Rest Induces a Distinct Transcriptional Program in the Nervous System of the Exercised L. stagnalis
Abstract
1. Introduction
2. Results
2.1. Transcriptome Assembly and Identification of Proteins Homologous to the Coding Sequences of L. stagnalis
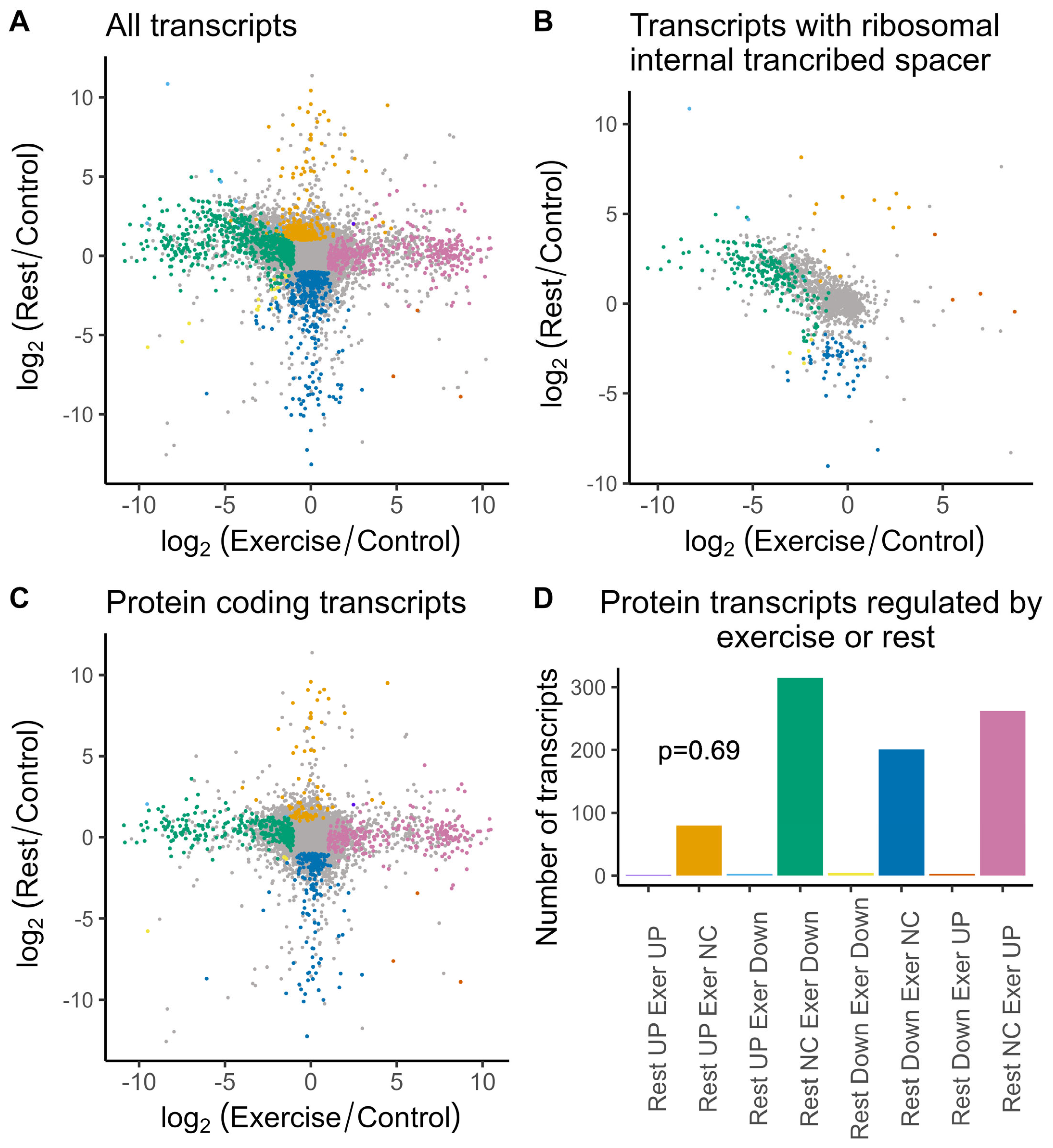
2.2. Differential Gene Expression
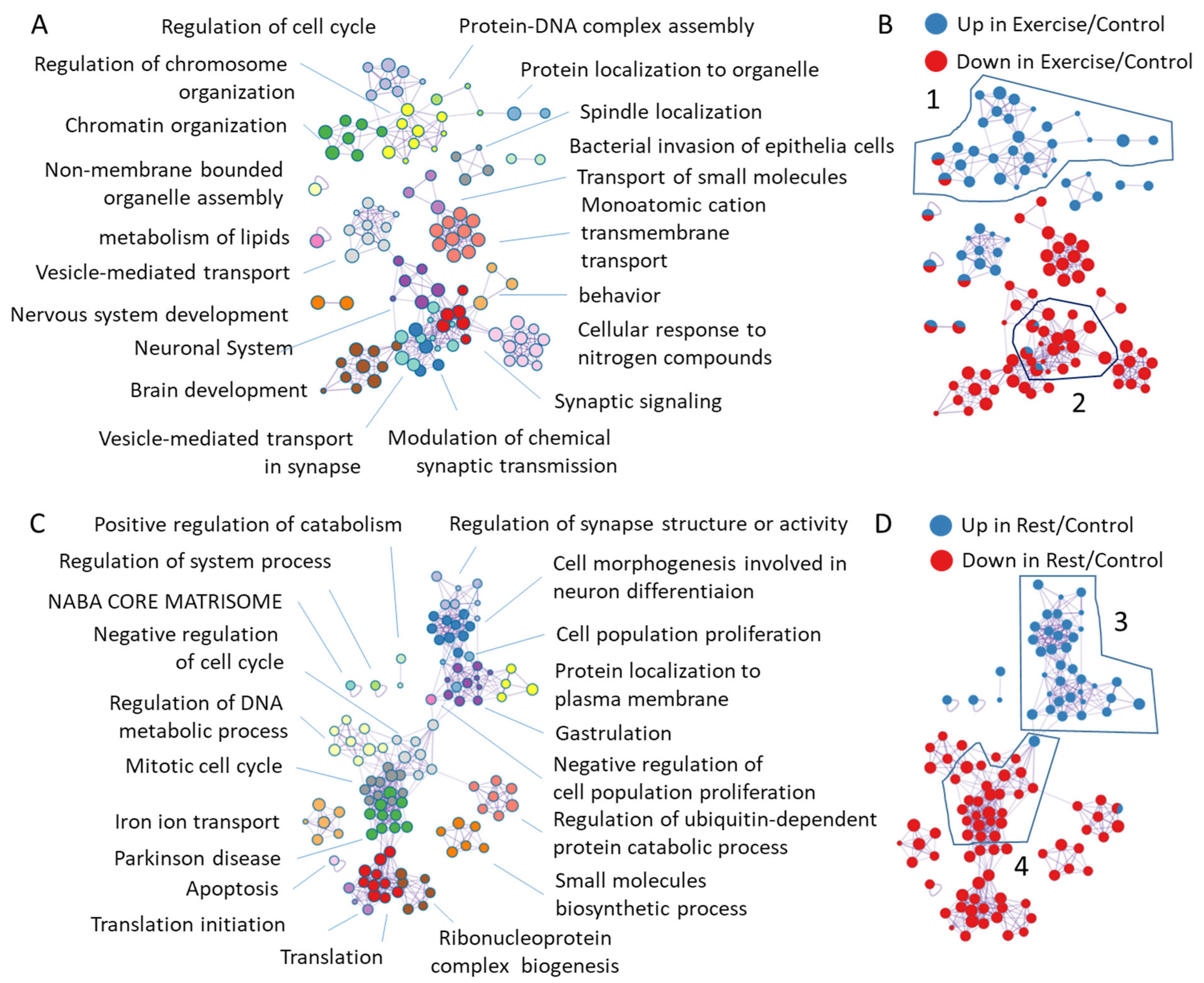
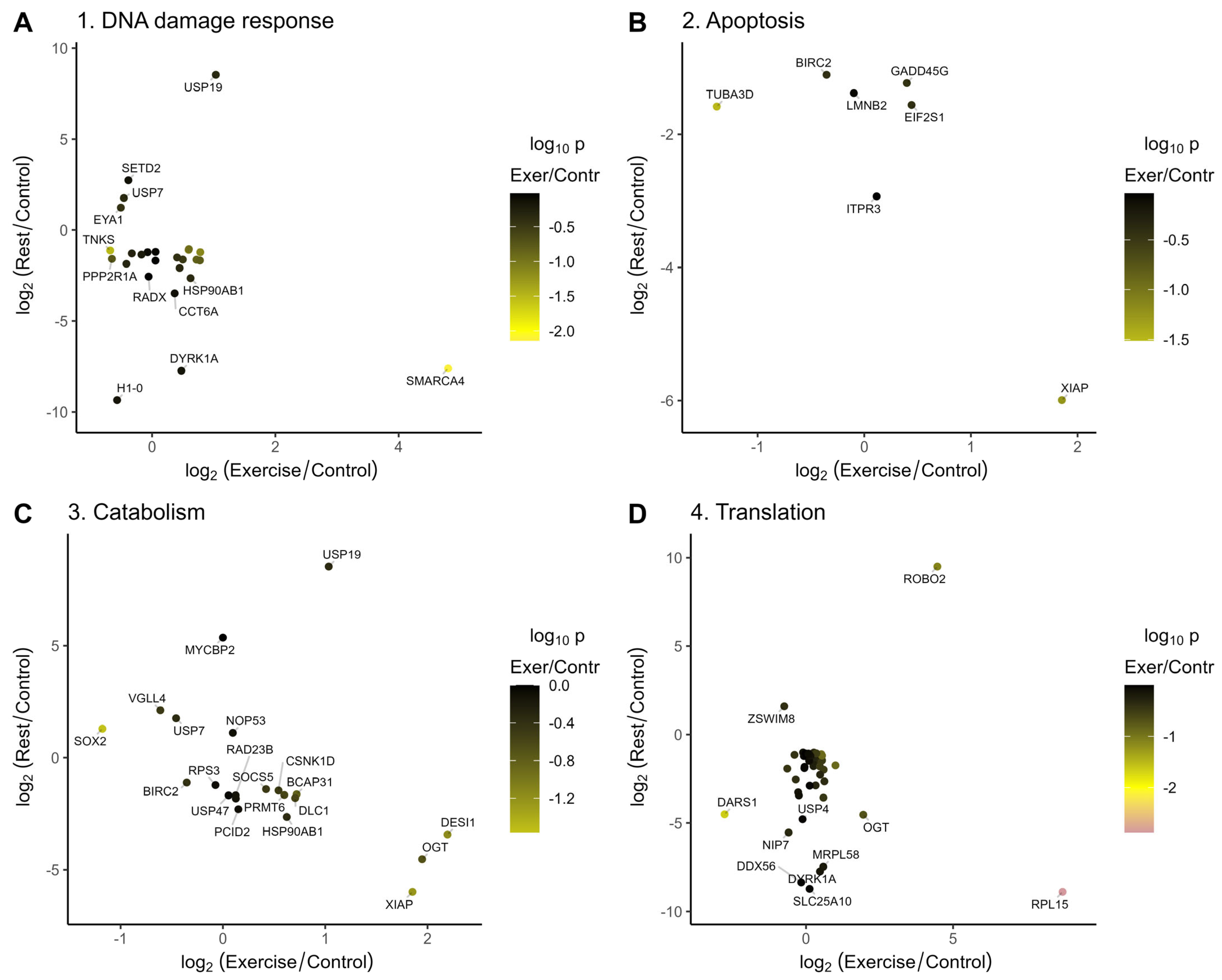
2.3. Annotation of Genes That Change Their Expression After Exercise and Rest
2.3.1. Annotation of Genes Induced or Repressed upon Exercise
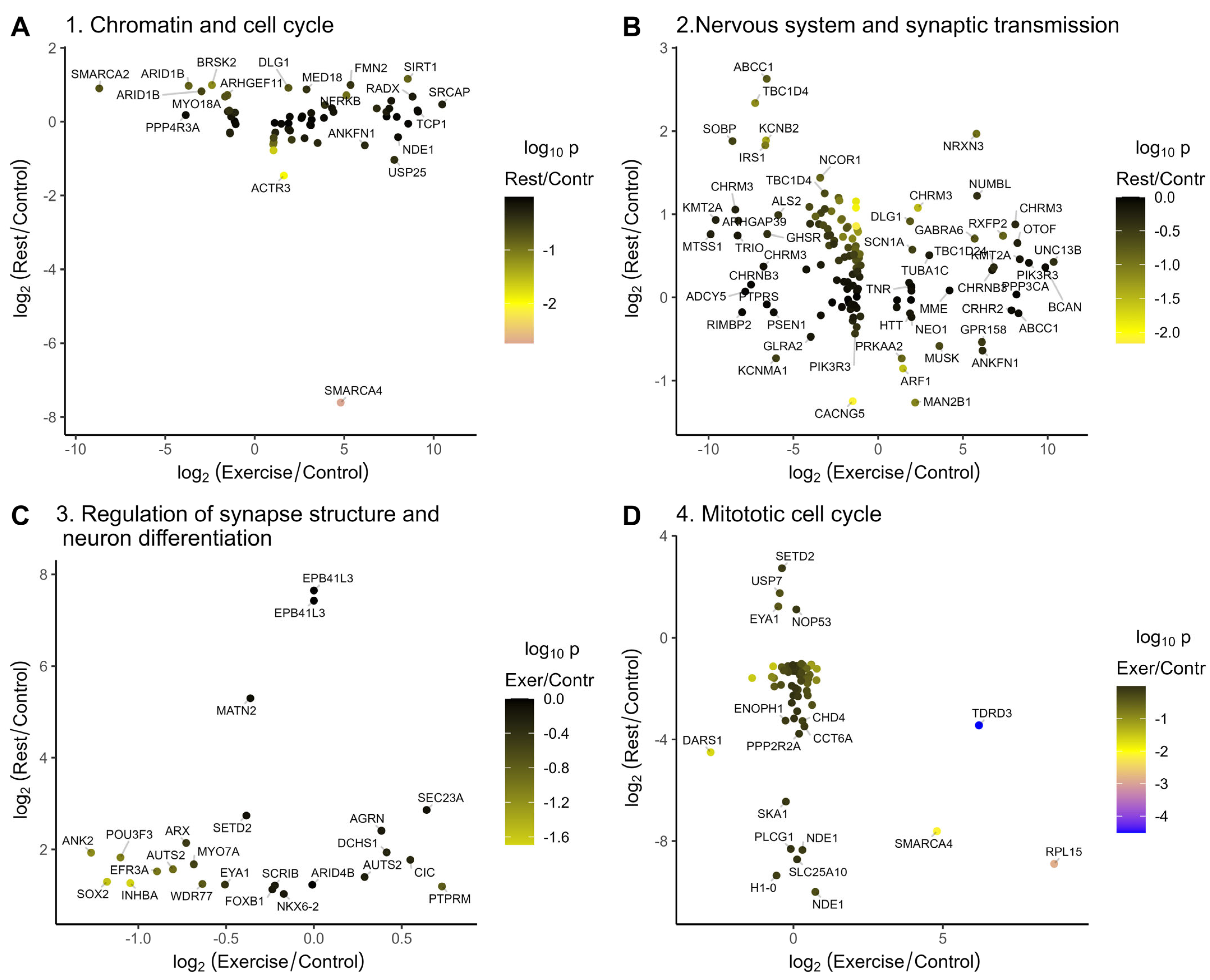
2.3.2. Annotation of Genes Induced or Repressed upon Rest
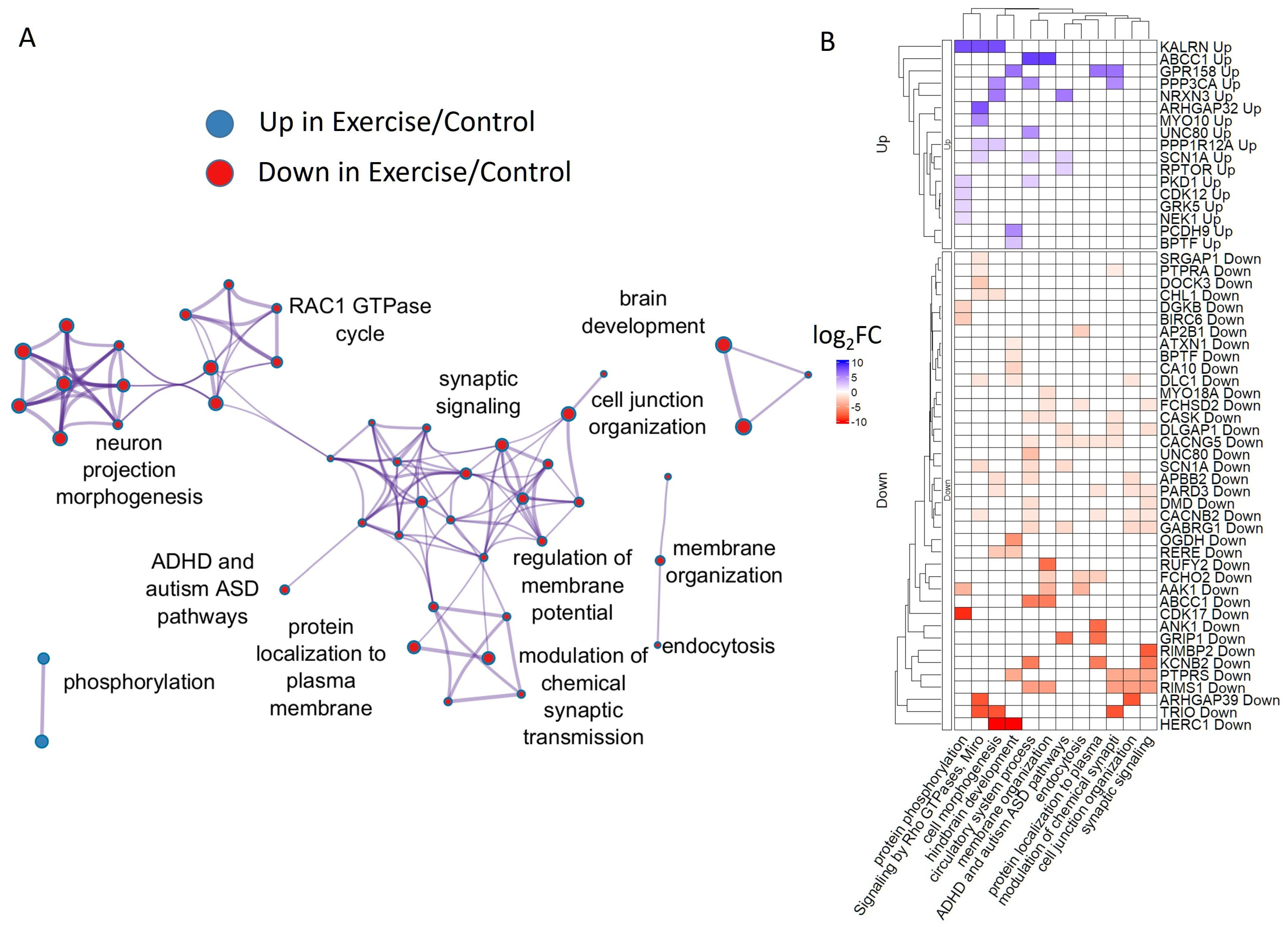
2.4. Genes That Are Similarly Regulated by Exercise in Snails and Mice Revealed Conserved Functional Annotations
| RefSeq Accession Mber | SYMBOL | log2FC Mouse | padj Mouse | log2FC Snail | FDR Snail | DEG Mouse | DEG Snail |
|---|---|---|---|---|---|---|---|
| NP_032939.1 | Ppp3ca | −90.6 | 3.65 × 10−4 | 8.2 | 5.85 × 10−6 | Down | UP Exer |
| NP_064450.3 | Fmn2 | −5.9 | 0.0126 | 3.9 | 0.000507 | Down | UP Exer |
| NP_032939.1 | Ppp3ca | −90.6 | 3.65 × 10−4 | 2.0 | 0.002533 | Down | UP Exer |
| NP_001078824.1 | Arid1b | −0.2 | 0.04065 | −3.7 | 0.002018 | Down | Down Exer |
| NP_055726.4 | Aak1 | −7.0 | 1.59 × 10−8 | −3.8 | 2.59 × 10−5 | Down | Down Exer |
| NP_055726.4 | Aak1 | −7.5 | 0.004973 | −3.8 | 2.59 × 10−5 | Down | Down Exer |
| NP_084542.2 | Inpp4a | −4.6 | 6.46 × 10−4 | −5.3 | 4.17 × 10−7 | Down | Down Exer |
| NP_001366215.1 | Zfp423 | −8.0 | 4.19 × 10−11 | −9.8 | 0.001463 | Down | Down Exer |
| NP_001078824.1 | Arid1b | 4.6 | 2.24 × 10−5 | −3.7 | 0.002018 | UP | Down Exer |
| NP_663592.3 | Herc1 | 13.7 | 1.62 × 10−4 | −10.9 | 3.54 × 10−15 | UP | Down Exer |
| NP_899233.1 | Asic2 | −62.4 | 5.07 × 10−4 | 9.1 | 0.000237 | Down | UP Rest |
| NP_036439.2 | Epb41l3 | −19.9 | 2.42 × 10−10 | 7.7 | 0.014063 | Down | UP Rest |
| NP_036439.2 | Epb41l3 | −19.9 | 2.42 × 10−10 | 7.4 | 6.6 × 10−5 | Down | UP Rest |
| NP_055847.1 | Pds5b | −6.7 | 0.026451 | 2.1 | 0.001158 | Down | UP Rest |
| NP_001103785.1 | Kif1a | −4.5 | 1.65 × 10−6 | 1.5 | 0.028579 | Down | UP Rest |
| NP_001161760.2 | Arhgap39 | 4.6 | 5.13 × 10−4 | 7.6 | 0.037305 | UP | UP Rest |
| NP_055952.2 | Efr3a | 16.1 | 4.22 × 10−6 | 1.5 | 0.01432 | UP | UP Rest |
| NP_058616.1 | Atp6v0a1 | −10.9 | 0.017536 | −1.4 | 0.040801 | Down | Down Rest |
| NP_085098.1 | Nbea | −36.4 | 9.15 × 10−5 | −10.1 | 8.51 × 10−7 | Down | Down Rest |
| NP_004761.2 | Kcnb2 | 60.9 | 0.001578 | −9.1 | 0.000163 | UP | Down Rest |
3. Discussion
3.1. Exercise and Rest After Exercise Cause Remarkably Different Gene Expression Programs
3.2. Down- and Up-Regulation of Ribosomal Genes by Exercise and Rest: Unexpected Findings of Differential Analysis
3.3. Rest After Exercise Is Important for Neurodevelopmental Gene Activation Across Species
4. Methods
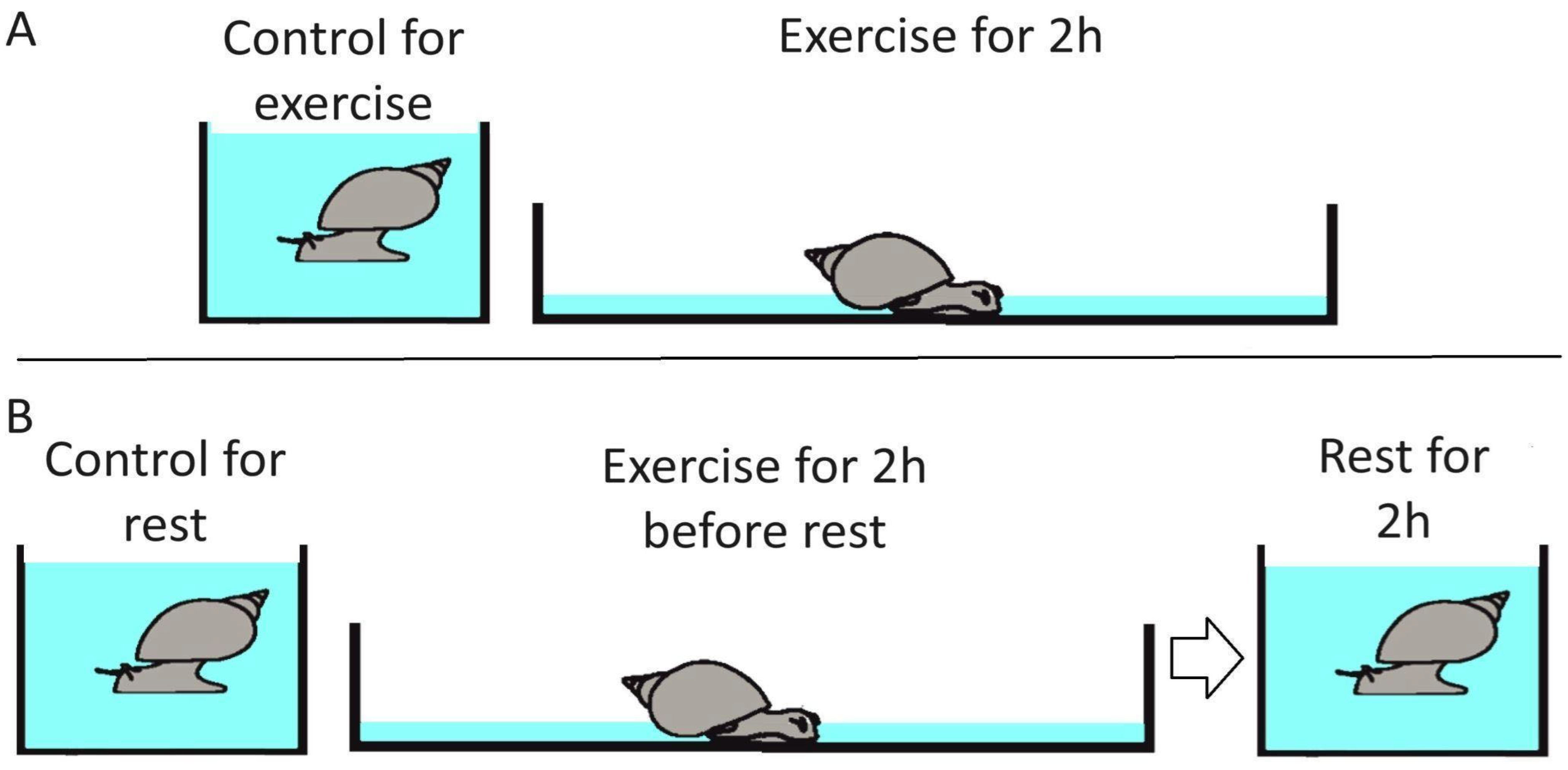
4.1. L. stagnalis Rest and Exercise Trials
4.2. RNA Extraction, Library Preparation, and Sequencing
4.3. RT-PCR
| TRINITY_DN87174_c0_g1_i3: | |
| CCATCATTCCATGCACAATC, | GGAGTTTGACTGGGGTGGTA; |
| TRINITY_DN1288_c1_g1_i2: | |
| CAGTGAGCTGAACCAGGACA, | CACCACTTTTTGGCTGGATT: |
| TRINITY_DN55636_c0_g1_i5: | |
| TGATAGCTCCCCCTCGAATA, | CGAGATTCCCACTGTCCCTA. |
4.4. Transcriptome Assembly and Differential Gene Expression
4.4.1. Transcriptome Assembly
4.4.2. Annotations of Transcripts by Known Homologous Sequences
4.4.3. Quantification of Differential Transcript Expression
4.4.4. Gene Ontology and Pathway Annotations of Differentially Expressed Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- da Costa Daniele, T.M.; de Bruin, P.F.C.; de Matos, R.S.; de Bruin, G.S.; Maia Chaves, C.; de Bruin, V.M.S. Exercise effects on brain and behavior in healthy mice, Alzheimer’s disease and Parkinson’s disease model-A systematic review and meta-analysis. Behav. Brain Res. 2020, 383, 112488. [Google Scholar] [CrossRef] [PubMed]
- Dyakonova, V.; Mezheritskiy, M.; Boguslavsky, D.; Dyakonova, T.; Chistopolsky, I.; Ito, E.; Zakharov, I. Exercise and the brain: Lessons from invertebrate studies. Front. Behav. Neurosci. 2022, 16, 928093. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, S.; Hommel, B.; Kibele, A.; Colzato, L.S. Neuromodulation of Aerobic Exercise-A Review. Front. Psychol. 2015, 6, 1890. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lagisz, M.; Foo, Y.Z.; Noble, D.W.A.; Anwer, H.; Nakagawa, S. Beneficial intergenerational effects of exercise on brain and cognition: A multilevel meta-analysis of mean and variance. Biol. Rev. Camb. Philos. Soc. 2021, 96, 1504–1527. [Google Scholar] [CrossRef] [PubMed]
- Mezheritskiy, M.I.; Dyakonova, V.E. Direct and inherited epigenetic changes in the nervous system caused by intensive locomotion: Possible adaptive significance. Russ. J. Dev. Biol. 2022, 53, 295–308. [Google Scholar] [CrossRef]
- Laranjeiro, R.; Harinath, G.; Hewitt, J.E.; Hartman, J.H.; Royal, M.A.; Meyer, J.N.; Vanapalli, S.A.; Driscoll, M. Swim exercise in Caenorhabditis elegans extends neuromuscular and gut healthspan, enhances learning ability, and protects against neurodegeneration. Proc. Natl. Acad. Sci. USA 2019, 116, 23829–23839. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, L.P.; Riddle, N.C. New opportunities: Drosophila as a model system for exercise research. J. Appl. Physiol. 2019, 127, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Korshunova, T.A.; Vorontsov, D.D.; Dyakonova, V.E. Previous motor activity affects the transition from uncertainty to decision making in snails. J. Exp. Biol. 2016, 219, 3635–3641. [Google Scholar] [CrossRef] [PubMed]
