
- Carle-Urioste, J.C.; Escobar-Vera, J.; El-Gogary, S.; Henrique-Silva, F.; Torigoi, E.; Crivellaro, O.; Herrera-Estrella, A.; El-Dorry, H. Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J. Biol. Chem. 1997, 272, 10169–10174. [Google Scholar] [CrossRef] [PubMed]
- Ilmen, M.; Saloheimo, A.; Onnela, M.; Penttilä, M.E. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 1997, 63, 1298–1306. [Google Scholar] [CrossRef]
- Seiboth, B.; Hakola, S.; Mach, R.L.; Suominen, P.L.; Kubicek, C.P. Role of four major cellulases in triggering of cellulase gene expression by cellulose in Trichoderma reesei. J. Bacteriol. 1997, 179, 5318–5320. [Google Scholar] [CrossRef]
- Teeri, T.T. Crystalline cellulose degradation: New insight into the function of cellobiohydrolases. Trends Biotechnol. 1997, 15, 160–167. [Google Scholar] [CrossRef]
- Kubicek, C.P. Systems biological approaches towards understanding cellulase production by Trichoderma reesei. J. Biotechnol. 2013, 163, 133–142. [Google Scholar] [CrossRef]
- Aro, N.; Pakula, T.; Penttilä, M. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol. Rev. 2005, 29, 719–739. [Google Scholar] [CrossRef]
- Mandels, M.; Reese, E.T. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J. Bacteriol. 1957, 73, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Shankar, T.; Isaiarasu, L. Cellulase production by Bacillus pumilus EWBCM1 under varying cultural conditions. Middle East J. Sci. Res. 2011, 8, 40–45. [Google Scholar]
- Gautam, S.P.; Bundela, P.S.; Pandey, A.K.; Awasthi, M.K.; Sarsaiya, S. Optimization of the medium for the production of cellulase by the Trichoderma viride using submerged fermentation. Int. J. Environ. Sci. 2010, 1, 656–665. [Google Scholar]
- Maeda, R.N.; da Silva, M.M.P.; Santa Anna, L.M.M.; Pereira, N. Nitrogen source optimization for cellulase production by Penicillium funiculosum, using a sequential experimental design methodology and the desirability function. Appl. Biochem. Biotechnol. 2010, 161, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Nanjundaswamy, A.; Okeke, B.C. Comprehensive optimization of culture conditions for production of biomass-hydrolyzing enzymes of Trichoderma SG2 in submerged and solid-state fermentation. Appl. Biochem. Biotechnol. 2020, 191, 444–462. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Amano, Y.; Nozaki, K. Improvements in glucose sensitivity and stability of Trichoderma reesei β-glucosidase using site-directed mutagenesis. PLoS ONE 2016, 11, e0147301. [Google Scholar] [CrossRef]
- Qian, Y.; Zhong, L.; Sun, Y.; Sun, N.; Zhang, L.; Liu, W.; Qu, Y.; Zhong, Y. Enhancement of cellulase production in Trichoderma reesei via disruption of multiple protease genes identified by comparative secretomics. Front. Microbiol. 2019, 10, 2784. [Google Scholar] [CrossRef]
- Dembek, M.; Stabler, R.A.; Witney, A.A.; Wren, B.W.; Fairweather, N.F. Transcriptional analysis of temporal gene expression in germinating Clostridium difficile 630 endospores. PLoS ONE 2013, 8, e64011. [Google Scholar] [CrossRef]
- Goswami, G.; Panda, D.; Samanta, R.; Boro, R.C.; Modi, M.K.; Bujarbaruah, K.M.; Barooah, M. Bacillus megaterium adapts to acid stress condition through a network of genes: Insight from a genome-wide transcriptome analysis. Sci. Rep. 2018, 8, 16105. [Google Scholar] [CrossRef]
- Martins, M.P.; Martinez-Rossi, N.M.; Sanches, P.R.; Gomes, E.V.; Bertolini, M.C.; Pedersoli, W.R.; Silva, R.N.; Rossi, A. The pH signaling transcription factor PAC-3 regulates metabolic and developmental processes in pathogenic fungi. Front. Microbiol. 2019, 10, 2076. [Google Scholar] [CrossRef]
- Peñalva, M.A.; Tilburn, J.; Bignell, E.; Arst, H.N. Ambient pH gene regulation in fungi: Making connections. Trends Microbiol. 2008, 16, 291–300. [Google Scholar] [CrossRef]
- Goswami, G.; Hazarika, D.J.; Chowdhury, N.; Bora, S.S.; Sarmah, U.; Naorem, R.S. Proline confers acid stress tolerance to Bacillus megaterium G18. Sci. Rep. 2022, 12, 8875. [Google Scholar] [CrossRef]
