Salicylic Acid Treatment and Its Effect on Seed Yield and Seed Molecular Composition of Pisum sativum under Abiotic Stress
Abstract
1. Introduction
2. Results
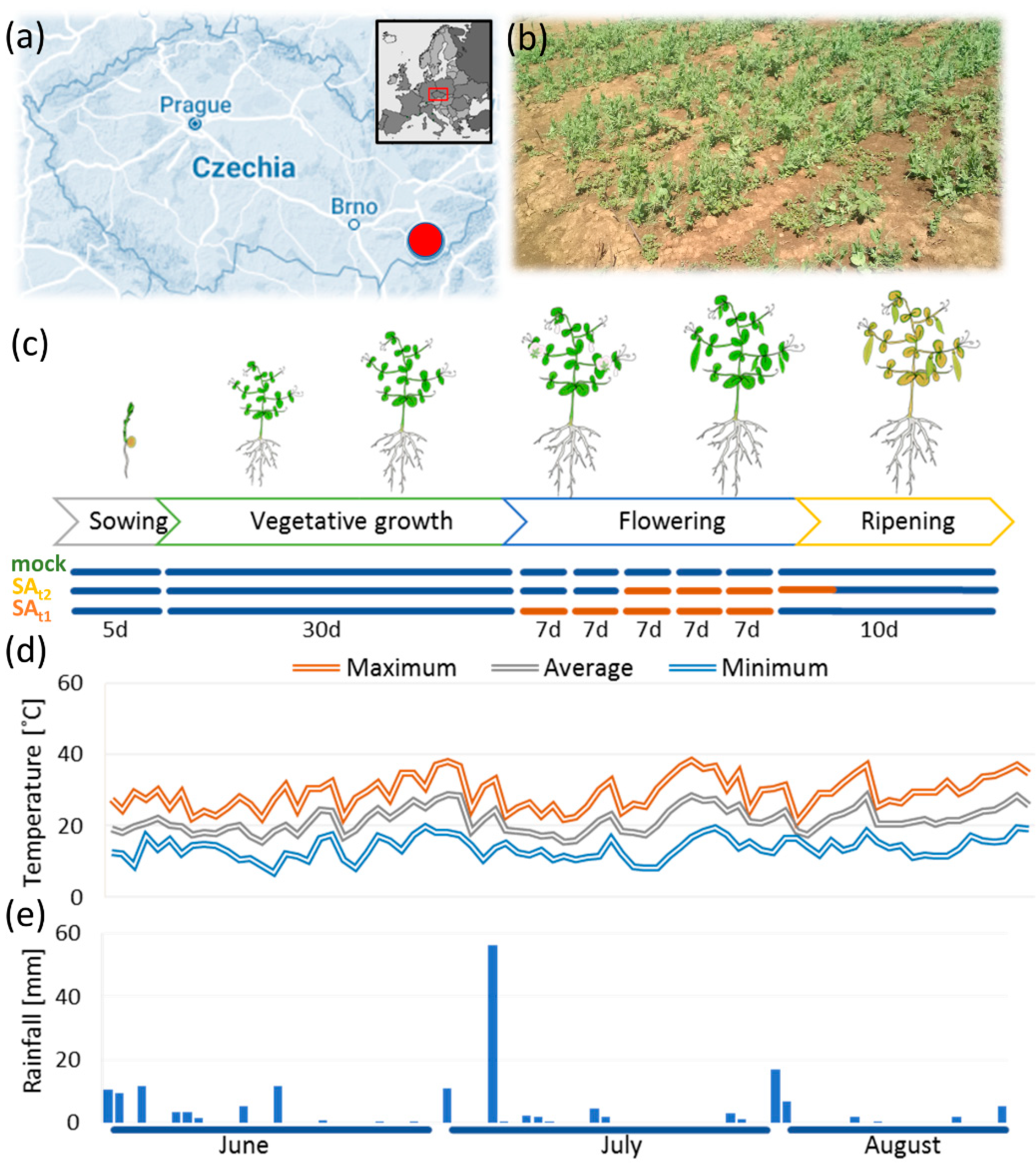
2.1. Field Experiment Results
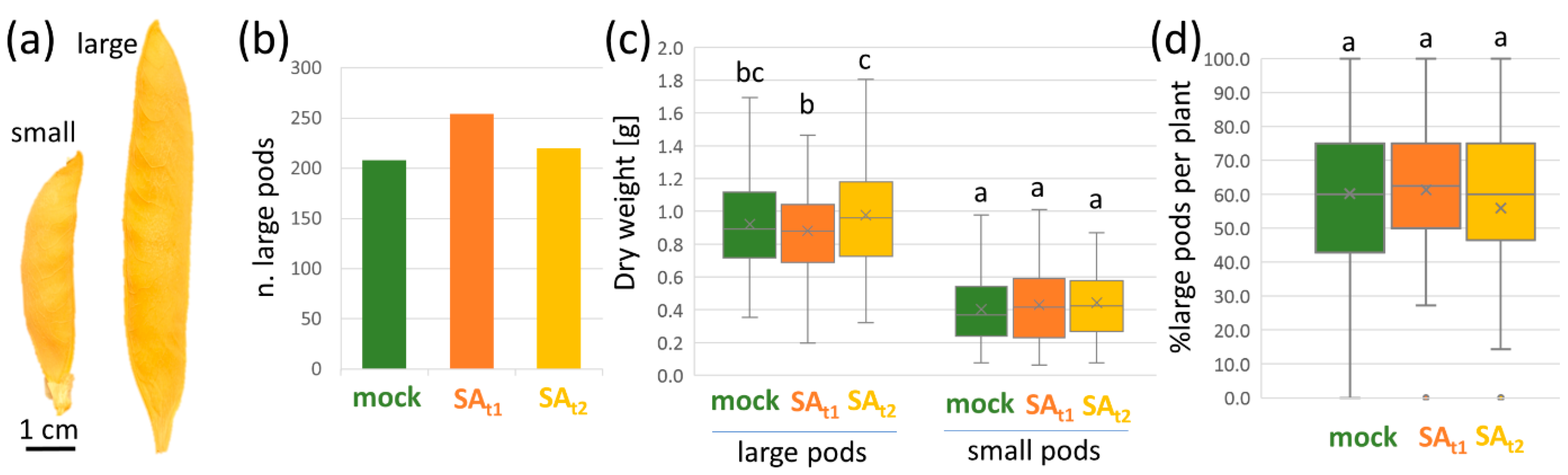
2.2. Salicylic Acid Treatment Increased the Mean Size of Seed Pods
2.3. Seed Proteome Analysis Did Not Show Striking Differences in Total Proteome Composition
2.4. Differentially Abundant Proteins Indicated a Decrease in Abscisic Acid Sensitivity
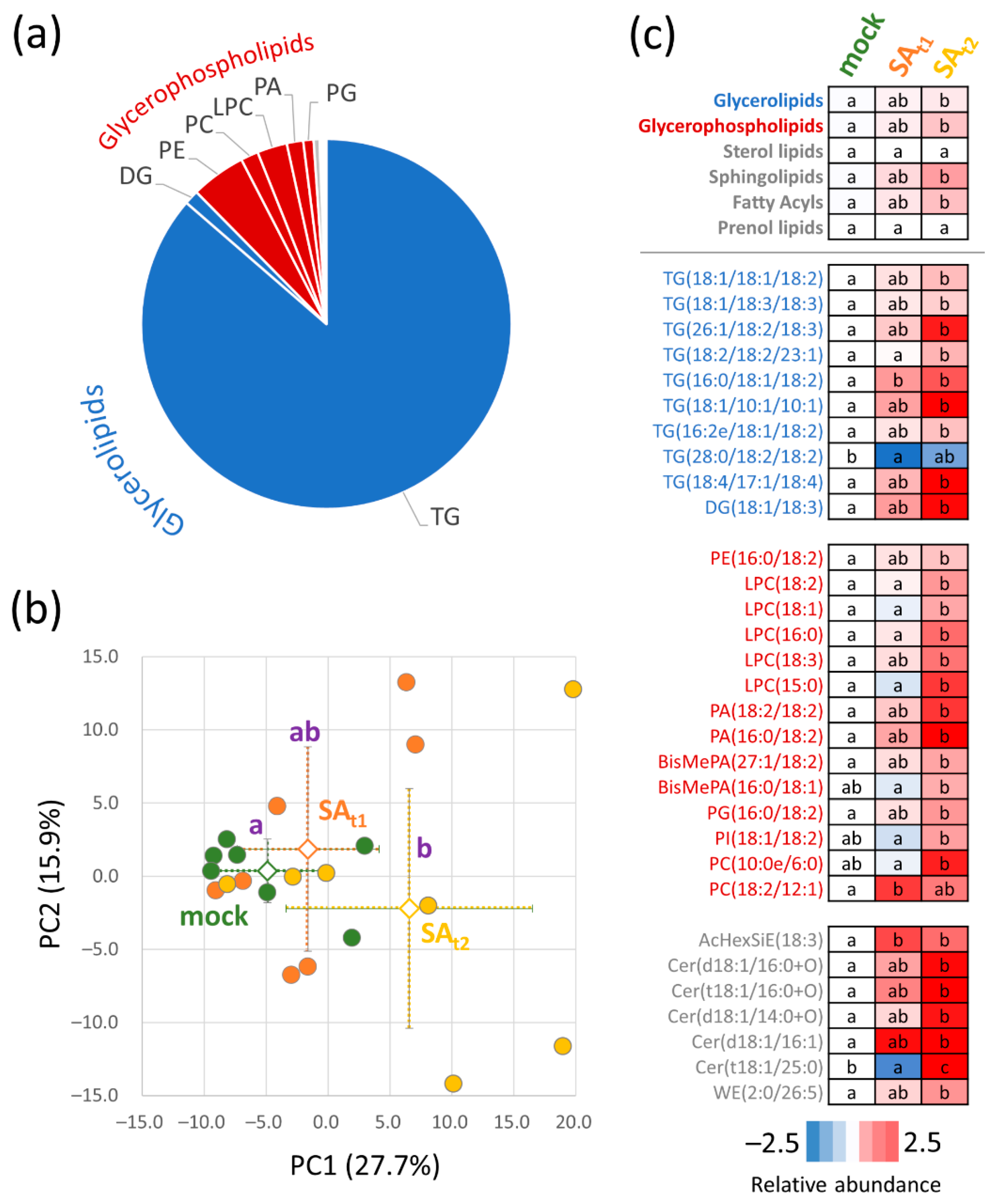
2.5. Seeds of Plants Treated with Salicylic Acid Showed a Higher Accumulation of Storage Lipids
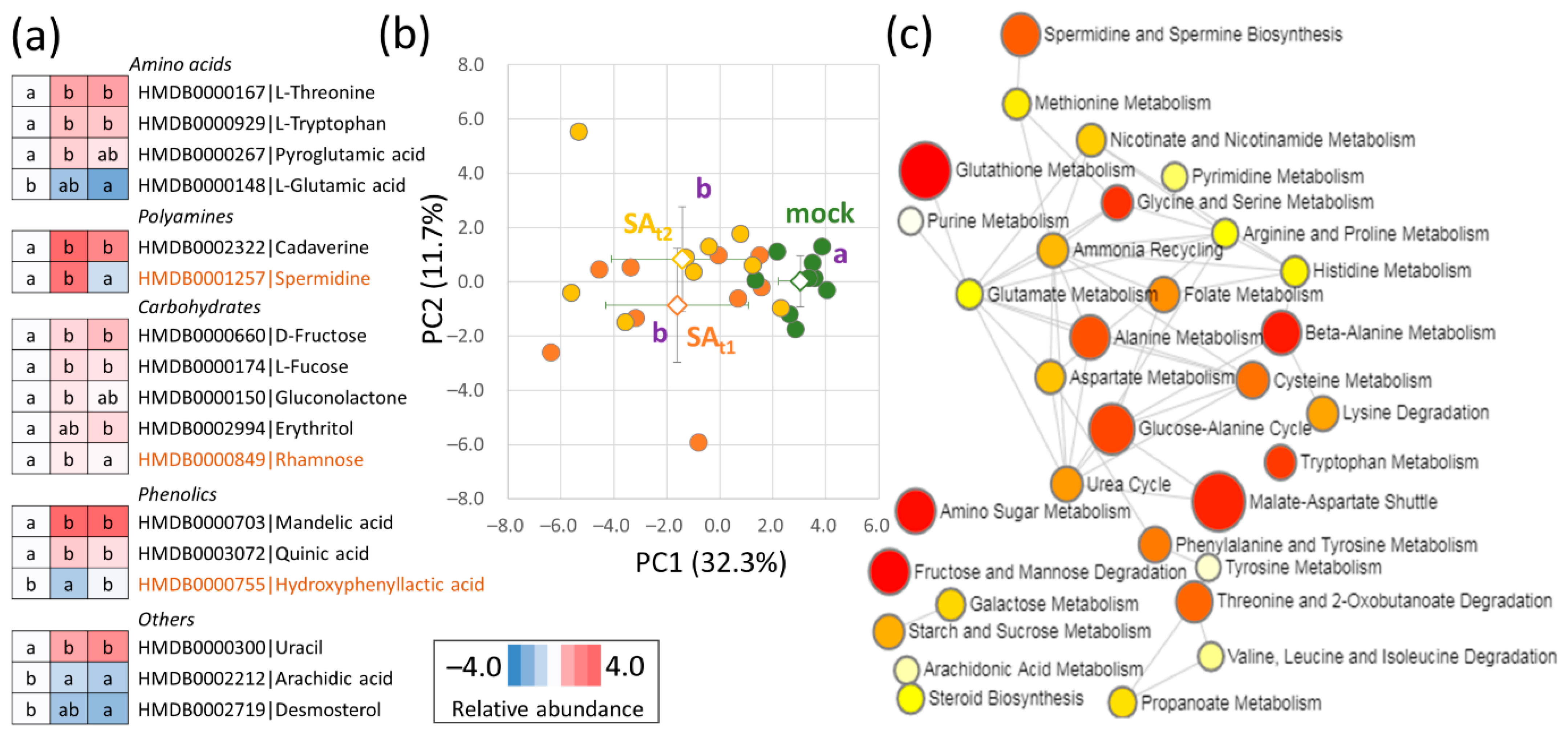
2.6. The Seed Metabolome Confirmed an Increase in Polyamine Production and Indicated the Accumulation of Phenolics
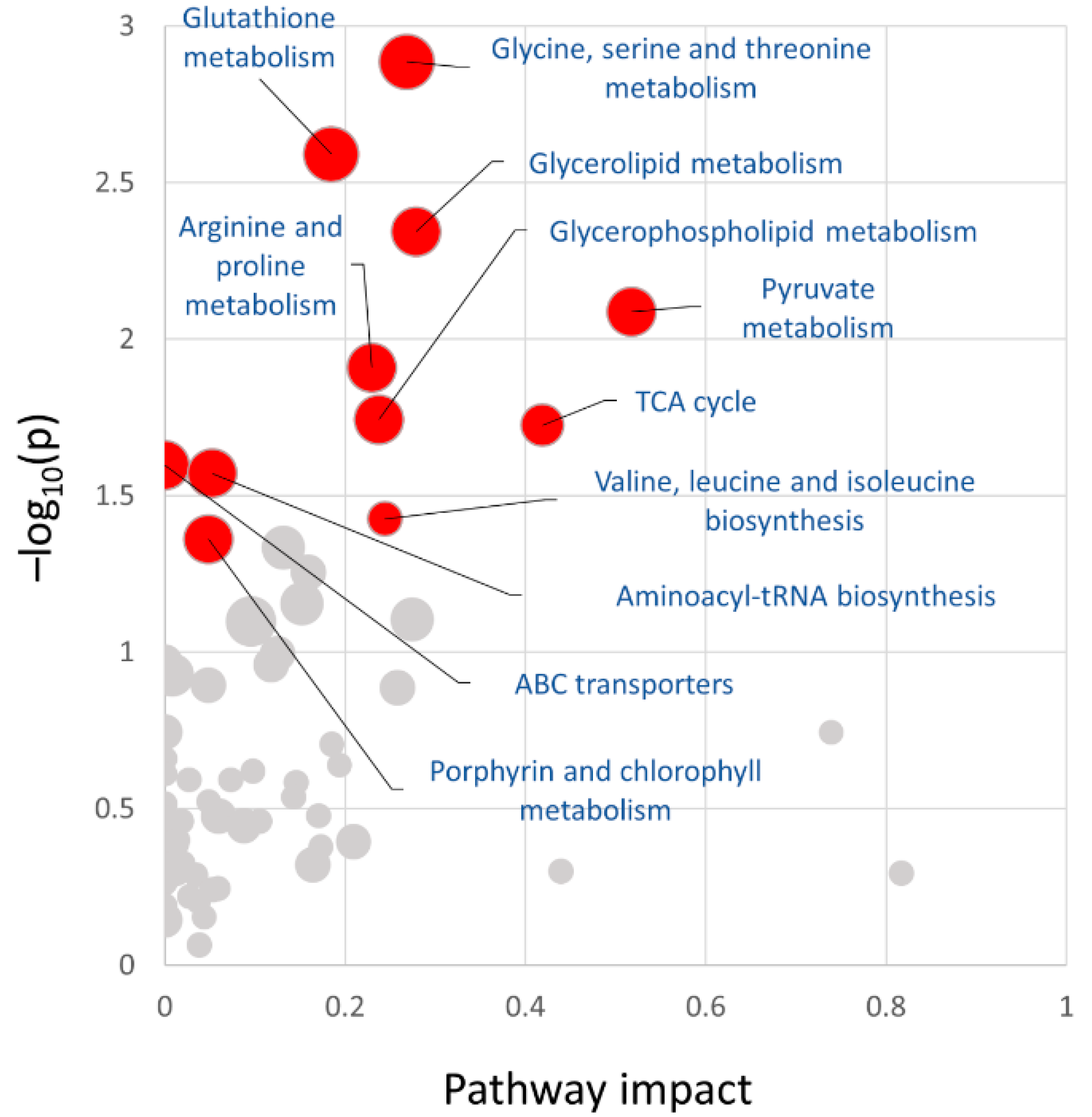
2.7. Integrative Analysis of Omics Data Indicated an Impact on the Citric Acid Cycle and Pyruvate Metabolism in Seeds of Plants Treated with Salicylic Acid
3. Discussion
3.1. Positive Effects of Salicylic Acid Treatment on Seed Yield
3.2. Polyamine Metabolism Could Correlate with the Increase in Number of Large Seed Pods
3.3. Putative Protein Candidates Responsible for a Better Performance of SAt2 Plants
3.4. Putative Lipid Candidates Responsible for a Better Performance of SAt2 Plants
4. Materials and Methods
4.1. Plant Material and Cultivation
4.2. Proteome Analysis
4.3. Metabolome Analysis
4.4. Lipidome Analysis
4.5. Data Analysis and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sita, K.; Sehgal, A.; HanumanthaRao, B.; Nair, R.M.; Vara Prasad, P.V.; Kumar, S.; Gaur, P.M.; Farooq, M.; Siddique, K.H.M.; Varshney, R.K.; et al. Food legumes and rising temperatures: Effects, adaptive functional mechanisms specific to reproductive growth stage and strategies to improve heat tolerance. Front. Plant Sci. 2017, 8, 1658. [Google Scholar] [CrossRef]
- Barnabás, B.; Jäger, K.; Fehér, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008, 31, 11–38. [Google Scholar] [CrossRef]