- Korneev, S.A.; Vavoulis, D.V.; Naskar, S.; Dyakonova, V.E.; Kemenes, I.; Kemenes, G. A CREB2-targeting microRNA is required for long-term memory after single-trial learning. Sci. Rep. 2018, 8, 3950. [Google Scholar] [CrossRef] [PubMed]
- Aonuma, H.; Mezheritskiy, M.; Boldyshev, B.; Totani, Y.; Vorontsov, D.; Zakharov, I.; Ito, E.; Dyakonova, V. The Role of Serotonin in the Influence of Intense Locomotion on the Behavior Under Uncertainty in the Mollusk Lymnaea stagnalis. Front. Physiol. 2020, 11, 221. [Google Scholar] [CrossRef] [PubMed]
- Dyakonova, T.L.; Sultanakhmetov, G.S.; Mezheritskiy, M.I.; Sakharov, D.A.; Dyakonova, V.E. Storage and erasure of behavioural experiences at the single neuron level. Sci. Rep. 2019, 9, 14733. [Google Scholar] [CrossRef] [PubMed]
- Rosato, M.; Hoelscher, B.; Lin, Z.; Agwu, C.; Xu, F. Transcriptome analysis provides genome annotation and expression profiles in the central nervous system of Lymnaea stagnalis at different ages. BMC Genom. 2021, 22, 637. [Google Scholar] [CrossRef] [PubMed]
- Slomovic, S.; Laufer, D.; Geiger, D.; Schuster, G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006, 34, 2966–2975. [Google Scholar] [CrossRef] [PubMed]
- An, W.; Yan, Y.; Ye, K. High resolution landscape of ribosomal RNA processing and surveillance. Nucleic Acids Res. 2024, 52, 10630–10644. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Huang, S.-Y.N.; Yang, X.; Saha, L.K.; Sun, Y.; Khandagale, P.; Jenkins, L.M.; Pommier, Y. The TDRD3-USP9X complex and MIB1 regulate TOP3B homeostasis and prevent deleterious TOP3B cleavage complexes. Nat. Commun. 2023, 14, 7524. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Joo, Y.; Bossi, S.; McDevitt, R.; Xie, A.; Wang, Y.; Xue, Y.; Su, S.; Lee, S.K.; Sah, N.; et al. Tdrd3-null mice show post-transcriptional and behavioral impairments associated with neurogenesis and synaptic plasticity. Res. Sq. 2023. [Google Scholar] [CrossRef] [PubMed]
- Joo, Y.; Xue, Y.; Wang, Y.; McDevitt, R.A.; Sah, N.; Bossi, S.; Su, S.; Lee, S.K.; Peng, W.; Xie, A.; et al. Topoisomerase 3β knockout mice show transcriptional and behavioural impairments associated with neurogenesis and synaptic plasticity. Nat. Commun. 2020, 11, 3143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cao, M.; Chang, C.-W.; Wang, C.; Shi, X.; Zhan, X.; Birnbaum, S.G.; Bezprozvanny, I.; Huber, K.M.; Wu, J.I. Autism-Associated Chromatin Regulator Brg1/SmarcA4 Is Required for Synapse Development and Myocyte Enhancer Factor 2-Mediated Synapse Remodeling. Mol. Cell. Biol. 2016, 36, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Zhou, Y.; Wu, B.; Chen, H.; Xu, S.; Wang, Y.; Zhang, P.; Li, G.; Xu, Q.; Zhou, W.; et al. Novel Variants of the SMARCA4 Gene Associated with Autistic Features Rather Than Typical Coffin-Siris Syndrome in Eight Chinese Pediatric Patients. J. Autism Dev. Disord. 2022, 52, 5033–5041. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Wei, Y.; Zhou, B.; Tang, T.; Wang, M.; Luan, R.; Bai, Y.; Li, H.; Wang, S.; Wang, C.; et al. The BAP18/CTCF/NURF complex contributes to modulation of enhancer rnas and endocrine resistance in breast cancer. SSRN J. 2022. [Google Scholar] [CrossRef]
- Sun, S.; Zhong, X.; Wang, C.; Sun, H.; Wang, S.; Zhou, T.; Zou, R.; Lin, L.; Sun, N.; Sun, G.; et al. BAP18 coactivates androgen receptor action and promotes prostate cancer progression. Nucleic Acids Res. 2016, 44, 8112–8128. [Google Scholar] [CrossRef] [PubMed]
- Jakovcevski, M.; Ruan, H.; Shen, E.Y.; Dincer, A.; Javidfar, B.; Ma, Q.; Peter, C.J.; Cheung, I.; Mitchell, A.C.; Jiang, Y.; et al. Neuronal Kmt2a/Mll1 histone methyltransferase is essential for prefrontal synaptic plasticity and working memory. J. Neurosci. 2015, 35, 5097–5108. [Google Scholar] [CrossRef] [PubMed]
- Breindl, M.; Spitzer, D.; Gerasimaitė, R.; Kairys, V.; Schubert, T.; Henfling, R.; Schwartz, U.; Lukinavičius, G.; Manelytė, L. Biochemical and cellular insights into the Baz2B protein, a non-catalytic subunit of the chromatin remodeling complex. Nucleic Acids Res. 2024, 52, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.M.; Guo, H.; Eichler, E.E.; Rosenfeld, J.A.; Pang, K.; Liu, Z.; Lalani, S.; Bi, W.; Yang, Y.; Bacino, C.A.; et al. BAZ2B haploinsufficiency as a cause of developmental delay, intellectual disability, and autism spectrum disorder. Hum. Mutat. 2020, 41, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Sewani, S.; Azamian, M.S.; Mendelsohn, B.A.; Mau-Them, F.T.; Réda, M.; Nambot, S.; Isidor, B.; van der Smagt, J.J.; Shen, J.J.; Shillington, A.; et al. Neurodevelopmental and other phenotypes recurrently associated with heterozygous BAZ2B loss-of-function variants. Am. J. Med. Genet. A 2024, 194, e63445. [Google Scholar] [CrossRef] [PubMed]
- Seczynska, M.; Lehner, P.J. The sound of silence: Mechanisms and implications of HUSH complex function. Trends Genet. 2023, 39, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Douse, C.H.; Tchasovnikarova, I.A.; Timms, R.T.; Protasio, A.V.; Seczynska, M.; Prigozhin, D.M.; Albecka, A.; Wagstaff, J.; Williamson, J.C.; Freund, S.M.V.; et al. TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control. Nat. Commun. 2020, 11, 4940. [Google Scholar] [CrossRef] [PubMed]
- Hagelkruys, A.; Horrer, M.; Taubenschmid-Stowers, J.; Kavirayani, A.; Novatchkova, M.; Orthofer, M.; Pai, T.-P.; Cikes, D.; Zhuk, S.; Balmaña, M.; et al. The HUSH complex controls brain architecture and protocadherin fidelity. Sci. Adv. 2022, 8, eabo7247. [Google Scholar] [CrossRef] [PubMed]