- Peñalva, M.A.; Arst, H.N., Jr. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 426–446. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, M.; Hong, D.; Fang, Z.; Xiao, Y.; Fang, W.; Zhang, X. Improving the cellobiose hydrolysis activity of glucose-stimulating β-glucosidase Bgl2A. Enzym. Microb. Technol. 2023, 169, 110289. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Li, C.; Ma, L.; Zhang, D.; Chen, S. Effect of highly branched hyphal morphology on the enhanced production of cellulase in Trichoderma reesei DES-15. 3 Biotech 2016, 6, 214. [Google Scholar] [CrossRef] [PubMed]
- Reis, C.E.R.; Junior, N.L.; Bento, H.B.; de Carvalho, A.K.F.; de Souza Vandenberghe, L.P.; Soccol, C.R.; Aminabhavi, T.M.; Chandel, A.K. Process strategies to reduce cellulase enzyme loading for renewable sugar production in biorefineries. Chem. Eng. J. 2023, 451, 138690. [Google Scholar] [CrossRef]
- de Albuquerque de Carvalho, M.L.; Carvalho, D.F.; de Barros Gomes, E.; Nobuyuki Maeda, R.; Melo Santa Anna, L.M.; Castro, A.M.D.; Pereira, N., Jr. Optimisation of cellulase production by Penicillium funiculosum in a stirred tank bioreactor using multivariate response surface analysis. Enzym. Res. 2014, 2014, 703291. [Google Scholar] [CrossRef] [PubMed]
- Ellilä, S.; Fonseca, L.; Uchima, C.; Cota, J.; Goldman, G.H.; Saloheimo, M.; Sacon, V.; Siika-Aho, M. Development of a low-cost cellulase production process using Trichoderma reesei for Brazilian biorefineries. Biotechnol. Biofuels 2017, 10, 30. [Google Scholar] [CrossRef]
- Gnansounou, E.; Dauriat, A. Techno-economic analysis of lignocellulosic ethanol: A review. Bioresour. Technol. 2010, 101, 4980–4991. [Google Scholar] [CrossRef]
- Nelson, N. A photometric adaptation of the Somogyi method for the determination of glucose. J. Biol. Chem. 1944, 153, 375–380. [Google Scholar] [CrossRef]
- Joo, A.; Jeya, M.; Lee, K.; Sim, W.; Kim, J.; Kim, I.; Kim, Y.; Oh, D.; Gunasekaran, P.; Lee, J. Purification and characterization of a β-1, 4-glucosidase from a newly isolated strain of Fomitopsis pinicola. Appl. Microbiol. Biotechnol. 2009, 83, 285–294. [Google Scholar] [CrossRef]
- Agarwal, U.P.; Ralph, S.A.; Reiner, R.S.; Baez, C. New cellulose crystallinity estimation method that differentiates between organized and crystalline phases. Carbohydr. Polym. 2018, 190, 262–270. [Google Scholar] [CrossRef]


| Scale | Parameter | Experiment | Experimental Condition | |
|---|---|---|---|---|
| Experimental Factor | Fixed Factor | |||
| Flask | Blending ratio of the C source | A4:C0 | Avicel:Cellulose = 4:0 | The N source is yeast extract and the initial pH is 5.0 |
| A3:C1 | Avicel:Cellulose = 3:1 | |||
| A0:C4 | Avicel:Cellulose = 0:4 | |||
| Type of N source | Tryptone | The N source is tryptone | The C source is A3:C1 and the initial pH is 5.0 | |
| Yeast extract | The N source is the yeast extract | |||
| Peptone | The N source is peptone | |||
| Initial pH | pH 5.5 | Initial pH is 5.5 | The C source is A3:C1 and the N source is the yeast extract | |
| pH 5.0 | Initial pH is 5.0 | |||
| pH 4.5 | Initial pH is 4.5 | |||
| Reactor | Initial pH | OPT. | Optimal condition (pH 5.5) | The C source is A3:C1 and the N source is the yeast extract |
| ORI. | Original condition (pH 5.0) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, I.; Kim, S.; Lee, Y.-Y.; Lee, J.; Yun, J. Cellulase Production by Ultraviolet-Derived Mutant Trichoderma sp. Mut-4 Under Submerged Fermentation: Parameter Optimization and Large-Scale Application. Int. J. Mol. Sci. 2025, 26, 8000. https://doi.org/10.3390/ijms26168000
Ha I, Kim S, Lee Y-Y, Lee J, Yun J. Cellulase Production by Ultraviolet-Derived Mutant Trichoderma sp. Mut-4 Under Submerged Fermentation: Parameter Optimization and Large-Scale Application. International Journal of Molecular Sciences. 2025; 26(16):8000. https://doi.org/10.3390/ijms26168000
Chicago/Turabian StyleHa, Iksu, Seungjun Kim, Yun-Yeong Lee, Junseo Lee, and Jeonghee Yun. 2025. "Cellulase Production by Ultraviolet-Derived Mutant Trichoderma sp. Mut-4 Under Submerged Fermentation: Parameter Optimization and Large-Scale Application" International Journal of Molecular Sciences 26, no. 16: 8000. https://doi.org/10.3390/ijms26168000
APA StyleHa, I., Kim, S., Lee, Y.-Y., Lee, J., & Yun, J. (2025). Cellulase Production by Ultraviolet-Derived Mutant Trichoderma sp. Mut-4 Under Submerged Fermentation: Parameter Optimization and Large-Scale Application. International Journal of Molecular Sciences, 26(16), 8000. https://doi.org/10.3390/ijms26168000





