Role of Monomer/Tetramer Equilibrium of Rod Visual Arrestin in the Interaction with Phosphorylated Rhodopsin
Abstract
1. Introduction
2. Results
2.1. Light-Induced Structural Change of Phosphorylated Rhodopsin
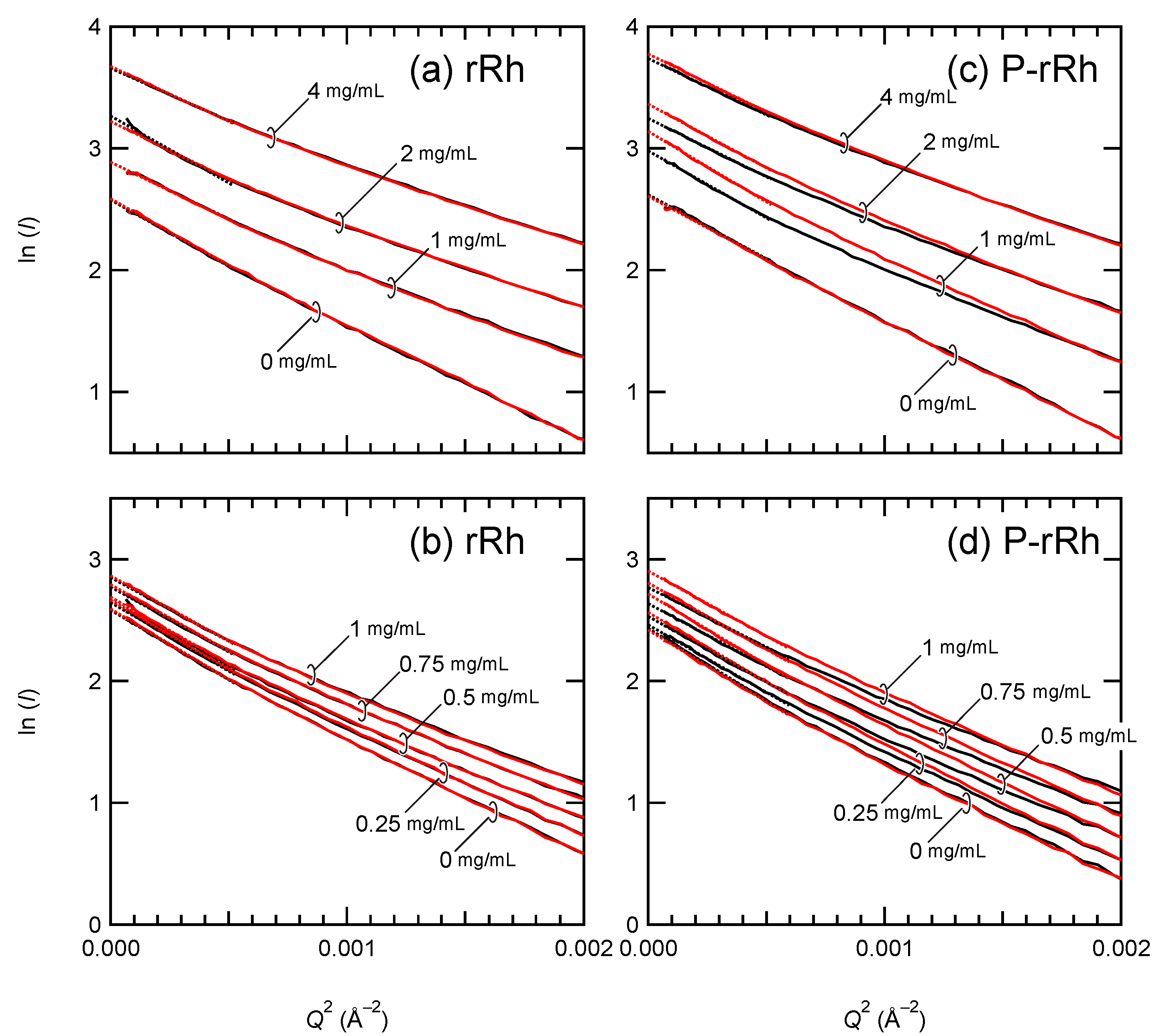
2.2. Small-Angle X-ray Scattering of Nanodiscs
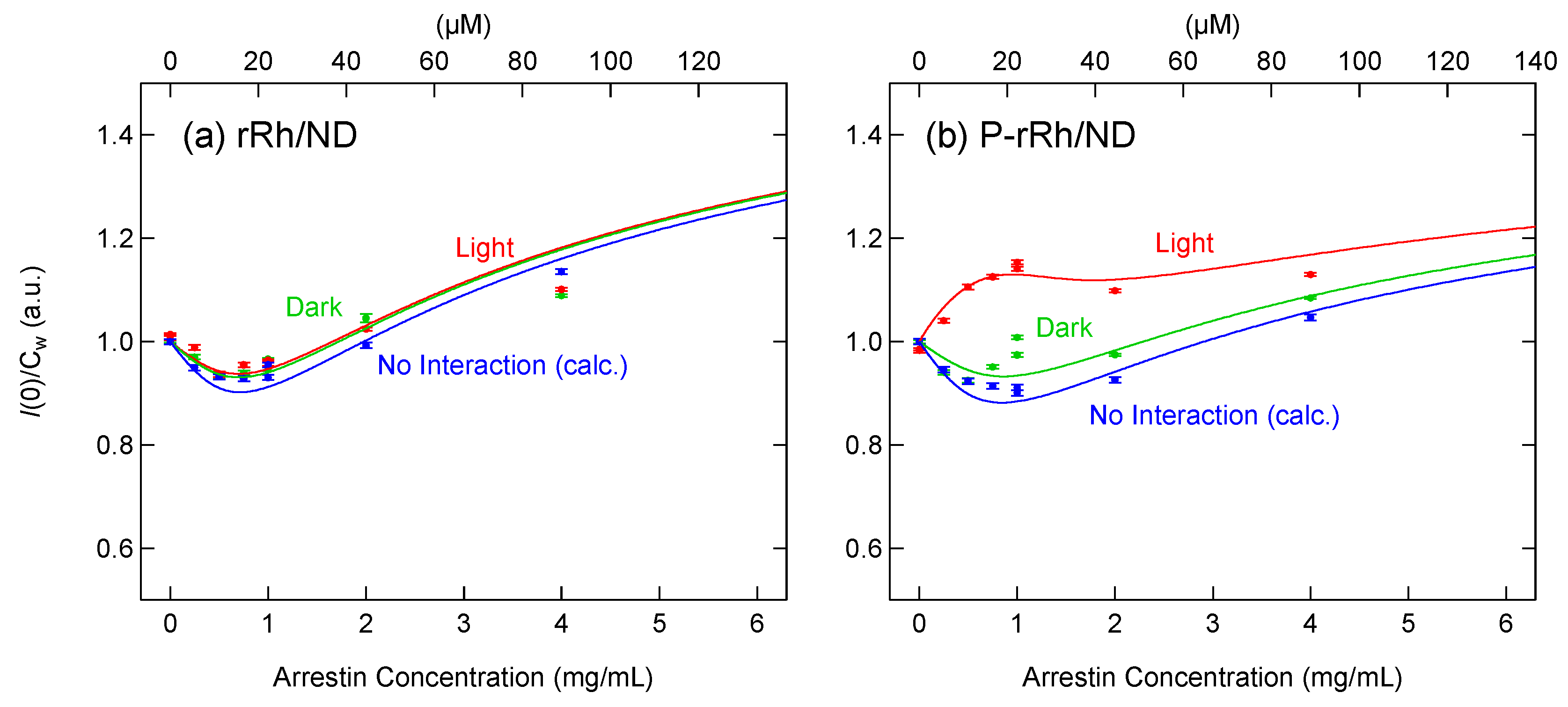
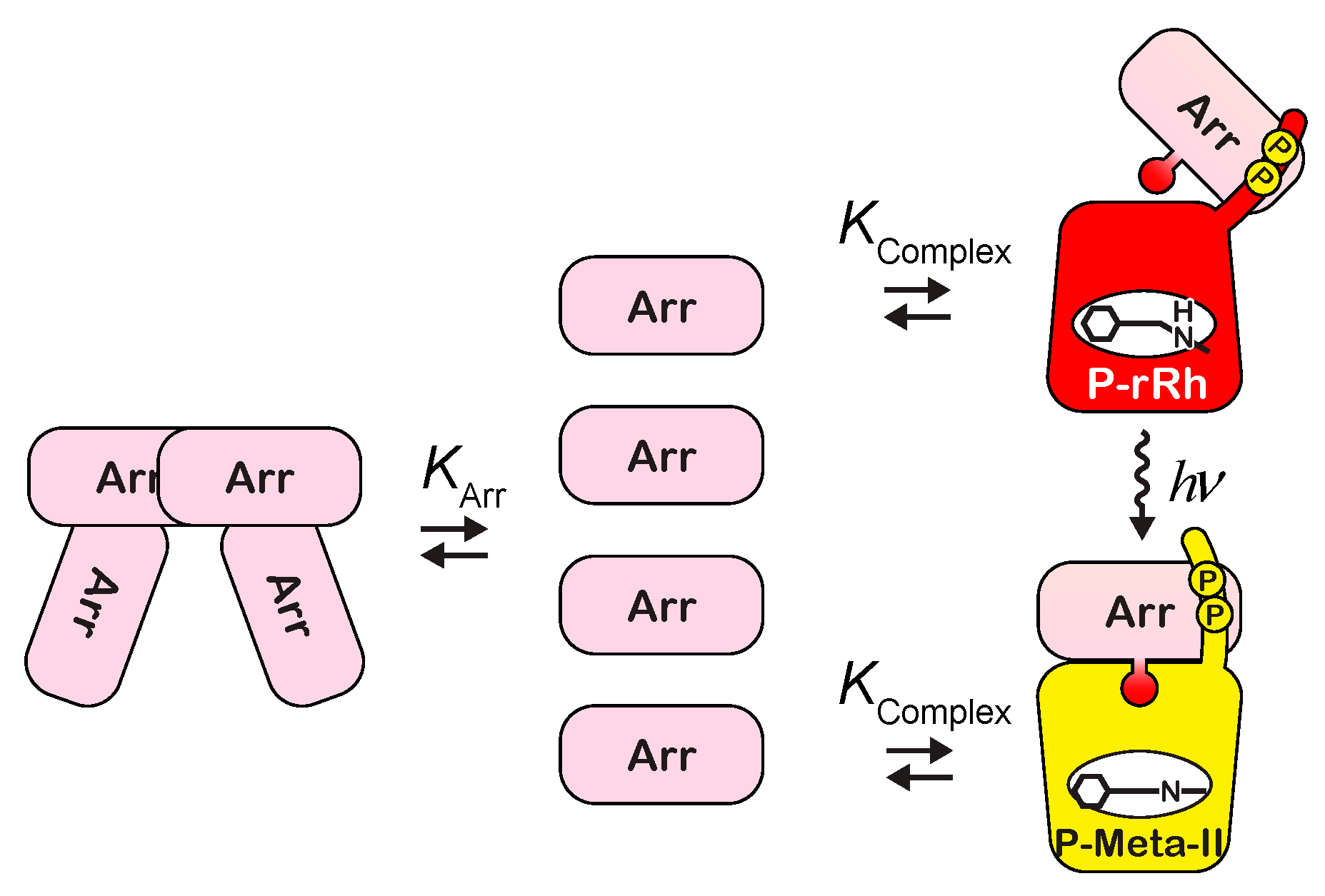
2.3. Formation of the Complex of Photoactivated Rhodopsin and Arrestin
2.4. Formation of Extra-Meta-II by Binding of Arrestin, Detected by UV-Visible Spectroscopy
3. Discussion
3.1. Interaction between Rhodopsin and Arrestin
3.2. Solution X-ray Scattering Experiments for the Analysis of Signal Transduction