- White-Brown, A.; Choufani, S.; Care4Rare Canada Consortium; Weksberg, R.; Dyment, D. Missense variant in SRCAP with distinct DNA methylation signature associated with non-FLHS SRCAP-related neurodevelopmental disorder. Am. J. Med. Genet. A 2023, 191, 2640–2646. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Madden, J.A.; Lin, J.; Berry, G.T.; Wojcik, M.H.; Zhao, X.; Brand, H.; Talkowski, M.; Lee, E.A.; Agrawal, P.B. A neurodevelopmental disorder caused by a novel de novo SVA insertion in exon 13 of the SRCAP gene. Eur. J. Hum. Genet. 2022, 30, 1083–1087. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.E.; McMahon, S.B.; Cole, M.D.; Hearing, P. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 2001, 276, 32627–32634. [Google Scholar] [CrossRef] [PubMed]
- Robert, F.; Hardy, S.; Nagy, Z.; Baldeyron, C.; Murr, R.; Déry, U.; Masson, J.-Y.; Papadopoulo, D.; Herceg, Z.; Tora, L. The transcriptional histone acetyltransferase cofactor TRRAP associates with the MRN repair complex and plays a role in DNA double-strand break repair. Mol. Cell. Biol. 2006, 26, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Cogné, B.; Ehresmann, S.; Beauregard-Lacroix, E.; Rousseau, J.; Besnard, T.; Garcia, T.; Petrovski, S.; Avni, S.; McWalter, K.; Blackburn, P.R.; et al. Missense variants in the histone acetyltransferase complex component gene TRRAP cause autism and syndromic intellectual disability. Am. J. Hum. Genet. 2019, 104, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Tapias, A.; Lázaro, D.; Yin, B.-K.; Rasa, S.M.M.; Krepelova, A.; Kelmer Sacramento, E.; Grigaravicius, P.; Koch, P.; Kirkpatrick, J.; Ori, A.; et al. HAT cofactor TRRAP modulates microtubule dynamics via SP1 signaling to prevent neurodegeneration. eLife 2021, 10, e61531. [Google Scholar] [CrossRef] [PubMed]
- Pratt, K.J.B.; Shea, J.M.; Remesal-Gomez, L.; Bieri, G.; Smith, L.K.; Couthouis, J.; Chen, C.P.; Roy, I.J.; Gontier, G.; Villeda, S.A. Loss of neuronal Tet2 enhances hippocampal-dependent cognitive function. Cell Rep. 2022, 41, 111612. [Google Scholar] [CrossRef] [PubMed]
- Gontier, G.; Iyer, M.; Shea, J.M.; Bieri, G.; Wheatley, E.G.; Ramalho-Santos, M.; Villeda, S.A. Tet2 Rescues Age-Related Regenerative Decline and Enhances Cognitive Function in the Adult Mouse Brain. Cell Rep. 2018, 22, 1974–1981. [Google Scholar] [CrossRef] [PubMed]
- Raab, J.R.; Runge, J.S.; Spear, C.C.; Magnuson, T. Co-regulation of transcription by BRG1 and BRM, two mutually exclusive SWI/SNF ATPase subunits. Epigenetics Chromatin 2017, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.J.; Jung, E.-M.; Ka, M.; Smith, A.L.; Jeon, B.T.; Santen, G.W.E.; Kim, W.-Y. The role of ARID1B, a BAF chromatin remodeling complex subunit, in neural development and behavior. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 89, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.J.; Jung, E.-M.; Ka, M.; Jeon, B.T.; Lee, H.; Kim, W.-Y. Differential roles of ARID1B in excitatory and inhibitory neural progenitors in the developing cortex. Sci. Rep. 2021, 11, 3856. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.J.; Smith, A.L.; Jung, E.-M.; Ka, M.; Kim, W.-Y. Neurobiology of ARID1B haploinsufficiency related to neurodevelopmental and psychiatric disorders. Mol. Psychiatry 2022, 27, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; He, Y.; Rehman, A.U.; Kong, Y.; Hong, S.; Ding, G.; Yalamanchili, H.K.; Wan, Y.-W.; Paul, B.; Wang, C.; et al. Loss of function of NCOR1 and NCOR2 impairs memory through a novel GABAergic hypothalamus-CA3 projection. Nat. Neurosci. 2019, 22, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.B.; Hayes, M.P.; Chen, D.H.; Raskind, W.H.; Watts, V.J. Functional characterization of AC5 gain-of-function variants: Impact on the molecular basis of ADCY5-related dyskinesia. Biochem. Pharmacol. 2019, 163, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Slawson, J.B.; Kuklin, E.A.; Mukherjee, K.; Pírez, N.; Donelson, N.C.; Griffith, L.C. Regulation of dopamine release by CASK-β modulates locomotor initiation in Drosophila melanogaster. Front. Behav. Neurosci. 2014, 8, 394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, C.; Kershberg, L.; Wang, J.; Schneeberger, S.; Kaeser, P.S. Dopamine Secretion Is Mediated by Sparse Active Zone-like Release Sites. Cell 2018, 172, 706–718.e15. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.G.; Cai, X.; Wang, J.; Bunzow, J.R.; Williams, J.T.; Kaeser, P.S. RIM is essential for stimulated but not spontaneous somatodendritic dopamine release in the midbrain. eLife 2019, 8, e47972. [Google Scholar] [CrossRef] [PubMed]
- Hobson, B.D.; Kong, L.; Angelo, M.F.; Lieberman, O.J.; Mosharov, E.V.; Herzog, E.; Sulzer, D.; Sims, P.A. Subcellular and regional localization of mRNA translation in midbrain dopamine neurons. Cell Rep. 2022, 38, 110208. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Lee, L.-J.; Fan, C.-C.; Chang, H.-C.; Shih, H.-A.; Min, M.-Y.; Chang, M.-S. Important roles of Vilse in dendritic architecture and synaptic plasticity. Sci. Rep. 2017, 7, 45646. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Villegas, E.M.; Pérez-Rodríguez, M.; Negrete-Díaz, J.V.; Ruiz, R.; Rosa, J.L.; de Toledo, G.A.; Rodríguez-Moreno, A.; Armengol, J.A. HERC1 ubiquitin ligase is required for hippocampal learning and memory. Front. Neuroanat. 2020, 14, 592797. [Google Scholar] [CrossRef] [PubMed]
- Birk, E.; Har-Zahav, A.; Manzini, C.M.; Pasmanik-Chor, M.; Kornreich, L.; Walsh, C.A.; Noben-Trauth, K.; Albin, A.; Simon, A.J.; Colleaux, L.; et al. SOBP is mutated in syndromic and nonsyndromic intellectual disability and is highly expressed in the brain limbic system. Am. J. Hum. Genet. 2010, 87, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.J.; Frankland, P.W.; Marowitz, Z.; Friedman, E.; Laszlo, G.S.; Cioffi, D.; Jacks, T.; Bourtchuladze, R. A mouse model for the learning and memory deficits associated with neurofibromatosis type I. Nat. Genet. 1997, 15, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Ottenhoff, M.J.; Dijkhuizen, S.; Ypelaar, A.C.H.; de Oude, N.L.; Koekkoek, S.K.E.; Wang, S.S.H.; De Zeeuw, C.I.; Elgersma, Y.; Boele, H.J. Cerebellum-dependent associative learning is not impaired in a mouse model of neurofibromatosis type 1. Sci. Rep. 2022, 12, 19041. [Google Scholar] [CrossRef] [PubMed]