- Osorio, E.E.; Davis, A.R.; Bueckert, R.A. High temperatures disturb ovule development in field pea (Pisum sativum). Botany 2022, 100, 47–61. [Google Scholar] [CrossRef]
- Mohapatra, C.; Chand, R.; Tiwari, J.K.; Singh, A.K. Effect of heat stress during flowering and pod formation in pea (Pisum sativum L.). Physiol. Mol. Biol. Plants 2020, 26, 1119–1125. [Google Scholar] [CrossRef]
- Lamichaney, A.; Parihar, A.K.; Hazra, K.K.; Dixit, G.P.; Katiyar, P.K.; Singh, D.; Singh, A.K.; Kumar, N.; Singh, N.P. Untangling the influence of heat stress on crop phenology, seed set, seed weight, and germination in field pea (Pisum sativum L.). Front. Plant Sci. 2021, 12, 635868. [Google Scholar] [CrossRef]
- Shi, M.; Wang, C.; Wang, P.; Zhang, M.; Liao, W. Methylation in DNA, histone, and RNA during flowering under stress condition: A review. Plant Sci. 2022, 324, 111431. [Google Scholar] [CrossRef]
- Chakraborty, A.; Chaudhury, R.; Dutta, S.; Basak, M.; Dey, S.; Schäffner, A.R.; Das, M. Role of metabolites in flower development and discovery of compounds controlling flowering time. Plant Physiol. Biochem. 2022, 190, 109–118. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Q.; Li, J.; Yan, X.; He, H.; Gao, X.; Jia, G. High temperature in the root zone repressed flowering in Lilium × formolongi by disturbing the photoperiodic pathway and reconfiguring hormones and primary metabolism. Environ. Exp. Bot. 2021, 192, 104644. [Google Scholar] [CrossRef]
- Takeno, K. Stress-induced flowering: The third category of flowering response. J. Exp. Bot. 2016, 67, 4925–4934. [Google Scholar] [CrossRef]
- Yamada, M.; Takeno, K. Stress and salicylic acid induce the expression of PnFT2 in the regulation of the stress-induced flowering of Pharbitis nil. J. Plant Physiol. 2014, 171, 205–212. [Google Scholar] [CrossRef]
- Banday, Z.Z.; Nandi, A.K. Interconnection between flowering time control and activation of systemic acquired resistance. Front. Plant Sci. 2015, 6, 174. [Google Scholar] [CrossRef]