- Methi, A.; Islam, M.R.; Kaurani, L.; Sakib, M.S.; Krüger, D.M.; Pena, T.; Burkhardt, S.; Liebetanz, D.; Fischer, A. A Single-Cell Transcriptomic Analysis of the Mouse Hippocampus After Voluntary Exercise. Mol. Neurobiol. 2024, 61, 5628–5645. [Google Scholar] [CrossRef] [PubMed]
- Niwa, Y.; Kanda, G.N.; Yamada, R.G.; Shi, S.; Sunagawa, G.A.; Ukai-Tadenuma, M.; Fujishima, H.; Matsumoto, N.; Masumoto, K.-H.; Nagano, M.; et al. Muscarinic acetylcholine receptors chrm1 and chrm3 are essential for REM sleep. Cell Rep. 2018, 24, 2231–2247.e7. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhu, Q.-Q.; Niu, M.-L.; Li, N.; Ren, B.-Q.; Yu, T.-B.; Zhou, Z.-S.; Guo, J.-D.; Zhou, Y. Ghrelin infusion into the basolateral amygdala suppresses CTA memory formation in rats via the PI3K/Akt/mTOR and PLC/PKC signaling pathways. Acta Pharmacol. Sin. 2022, 43, 2242–2252. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.D.; Kim, D.W.; Zhou, Y.; Jansen, D.; Slosky, L.M.; Clark, N.B.; Ray, C.R.; Hu, X.; Southall, N.; Wang, A.; et al. Discovery of a functionally selective ghrelin receptor (GHSR1a) ligand for modulating brain dopamine. Proc. Natl. Acad. Sci. USA 2022, 119, e2112397119. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Kubota, T.; Mariko, I.; Takamoto, I.; Aihara, M.; Sakurai, Y.; Wada, N.; Miki, T.; Yamauchi, T.; Kubota, N.; et al. Lack of Brain Insulin Receptor Substrate-1 Causes Growth Retardation, With Decreased Expression of Growth Hormone-Releasing Hormone in the Hypothalamus. Diabetes 2021, 70, 1640–1653. [Google Scholar] [CrossRef] [PubMed]
- Todd, P.A.C.; McCue, H.V.; Haynes, L.P.; Barclay, J.W.; Burgoyne, R.D. Interaction of ARF-1.1 and neuronal calcium sensor-1 in the control of the temperature-dependency of locomotion in Caenorhabditis elegans. Sci. Rep. 2016, 6, 30023. [Google Scholar] [CrossRef] [PubMed]
- Kononenko, N.L.; Diril, M.K.; Puchkov, D.; Kintscher, M.; Koo, S.J.; Pfuhl, G.; Winter, Y.; Wienisch, M.; Klingauf, J.; Breustedt, J.; et al. Compromised fidelity of endocytic synaptic vesicle protein sorting in the absence of stonin 2. Proc. Natl. Acad. Sci. USA 2013, 110, E526–E535. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.; Leventis, P.A.; Silvescu, C.I.; Reinhold, V.N.; Schachter, H.; Boulianne, G.L. Null mutations in Drosophila N-acetylglucosaminyltransferase I produce defects in locomotion and a reduced life span. J. Biol. Chem. 2006, 281, 12776–12785. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.; Iliadi, K.G.; Leventis, P.A.; Schachter, H.; Boulianne, G.L. Neuronal expression of Mgat1 rescues the shortened life span of Drosophila Mgat11 null mutants and increases life span. Proc. Natl. Acad. Sci. USA 2010, 107, 9677–9682. [Google Scholar] [CrossRef] [PubMed]
- Falace, A.; Buhler, E.; Fadda, M.; Watrin, F.; Lippiello, P.; Pallesi-Pocachard, E.; Baldelli, P.; Benfenati, F.; Zara, F.; Represa, A.; et al. TBC1D24 regulates neuronal migration and maturation through modulation of the ARF6-dependent pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Finelli, M.J.; Aprile, D.; Castroflorio, E.; Jeans, A.; Moschetta, M.; Chessum, L.; Degiacomi, M.T.; Grasegger, J.; Lupien-Meilleur, A.; Bassett, A.; et al. The epilepsy-associated protein TBC1D24 is required for normal development, survival and vesicle trafficking in mammalian neurons. Hum. Mol. Genet. 2019, 28, 584–597. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.V.; Seit-Nebi, A.S.; Marston, S.B.; Gusev, N.B. Some properties of human small heat shock protein Hsp22 (H11 or HspB8). Biochem. Biophys. Res. Commun. 2004, 315, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmus, M.M.M.; Boelens, W.C.; Otte-Höller, I.; Kamps, B.; Kusters, B.; Maat-Schieman, M.L.C.; de Waal, R.M.W.; Verbeek, M.M. Small heat shock protein HspB8: Its distribution in Alzheimer’s disease brains and its inhibition of amyloid-beta protein aggregation and cerebrovascular amyloid-beta toxicity. Acta Neuropathol. 2006, 111, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Crippa, V.; Sau, D.; Rusmini, P.; Boncoraglio, A.; Onesto, E.; Bolzoni, E.; Galbiati, M.; Fontana, E.; Marino, M.; Carra, S.; et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS). Hum. Mol. Genet. 2010, 19, 3440–3456. [Google Scholar] [CrossRef] [PubMed]
- Crippa, V.; D’Agostino, V.G.; Cristofani, R.; Rusmini, P.; Cicardi, M.E.; Messi, E.; Loffredo, R.; Pancher, M.; Piccolella, M.; Galbiati, M.; et al. Transcriptional induction of the heat shock protein B8 mediates the clearance of misfolded proteins responsible for motor neuron diseases. Sci. Rep. 2016, 6, 22827. [Google Scholar] [CrossRef] [PubMed]
- Chierichetti, M.; Cerretani, M.; Ciammaichella, A.; Crippa, V.; Rusmini, P.; Ferrari, V.; Tedesco, B.; Casarotto, E.; Cozzi, M.; Mina, F.; et al. Identification of HSPB8 modulators counteracting misfolded protein accumulation in neurodegenerative diseases. Life Sci. 2023, 322, 121323. [Google Scholar] [CrossRef] [PubMed]
- Irobi, J.; Van Impe, K.; Seeman, P.; Jordanova, A.; Dierick, I.; Verpoorten, N.; Michalik, A.; De Vriendt, E.; Jacobs, A.; Van Gerwen, V.; et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat. Genet. 2004, 36, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Luo, Z.; Shan, W.; Zuo, Z. Role of sox2 in learning, memory, and postoperative cognitive dysfunction in mice. Cells 2021, 10, 727. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, S.; Serra, L.; Pagin, M.; Nicolis, S.K. Deconstructing sox2 function in brain development and disease. Cells 2022, 11, 1604. [Google Scholar] [CrossRef] [PubMed]
- Teunissen, M.W.A.; Lewerissa, E.; van Hugte, E.J.H.; Wang, S.; Ockeloen, C.W.; Koolen, D.A.; Pfundt, R.; Marcelis, C.L.M.; Brilstra, E.; Howe, J.L.; et al. ANK2 loss-of-function variants are associated with epilepsy, and lead to impaired axon initial segment plasticity and hyperactive network activity in hiPSC-derived neuronal networks. Hum. Mol. Genet. 2023, 32, 2373–2385. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Wilson, S.J.; Hale, T.K.; Fitzsimons, H.L. Ankyrin2 is essential for neuronal morphogenesis and long-term courtship memory in Drosophila. Mol. Brain 2023, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Snijders Blok, L.; Kleefstra, T.; Venselaar, H.; Maas, S.; Kroes, H.Y.; Lachmeijer, A.M.A.; van Gassen, K.L.I.; Firth, H.V.; Tomkins, S.; Bodek, S.; et al. De novo variants disturbing the transactivation capacity of POU3F3 cause a characteristic neurodevelopmental disorder. Am. J. Hum. Genet. 2019, 105, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.-B.; Zeng, Y.-Q.; Liu, P.-P.; Mi, T.-W.; Zhang, S.-F.; Dai, S.-K.; Tang, Q.-Y.; Yang, L.; Xu, Y.-J.; Yan, H.-L.; et al. The histone H3K27 demethylase UTX regulates synaptic plasticity and cognitive behaviors in mice. Front. Mol. Neurosci. 2017, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Seward, C.H.; Chen, C.-Y.; LeBlanc, A.; Leddy, A.M.; Stubbs, L. Isolated loss of the AUTS2 long isoform, brain-wide or targeted to Calbindin -lineage cells, generates a specific suite of brain, behavioral and molecular pathologies. BioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Biel, A.; Castanza, A.S.; Rutherford, R.; Fair, S.R.; Chifamba, L.; Wester, J.C.; Hester, M.E.; Hevner, R.F. AUTS2 syndrome: Molecular mechanisms and model systems. Front. Mol. Neurosci. 2022, 15, 858582. [Google Scholar] [CrossRef] [PubMed]
- Kuo, P.H.; Chuang, L.C.; Liu, J.R.; Liu, C.M.; Huang, M.C.; Lin, S.K.; Sunny Sun, H.; Hsieh, M.H.; Hung, H.; Lu, R.B. Identification of novel loci for bipolar I disorder in a multi-stage genome-wide association study. Prog. Neuropsychopharmacol. Biol. Psychiatry 2014, 51, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Strauss, K.A.; Markx, S.; Georgi, B.; Paul, S.M.; Jinks, R.N.; Hoshi, T.; McDonald, A.; First, M.B.; Liu, W.; Benkert, A.R.; et al. A population-based study of KCNH7 p.Arg394His and bipolar spectrum disorder. Hum. Mol. Genet. 2014, 23, 6395–6406. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Quan, Y.; Duan, G.; Wu, H.; Bai, T.; Wang, Y.; Zhou, S.; Ou, J.; Shen, Y.; Hu, Z.; et al. Mutation pattern and genotype-phenotype correlations of SETD2 in neurodevelopmental disorders. Eur. J. Med. Genet. 2021, 64, 104200. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, B.; Thor, S.; Piper, M. Cellular and molecular functions of SETD2 in the central nervous system. J. Cell Sci. 2023, 136, jcs261406. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ferguson, C.J.; Mitchell, D.C.; Titus, A.; Paulo, J.A.; Hwang, A.; Lin, T.-H.; Yano, H.; Gu, W.; Song, S.-K.; et al. The Hao-Fountain syndrome protein USP7 regulates neuronal connectivity in the brain via a novel p53-independent ubiquitin signaling pathway. BioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Tian, Y.; Huo, Y.; Man, H.-Y. Role of the DUB enzyme USP7 in dendritic arborization, neuronal migration, and autistic-like behaviors in mice. iScience 2022, 25, 104595. [Google Scholar] [CrossRef] [PubMed]
- Gore, B.B.; Miller, S.M.; Jo, Y.S.; Baird, M.A.; Hoon, M.; Sanford, C.A.; Hunker, A.; Lu, W.; Wong, R.O.; Zweifel, L.S. Roundabout receptor 2 maintains inhibitory control of the adult midbrain. eLife 2017, 6, e23858. [Google Scholar] [CrossRef] [PubMed]
- Wurmser, M.; Muppavarapu, M.; Tait, C.M.; Laumonnerie, C.; González-Castrillón, L.M.; Wilson, S.I. Robo2 receptor gates the anatomical divergence of neurons derived from a common precursor origin. Front. Cell Dev. Biol. 2021, 9, 668175. [Google Scholar] [CrossRef] [PubMed]
- Blockus, H.; Rolotti, S.V.; Szoboszlay, M.; Peze-Heidsieck, E.; Ming, T.; Schroeder, A.; Apostolo, N.; Vennekens, K.M.; Katsamba, P.S.; Bahna, F.; et al. Synaptogenic activity of the axon guidance molecule Robo2 underlies hippocampal circuit function. Cell Rep. 2021, 37, 109828. [Google Scholar] [CrossRef] [PubMed]
- James, G.; Key, B.; Beverdam, A. The E3 ubiquitin ligase Mycbp2 genetically interacts with Robo2 to modulate axon guidance in the mouse olfactory system. Brain Struct. Funct. 2014, 219, 861–874. [Google Scholar] [CrossRef] [PubMed]
- AlAbdi, L.; Desbois, M.; Rusnac, D.-V.; Sulaiman, R.A.; Rosenfeld, J.A.; Lalani, S.; Murdock, D.R.; Burrage, L.C.; Undiagnosed Diseases Network; Billie Au, P.Y.; et al. Loss-of-function variants in MYCBP2 cause neurobehavioural phenotypes and corpus callosum defects. Brain 2023, 146, 1373–1387. [Google Scholar] [CrossRef] [PubMed]
- Lau, D.; Bengtson, C.P.; Buchthal, B.; Bading, H. BDNF Reduces Toxic Extrasynaptic NMDA Receptor Signaling via Synaptic NMDA Receptors and Nuclear-Calcium-Induced Transcription of inhba/Activin A. Cell Rep. 2015, 12, 1353–1366. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.R.; Pirruccello, M.; Cheng, F.; Kang, H.J.; Fernandez, T.V.; Baskin, J.M.; Choi, M.; Liu, L.; Ercan-Sencicek, A.G.; Murdoch, J.D.; et al. Rare deleterious mutations of the gene EFR3A in autism spectrum disorders. Mol. Autism 2014, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Ye, B.; Zhang, L.; Wang, Q.; Liu, Z.; Ji, S.; Liu, Q.; Lv, J.; Ma, Y.; Xu, Y.; et al. Efr3a Insufficiency Attenuates the Degeneration of Spiral Ganglion Neurons after Hair Cell Loss. Front. Mol. Neurosci. 2017, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Baig-Ward, K.M.; Chu, T.J.; Keil, M.; Mumford, S.; Polk, C.; Segars, J.H. Anxiety related phenotypes in a-kinase anchoring protein 13 (AKAP13) mice: A potential model for obsessive- compulsive disorder. Fertil. Steril. 2014, 102, e45. [Google Scholar] [CrossRef]