- Alonso-Ramírez, A.; Rodríguez, D.; Reyes, D.; Jiménez, J.A.; Nicolás, G.; López-Climent, M.; Gómez-Cadenas, A.; Nicolás, C. Evidence for a role of gibberellins in salicylic acid-modulated early plant responses to abiotic stress in Arabidopsis seeds. Plant Physiol. 2009, 150, 1335–1344. [Google Scholar] [CrossRef]
- Lin, K.-H.; Huang, S.-B.; Wu, C.-W.; Chang, Y.-S. Effects of salicylic acid and calcium chloride on heat tolerance of Poinsettia. HortScience 2019, 54, 499–504. [Google Scholar] [CrossRef]
- Wassie, M.; Zhang, W.; Zhang, Q.; Ji, K.; Cao, L.; Chen, L. Exogenous salicylic acid ameliorates heat stress-induced damages and improves growth and photosynthetic efficiency in alfalfa (Medicago sativa L.). Ecotoxicol. Environ. Saf. 2020, 191, 110206. [Google Scholar] [CrossRef]
- Fan, Y.; Lv, Z.; Li, Y.; Qin, B.; Song, Q.; Ma, L.; Wu, Q.; Zhang, W.; Ma, S.; Ma, C.; et al. Salicylic acid reduces wheat yield loss caused by high temperature stress by enhancing the photosynthetic performance of the flag leaves. Agronomy 2022, 12, 1386. [Google Scholar] [CrossRef]
- Szalai, G.; Horgosi, S.; Soós, V.; Majláth, I.; Balázs, E.; Janda, T. Salicylic acid treatment of pea seeds induces its de novo synthesis. J. Plant Physiol. 2011, 168, 213–219. [Google Scholar] [CrossRef]
- Meitzel, T.; Radchuk, R.; McAdam, E.L.; Thormählen, I.; Feil, R.; Munz, E.; Hilo, A.; Geigenberger, P.; Ross, J.J.; Lunn, J.E.; et al. Trehalose 6-phosphate promotes seed filling by activating auxin biosynthesis. New Phytol. 2021, 229, 1553–1565. [Google Scholar] [CrossRef]
- Hartmann, M.; Zeier, J. l-lysine metabolism to N-hydroxypipecolic acid: An integral immune-activating pathway in plants. Plant J. 2018, 96, 5–21. [Google Scholar] [CrossRef]
- Bhaskara, G.B.; Nguyen, T.T.; Verslues, P.E. Unique drought resistance functions of the highly ABA-induced clade a protein phosphatase 2cs. Plant Physiol. 2012, 160, 379–395. [Google Scholar] [CrossRef]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT and TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Chowdhury, Z.; Mohanty, D.; Giri, M.K.; Venables, B.J.; Chaturvedi, R.; Chao, A.; Petros, R.A.; Shah, J. Dehydroabietinal promotes flowering time and plant defense in Arabidopsis via the autonomous pathway genes FLOWERING LOCUS D, FVE, and RELATIVE OF EARLY FLOWERING 6. J. Exp. Bot. 2020, 71, 4903–4913. [Google Scholar] [CrossRef]
- Chen, T.; Cui, P.; Chen, H.; Ali, S.; Zhang, S.; Xiong, L. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 2013, 9, e1003875. [Google Scholar] [CrossRef]
- Kim, Y.-O.; Kim, J.S.; Kang, H. Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 2005, 42, 890–900. [Google Scholar] [CrossRef]
- Zhang, H.; Ransom, C.; Ludwig, P.; van Nocker, S. Genetic analysis of early flowering mutants in arabidopsis defines a class of pleiotropic developmental regulator required for expression of the flowering-time switch flowering locus C. Genetics 2003, 164, 347–358. [Google Scholar] [CrossRef]
- Murphy, A.M.; Otto, B.; Brearley, C.A.; Carr, J.P.; Hanke, D.E. A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 2008, 56, 638–652. [Google Scholar] [CrossRef]
- Černý, M.; Novák, J.; Habánová, H.; Cerna, H.; Brzobohatý, B. Role of the proteome in phytohormonal signaling. Biochim. Biophys. Acta-Proteins Proteom. 2016, 1864, 1003–1015. [Google Scholar] [CrossRef]
- Zhang, C.X.; Feng, B.H.; Chen, T.T.; Zhang, X.F.; Tao, L.X.; Fu, G.F. Sugars, antioxidant enzymes and IAA mediate salicylic acid to prevent rice spikelet degeneration caused by heat stress. Plant Growth Regul. 2017, 83, 313–323. [Google Scholar] [CrossRef]
- Liu, H.-T.; Huang, W.-D.; Pan, Q.-H.; Weng, F.-H.; Zhan, J.-C.; Liu, Y.; Wan, S.-B.; Liu, Y.-Y. Contributions of PIP2-specific-phospholipase C and free salicylic acid to heat acclimation-induced thermotolerance in pea leaves. J. Plant Physiol. 2006, 163, 405–416. [Google Scholar] [CrossRef]