- Doobin, D.J.; Kemal, S.; Dantas, T.J.; Vallee, R.B. Severe NDE1-mediated microcephaly results from neural progenitor cell cycle arrests at multiple specific stages. Nat. Commun. 2016, 7, 12551. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Tsuboi, D.; Wang, C.; Kushima, I.; Koide, T.; Ikeda, M.; Iwayama, Y.; Toyota, T.; Yamamoto, N.; Kunimoto, S.; et al. Identification of Rare, Single-Nucleotide Mutations in NDE1 and Their Contributions to Schizophrenia Susceptibility. Schizophr. Bull. 2015, 41, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Lee, J.; Kim, H.-J.; Lee, B.E.; Jeong, J.; Cho, E.J.; Jang, H.-J.; Shin, K.J.; Kim, M.J.; Chae, Y.C.; et al. PLCγ1 in dopamine neurons critically regulates striatal dopamine release via VMAT2 and synapsin III. Exp. Mol. Med. 2023, 55, 2357–2375. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Lee, H.; Argiropoulos, B.; Dorrani, N.; Mann, J.; Martinez-Agosto, J.A.; Gomez-Ospina, N.; Gallant, N.; Bernstein, J.A.; Hudgins, L.; et al. DYRK1A haploinsufficiency causes a new recognizable syndrome with microcephaly, intellectual disability, speech impairment, and distinct facies. Eur. J. Hum. Genet. 2015, 23, 1473–1481. [Google Scholar] [CrossRef] [PubMed]
- Guard, S.E.; Poss, Z.C.; Ebmeier, C.C.; Pagratis, M.; Simpson, H.; Taatjes, D.J.; Old, W.M. The nuclear interactome of DYRK1A reveals a functional role in DNA damage repair. Sci. Rep. 2019, 9, 6539. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Morris, J.L.; Yang, K.; Fu, Z.; Zhu, X.; Johnson, F.; Meehan, B.; Witkowski, L.; Yasmeen, A.; Golenar, T.; et al. SMARCA4/2 loss inhibits chemotherapy-induced apoptosis by restricting IP3R3-mediated Ca2+ flux to mitochondria. Nat. Commun. 2021, 12, 5404. [Google Scholar] [CrossRef] [PubMed]
- Cabello-Murgui, J.; Jiménez-Jiménez, J.; Vílchez, J.J.; Azorín, I.; Martí-Martínez, P.; Millet, E.; Lupo, V.; Sevilla, T.; Sivera, R. ITPR3-associated neuropathy: Report of a further family with adult onset intermediate Charcot-Marie-Tooth disease. Eur. J. Neurol. 2024, 31, e16485. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.; Srivastava, G.; Hsieh, W.-S.; Gao, Z.; Murray, P.; Liao, S.-K.; Ambinder, R.; Tao, Q. The stress-responsive gene GADD45G is a functional tumor suppressor, with its response to environmental stresses frequently disrupted epigenetically in multiple tumors. Clin. Cancer Res. 2005, 11, 6442–6449. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Marshall, P.R.; Leighton, L.J.; Zajaczkowski, E.L.; Wang, Z.; Madugalle, S.U.; Yin, J.; Bredy, T.W.; Wei, W. The DNA Repair-Associated Protein Gadd45γ Regulates the Temporal Coding of Immediate Early Gene Expression within the Prelimbic Prefrontal Cortex and Is Required for the Consolidation of Associative Fear Memory. J. Neurosci. 2019, 39, 970–983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Q.; Wang, H.; Wu, Y.; Ye, X.; Gong, Z.; Li, Q.; Xuan, A. Gadd45g, A Novel Antidepressant Target, Mediates Metformin-Induced Neuronal Differentiation of Neural Stem Cells Via DNA Demethylation. Stem Cells 2022, 40, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Kase, Y.; Sato, T.; Okano, Y.; Okano, H. The GADD45G/p38 MAPK/CDC25B signaling pathway enhances neurite outgrowth by promoting microtubule polymerization. iScience 2022, 25, 104089. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, X.; Yu, B.; Gu, Y.; Yuan, Y.; Yao, D.; Ding, F.; Gu, X. Gene network revealed involvements of Birc2, Birc3 and Tnfrsf1a in anti-apoptosis of injured peripheral nerves. PLoS ONE 2012, 7, e43436. [Google Scholar] [CrossRef] [PubMed]
- Reigada, D.; Maza, R.M.; Muñoz-Galdeano, T.; Barreda-Manso, M.A.; Soto, A.; Lindholm, D.; Navarro-Ruíz, R.; Nieto-Díaz, M. Overexpression of the X-Linked Inhibitor of Apoptosis Protein (XIAP) in Neurons Improves Cell Survival and the Functional Outcome after Traumatic Spinal Cord Injury. Int. J. Mol. Sci. 2023, 24, 2791. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Liang, S.; Yuan, T.; Liu, J. EIF2S1 silencing impedes neuroblastoma development through GPX4 inactivation and ferroptosis induction. Int. J. Genom. 2024, 2024, 6594426. [Google Scholar] [CrossRef] [PubMed]
- Solary, E.; Bernard, O.A.; Tefferi, A.; Fuks, F.; Vainchenker, W. The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia 2014, 28, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Boulard, M.; Rucli, S.; Edwards, J.R.; Bestor, T.H. Methylation-directed glycosylation of chromatin factors represses retrotransposon promoters. Proc. Natl. Acad. Sci. USA 2020, 117, 14292–14298. [Google Scholar] [CrossRef] [PubMed]
- Pravata, V.M.; Muha, V.; Gundogdu, M.; Ferenbach, A.T.; Kakade, P.S.; Vandadi, V.; Wilmes, A.C.; Borodkin, V.S.; Joss, S.; Stavridis, M.P.; et al. Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability. Proc. Natl. Acad. Sci. USA 2019, 116, 14961–14970. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Dong, X.; Cheng, X.; Zhu, Q.; Zhang, J.; Li, Q.; Huang, X.; Wang, M.; Li, L.; Guo, W.; et al. Ogt controls neural stem/progenitor cell pool and adult neurogenesis through modulating Notch signaling. Cell Rep. 2021, 34, 108905. [Google Scholar] [CrossRef] [PubMed]
- Pryszlak, M.; Wiggans, M.; Chen, X.; Jaramillo, J.E.; Burns, S.E.; Richards, L.M.; Pugh, T.J.; Kaplan, D.R.; Huang, X.; Dirks, P.B.; et al. The DEAD-box helicase DDX56 is a conserved stemness regulator in normal and cancer stem cells. Cell Rep. 2021, 34, 108903. [Google Scholar] [CrossRef] [PubMed]