- Cingoz, G.S.; Gurel, E. Effects of salicylic acid on thermotolerance and cardenolide accumulation under high temperature stress in Digitalis trojana Ivanina. Plant Physiol. Biochem. 2016, 105, 145–149. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Savka, M.A.; Hudson, A.O. The Synthesis and Role of β-Alanine in Plants. Front. Plant Sci. 2019, 10, 921. [Google Scholar] [CrossRef]
- Latrasse, D.; Germann, S.; Houba-Hérin, N.; Dubois, E.; Bui-Prodhomme, D.; Hourcade, D.; Juul-Jensen, T.; Le Roux, C.; Majira, A.; Simoncello, N.; et al. Control of flowering and cell fate by LIF2, an RNA binding partner of the polycomb complex component LHP1. PLoS ONE 2011, 6, e16592. [Google Scholar] [CrossRef]
- Le Roux, C.; Del Prete, S.; Boutet-Mercey, S.; Perreau, F.; Balagué, C.; Roby, D.; Fagard, M.; Gaudin, V. The hnRNP-Q protein LIF2 participates in the plant immune response. PLoS ONE 2014, 9, e99343. [Google Scholar] [CrossRef]
- Joshi, V.; Joung, J.-G.; Fei, Z.; Jander, G. Interdependence of threonine, methionine and isoleucine metabolism in plants: Accumulation and transcriptional regulation under abiotic stress. Amino Acids 2010, 39, 933–947. [Google Scholar] [CrossRef]
- Sechet, J.; Marion-Poll, A.; North, H. Emerging Functions for cell wall polysaccharides accumulated during eudicot seed development. Plants 2018, 7, 81. [Google Scholar] [CrossRef]
- Dufková, H.; Berka, M.; Psota, V.; Brzobohatý, B.; Černý, M. Environmental impacts on barley grain composition and longevity. J. Exp. Bot. 2023, erac498. [Google Scholar] [CrossRef]
- Song, C.-P.; Galbraith, D.W. AtSAP18, An orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 2006, 60, 241–257. [Google Scholar] [CrossRef]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.N.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef]
- Virdi, A.S.; Singh, S.; Singh, P. Abiotic stress responses in plants: Roles of calmodulin-regulated proteins. Front. Plant Sci. 2015, 6, 809. [Google Scholar] [CrossRef]
- Raina, M.; Kisku, A.V.; Joon, S.; Kumar, S.; Kumar, D. Calmodulin and calmodulin-like Ca2+ binding proteins as molecular players of abiotic stress response in plants. In Calcium Transport Elements in Plants; Elsevier: Amsterdam, The Netherlands, 2021; pp. 231–248. [Google Scholar]
- Donahue, J.L.; Alford, S.R.; Torabinejad, J.; Kerwin, R.E.; Nourbakhsh, A.; Ray, W.K.; Hernick, M.; Huang, X.; Lyons, B.M.; Hein, P.P.; et al. The Arabidopsis thaliana myo- Inositol 1-Phosphate Synthase 1 gene is required for myo -inositol synthesis and suppression of cell death. Plant Cell 2010, 22, 888–903. [Google Scholar] [CrossRef]
- Liang, Y.; Huang, Y.; Liu, C.; Chen, K.; Li, M. Functions and interaction of plant lipid signaling under abiotic stresses. Plant Biol. 2023. [Google Scholar] [CrossRef]
- Zienkiewicz, K.; Zienkiewicz, A. Degradation of lipid droplets in plants and algae-right time, many paths, one goal. Front. Plant Sci. 2020, 11, 579019. [Google Scholar] [CrossRef] [PubMed]
- Thürmer, M.; Gollowitzer, A.; Pein, H.; Neukirch, K.; Gelmez, E.; Waltl, L.; Wielsch, N.; Winkler, R.; Löser, K.; Grander, J.; et al. PI(18:1/18:1) is a SCD1-derived lipokine that limits stress signaling. Nat. Commun. 2022, 13, 2982. [Google Scholar] [CrossRef] [PubMed]
- Hallmark, H.T.; Černý, M.; Brzobohatý, B.; Rashotte, A.M.A.M. trans-Zeatin-N-glucosides have biological activity in Arabidopsis thaliana. PLoS ONE 2020, 15, e0232762. [Google Scholar] [CrossRef]
- Dufková, H.; Berka, M.; Luklová, M.; Rashotte, A.M.; Brzobohatý, B.; Černý, M. Eggplant germination is promoted by hydrogen peroxide and temperature in an independent but overlapping manner. Molecules 2019, 24, 4270. [Google Scholar] [CrossRef] [PubMed]
- Luklová, M.; Novák, J.; Kopecká, R.; Kameniarová, M.; Gibasová, V.; Brzobohatý, B.; Černý, M. Phytochromes and their role in diurnal variations of ROS metabolism and plant proteome. Int. J. Mol. Sci. 2022, 23, 14134. [Google Scholar] [CrossRef]