- Chicherin, I.V.; Dukhalin, S.V.; Khannanov, R.A.; Baleva, M.V.; Levitskii, S.A.; Patrushev, M.V.; Sergiev, P.V.; Kamenski, P. Functional Diversity of Mitochondrial Peptidyl-tRNA Hydrolase ICT1 in Human Cells. Front. Mol. Biosci. 2021, 8, 716885. [Google Scholar] [CrossRef] [PubMed]
- Morello, L.G.; Hesling, C.; Coltri, P.P.; Castilho, B.A.; Rimokh, R.; Zanchin, N.I.T. The NIP7 protein is required for accurate pre-rRNA processing in human cells. Nucleic Acids Res. 2011, 39, 648–665. [Google Scholar] [CrossRef] [PubMed]
- Di Vona, C.; Barba, L.; Ferrari, R.; de la Luna, S. Loss of the DYRK1A protein kinase results in the reduction in ribosomal protein gene expression, ribosome mass and reduced translation. Biomolecules 2023, 14, 31. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Irannejad, R.; Bowman, S.L.; Du, Y.; Puthenveedu, M.A.; von Zastrow, M.; Benovic, J.L. The α-Arrestin ARRDC3 Regulates the Endosomal Residence Time and Intracellular Signaling of the β2-Adrenergic Receptor. J. Biol. Chem. 2016, 291, 14510–14525. [Google Scholar] [CrossRef] [PubMed]
- Dores, M.R.; Lin, H.; Grimsey, N.J.; Mendez, F.; Trejo, J. The α-arrestin ARRDC3 mediates ALIX ubiquitination and G protein-coupled receptor lysosomal sorting. Mol. Biol. Cell 2015, 26, 4660–4673. [Google Scholar] [CrossRef] [PubMed]
- Gordon, B.S.; Rossetti, M.L.; Eroshkin, A.M. Arrdc2 and Arrdc3 elicit divergent changes in gene expression in skeletal muscle following anabolic and catabolic stimuli. Physiol. Genom. 2019, 51, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Laskin, G.R.; Cabrera, A.R.; Greene, N.P.; Tomko, R.J.; Vied, C.; Gordon, B.S. The mechanosensitive gene arrestin domain containing 2 regulates myotube diameter with direct implications for disuse atrophy with aging. Am. J. Physiol, Cell Physiol. 2024, 326, C768–C783. [Google Scholar] [CrossRef] [PubMed]
- Pierce-Shimomura, J.T.; Chen, B.L.; Mun, J.J.; Ho, R.; Sarkis, R.; McIntire, S.L. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc. Natl. Acad. Sci. USA 2008, 105, 20982–20987. [Google Scholar] [CrossRef] [PubMed]
- Parra-Díaz, P.; Monteil, A.; Calame, D.; Hadouiri, N.; Soliani, L.; Spinelli, E.; Caron, E.J.; Dieterich, K.; Kritzer, A.; Riley, K.; et al. Genotype-Phenotype Landscape of NALCN and UNC80-Related Disorders. Neurology 2025, 104, e213429. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, J.; Kofuji, S.; Itoh, R.; Momiyama, T.; Takayama, K.; Murakami, H.; Chida, S.; Tsuya, Y.; Takasuga, S.; Eguchi, S.; et al. The PtdIns(3,4)P(2) phosphatase INPP4A is a suppressor of excitotoxic neuronal death. Nature 2010, 465, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Knop, F.; Zounarová, A.; Šabata, V.; Middelkoop, T.C.; Macůrková, M. Caenorhabditis elegans SEL-5/AAK1 regulates cell migration and cell outgrowth independently of its kinase activity. eLife 2024, 13, e91054. [Google Scholar] [CrossRef] [PubMed]
- Kostich, W.; Hamman, B.D.; Li, Y.-W.; Naidu, S.; Dandapani, K.; Feng, J.; Easton, A.; Bourin, C.; Baker, K.; Allen, J.; et al. Inhibition of AAK1 kinase as a novel therapeutic approach to treat neuropathic pain. J. Pharmacol. Exp. Ther. 2016, 358, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, R.; Du, X.; Huang, Y.; Gao, Y.; Wen, Y.; Qiao, D.; Sun, N.; Liu, Z. Identification of aberrant plasma vesicles containing AAK1 and CCDC18-AS1 in adolescents with major depressive disorder and preliminary exploration of treatment efficacy. Genomics 2025, 117, 110993. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Guo, T.; Pei, F.; Feng, J.; Jing, J.; Xu, J.; Yamada, T.; Ho, T.-V.; Du, J.; Sehgal, P.; et al. ARID1B maintains mesenchymal stem cell quiescence via inhibition of BCL11B-mediated non-canonical Activin signaling. Nat. Commun. 2024, 15, 4614. [Google Scholar] [CrossRef] [PubMed]
- Bosch, E.; Güse, E.; Kirchner, P.; Winterpacht, A.; Walther, M.; Alders, M.; Kerkhof, J.; Ekici, A.B.; Sticht, H.; Sadikovic, B.; et al. The missing link: ARID1B non-truncating variants causing Coffin-Siris syndrome due to protein aggregation. Hum. Genet. 2024, 143, 965–978. [Google Scholar] [CrossRef] [PubMed]
- Vogel-Ciernia, A.; Wood, M.A. Neuron-specific chromatin remodeling: A missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders. Neuropharmacology 2014, 80, 18–27. [Google Scholar] [CrossRef] [PubMed]
- van der Sluijs, P.J.; Gösgens, M.; Dingemans, A.J.M.; Striano, P.; Riva, A.; Mignot, C.; Faudet, A.; Vasileiou, G.; Walther, M.; Schrier Vergano, S.A.; et al. ARID1B-related disorder in 87 adults: Natural history and self-sustainability. Genet. Med. Open 2024, 2, 101873. [Google Scholar] [CrossRef] [PubMed]
- Meyers, E.A.; Kessler, J.A. TGF-β Family Signaling in Neural and Neuronal Differentiation, Development, and Function. Cold Spring Harb. Perspect. Biol. 2017, 9, a022244. [Google Scholar] [CrossRef] [PubMed]
- Kakefuda, K.; Oyagi, A.; Ishisaka, M.; Tsuruma, K.; Shimazawa, M.; Yokota, K.; Shirai, Y.; Horie, K.; Saito, N.; Takeda, J.; et al. Diacylglycerol kinase β knockout mice exhibit lithium-sensitive behavioral abnormalities. PLoS ONE 2010, 5, e13447. [Google Scholar] [CrossRef] [PubMed]
- Shirai, Y.; Kouzuki, T.; Kakefuda, K.; Moriguchi, S.; Oyagi, A.; Horie, K.; Morita, S.; Shimazawa, M.; Fukunaga, K.; Takeda, J.; et al. Essential role of neuron-enriched diacylglycerol kinase (DGK), DGKbeta in neurite spine formation, contributing to cognitive function. PLoS ONE 2010, 5, e11602. [Google Scholar] [CrossRef] [PubMed]
- Pintacuda, G.; Hsu, Y.-H.H.; Tsafou, K.; Li, K.W.; Martín, J.M.; Riseman, J.; Biagini, J.C.; Ching, J.K.T.; Mena, D.; Gonzalez-Lozano, M.A.; et al. Protein interaction studies in human induced neurons indicate convergent biology underlying autism spectrum disorders. Cell Genom. 2023, 3, 100250. [Google Scholar] [CrossRef] [PubMed]
- Murakami, N.; Bolton, D.; Hwang, Y.-W. Dyrk1A binds to multiple endocytic proteins required for formation of clathrin-coated vesicles. Biochemistry 2009, 48, 9297–9305. [Google Scholar] [CrossRef] [PubMed]
- Peron, A.; D’Arco, F.; Aldinger, K.A.; Smith-Hicks, C.; Zweier, C.; Gradek, G.A.; Bradbury, K.; Accogli, A.; Andersen, E.F.; Au, P.Y.B.; et al. BCL11A intellectual developmental disorder: Defining the clinical spectrum and genotype-phenotype correlations. Eur. J. Hum. Genet. 2025, 33, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Shibutani, M.; Horii, T.; Shoji, H.; Morita, S.; Kimura, M.; Terawaki, N.; Miyakawa, T.; Hatada, I. Arid1b Haploinsufficiency Causes Abnormal Brain Gene Expression and Autism-Related Behaviors in Mice. Int. J. Mol. Sci. 2017, 18, 1872. [Google Scholar] [CrossRef] [PubMed]
- Chistopolsky, I.; Leonova, A.; Mezheritskiy, M.; Boguslavsky, D.; Kristinina, A.; Zakharov, I.; Sorminskiy, A.; Vorontsov, D.; Dyakonova, V. Intense Locomotion Enhances Oviposition in the Freshwater Mollusc Lymnaea stagnalis: Cellular and Molecular Correlates. Biology 2023, 12, 764. [Google Scholar] [CrossRef] [PubMed]
- Carra, S.; Boncoraglio, A.; Kanon, B.; Brunsting, J.F.; Minoia, M.; Rana, A.; Vos, M.J.; Seidel, K.; Sibon, O.C.M.; Kampinga, H.H. Identification of the Drosophila ortholog of HSPB8: Implication of HSPB8 loss of function in protein folding diseases. J. Biol. Chem. 2010, 285, 37811–37822. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384–6391. [Google Scholar] [CrossRef] [PubMed]
- Bhatter, N.; Dmitriev, S.E.; Ivanov, P. Cell death or survival: Insights into the role of mRNA translational control. Semin. Cell Dev. Biol. 2024, 154, 138–154. [Google Scholar] [CrossRef] [PubMed]
- Dastidar, S.G.; Nair, D. A ribosomal perspective on neuronal local protein synthesis. Front. Mol. Neurosci. 2022, 15, 823135. [Google Scholar] [CrossRef] [PubMed]
- Bocharova, L.S.; Borovyagin, V.L.; Dyakonova, T.L.; Warton, S.S.; Veprintsev, B.N. Ultrastructure and RNA synthesis in a molluscan giant neuron under electrical stimulation. Brain Res. 1972, 36, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Beletskiy, A.; Zolotar, A.; Fortygina, P.; Chesnokova, E.; Uroshlev, L.; Balaban, P.; Kolosov, P. Downregulation of Ribosomal Protein Genes Is Revealed in a Model of Rat Hippocampal Neuronal Culture Activation with GABA(A)R/GlyRa2 Antagonist Picrotoxin. Cells 2024, 13, 383. [Google Scholar] [CrossRef] [PubMed]
- Dyakonova, V.E. DNA instability in neurons: Lifespan clock and driver of evolution. Biochemistry 2023, 88, 1719–1731. [Google Scholar] [CrossRef] [PubMed]
- Espeso-Gil, S.; Holik, A.Z.; Bonnin, S.; Jhanwar, S.; Chandrasekaran, S.; Pique-Regi, R.; Albaigès-Ràfols, J.; Maher, M.; Permanyer, J.; Irimia, M.; et al. Environmental enrichment induces epigenomic and genome organization changes relevant for cognition. Front. Mol. Neurosci. 2021, 14, 664912. [Google Scholar] [CrossRef] [PubMed]
- Borodinova, A.A.; Balaban, P.M. Epigenetic Regulation as a Basis for Long-Term Changes in the Nervous System: In Search of Specificity Mechanisms. Biochemistry 2020, 85, 994–1010. [Google Scholar] [CrossRef] [PubMed]
- Borodinova, A.A.; Kuznetsova, M.A.; Alekseeva, V.S.; Balaban, P.M. Histone acetylation determines transcription of atypical protein kinases in rat neurons. Sci. Rep. 2019, 9, 4332. [Google Scholar] [CrossRef] [PubMed]
- LaRese, T.P.; Yan, Y.; Eipper, B.A.; Mains, R.E. Using Kalirin conditional knockout mice to distinguish its role in dopamine receptor mediated behaviors. BMC Neurosci. 2017, 18, 45. [Google Scholar] [CrossRef] [PubMed]
- Sahraei, H.; Askaripour, M.; Esmaeilpour, K.; Shahsavari, F.; Rajabi, S.; Moradi-Kor, N. GABAB receptor activation ameliorates spatial memory impairments in stress-exposed rats. Neuropsychiatr. Dis. Treat. 2019, 15, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, H.; Hall, F.S.; Horiuchi, Y.; Sakurai, T.; Hishimoto, A.; Grumet, M.; Uhl, G.R.; Onaivi, E.S.; Arinami, T. NrCAM-regulating neural systems and addiction-related behaviors. Addict. Biol. 2014, 19, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Y.; Worley, P.F.; Mattson, M.P.; Gaiano, N. The canonical Notch pathway effector RBP-J regulates neuronal plasticity and expression of GABA transporters in hippocampal networks. Hippocampus 2015, 25, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Young, A.P.; Landry, C.F.; Jackson, D.J.; Wyeth, R.C. Tissue-specific evaluation of suitable reference genes for RT-qPCR in the pond snail, Lymnaea stagnalis. PeerJ 2019, 7, e7888. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rozenberg, J.M.; Boguslavsky, D.; Chistopolsky, I.; Zakharov, I.; Dyakonova, V. Rest Induces a Distinct Transcriptional Program in the Nervous System of the Exercised L. stagnalis. Int. J. Mol. Sci. 2025, 26, 6970. https://doi.org/10.3390/ijms26146970
Rozenberg JM, Boguslavsky D, Chistopolsky I, Zakharov I, Dyakonova V. Rest Induces a Distinct Transcriptional Program in the Nervous System of the Exercised L. stagnalis. International Journal of Molecular Sciences. 2025; 26(14):6970. https://doi.org/10.3390/ijms26146970
Chicago/Turabian StyleRozenberg, Julian M., Dmitri Boguslavsky, Ilya Chistopolsky, Igor Zakharov, and Varvara Dyakonova. 2025. "Rest Induces a Distinct Transcriptional Program in the Nervous System of the Exercised L. stagnalis" International Journal of Molecular Sciences 26, no. 14: 6970. https://doi.org/10.3390/ijms26146970
APA StyleRozenberg, J. M., Boguslavsky, D., Chistopolsky, I., Zakharov, I., & Dyakonova, V. (2025). Rest Induces a Distinct Transcriptional Program in the Nervous System of the Exercised L. stagnalis. International Journal of Molecular Sciences, 26(14), 6970. https://doi.org/10.3390/ijms26146970