- Kreplak, J.; Madoui, M.-A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Dorfer, V.; Pichler, P.; Stranzl, T.; Stadlmann, J.; Taus, T.; Winkler, S.; Mechtler, K. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J. Proteome Res. 2014, 13, 3679–3684. [Google Scholar] [CrossRef]
- Berka, M.; Luklová, M.; Dufková, H.; Berková, V.; Novák, J.; Saiz-Fernández, I.; Rashotte, A.M.; Brzobohaty, B.; Cerny, M. Barley root proteome and metabolome in response to cytokinin and abiotic stimuli. Front. Plant Sci. 2020, 11, 590337. [Google Scholar] [CrossRef]
- Berková, V.; Berka, M.; Griga, M.; Kopecká, R.; Prokopová, M.; Luklová, M.; Horáček, J.; Smýkalová, I.; Čičmanec, P.; Novák, J.; et al. Molecular mechanisms underlying flax (Linum usitatissimum L.) tolerance to cadmium: A case study of proteome and metabolome of four different flax genotypes. Plants 2022, 11, 2931. [Google Scholar] [CrossRef]
- Hampejsová, R.; Berka, M.; Berková, V.; Jersáková, J.; Domkářová, J.; von Rundstedt, F.; Frary, A.; Saiz-Fernández, I.; Brzobohatý, B.; Černý, M. Interaction with fungi promotes the accumulation of specific defense molecules in orchid tubers and may increase the value of tubers for biotechnological and medicinal applications: The case study of interaction between Dactylorhiza sp. and Tulasnella calospora. Front. Plant Sci. 2022, 13, 757852. [Google Scholar] [CrossRef]
- Pino, L.K.; Searle, B.C.; Bollinger, J.G.; Nunn, B.; MacLean, B.; MacCoss, M.J. The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev. 2020, 39, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Dufková, H.; Berka, M.; Greplová, M.; Shejbalová, Š.; Hampejsová, R.; Luklová, M.; Domkářová, J.; Novák, J.; Kopačka, V.; Brzobohatý, B.; et al. The omics hunt for novel molecular markers of resistance to Phytophthora infestans. Plants 2022, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Černý, M.; Berka, M.; Dvořák, M.; Milenković, I.; Saiz-Fernández, I.; Brzobohatý, B.; Ďurkovič, J. Defense mechanisms promoting tolerance to aggressive Phytophthora species in hybrid poplar. Front. Plant Sci. 2022, 13, 1018272. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Liebermeister, W.; Noor, E.; Flamholz, A.; Davidi, D.; Bernhardt, J.; Milo, R. Visual account of protein investment in cellular functions. Proc. Natl. Acad. Sci. USA 2014, 111, 8488–8493. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Hewapathirana, S.; García-Seisdedos, D.; Kamatchinathan, S.; Kundu, D.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A Hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berková, V.; Berka, M.; Kameniarová, M.; Kopecká, R.; Kuzmenko, M.; Shejbalová, Š.; Abramov, D.; Čičmanec, P.; Frejlichová, L.; Jan, N.; et al. Salicylic Acid Treatment and Its Effect on Seed Yield and Seed Molecular Composition of Pisum sativum under Abiotic Stress. Int. J. Mol. Sci. 2023, 24, 5454. https://doi.org/10.3390/ijms24065454
Berková V, Berka M, Kameniarová M, Kopecká R, Kuzmenko M, Shejbalová Š, Abramov D, Čičmanec P, Frejlichová L, Jan N, et al. Salicylic Acid Treatment and Its Effect on Seed Yield and Seed Molecular Composition of Pisum sativum under Abiotic Stress. International Journal of Molecular Sciences. 2023; 24(6):5454. https://doi.org/10.3390/ijms24065454
Chicago/Turabian StyleBerková, Veronika, Miroslav Berka, Michaela Kameniarová, Romana Kopecká, Marharyta Kuzmenko, Šarlota Shejbalová, Dmytro Abramov, Petr Čičmanec, Lucie Frejlichová, Novák Jan, and et al. 2023. "Salicylic Acid Treatment and Its Effect on Seed Yield and Seed Molecular Composition of Pisum sativum under Abiotic Stress" International Journal of Molecular Sciences 24, no. 6: 5454. https://doi.org/10.3390/ijms24065454
APA StyleBerková, V., Berka, M., Kameniarová, M., Kopecká, R., Kuzmenko, M., Shejbalová, Š., Abramov, D., Čičmanec, P., Frejlichová, L., Jan, N., Brzobohatý, B., & Černý, M. (2023). Salicylic Acid Treatment and Its Effect on Seed Yield and Seed Molecular Composition of Pisum sativum under Abiotic Stress. International Journal of Molecular Sciences, 24(6), 5454. https://doi.org/10.3390/ijms24065454