4. Materials and Methods
4.1. Sample Preparation
4.2. Analysis of Phosphorylation Levels
4.3. Solution X-ray Scattering
4.4. SAXS Data Analysis
4.5. Time-Resolved UV-Visible Spectroscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kobilka, B. The structural basis of G-protein-coupled receptor signaling (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 2013, 52, 6380–6388. [Google Scholar] [CrossRef] [PubMed]
- Deupi, X.; Standfuss, J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr. Opin. Struct. Biol. 2011, 21, 541–551. [Google Scholar] [CrossRef]
- Gurevich, V.V.; Gurevich, E.V. The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol. Ther. 2006, 110, 465–502. [Google Scholar] [CrossRef] [PubMed]
- Terakita, A. The opsins. Genome Biol. 2005, 6, 213. [Google Scholar] [CrossRef]
- Sutton, R.B.; Vishnivetskiy, S.A.; Robert, J.; Hanson, S.M.; Raman, D.; Knox, B.E.; Kono, M.; Navarro, J.; Gurevich, V.V. Crystal structure of cone arrestin at 2.3 Å: Evolution of receptor specificity. J. Mol. Biol. 2005, 354, 1069–1080. [Google Scholar] [CrossRef]
- Granzin, J.; Wilden, U.; Choe, H.W.; Labahn, J.; Krafft, B.; Büldt, G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature 1998, 391, 918–921. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, J.A.; Schubert, C.; Gurevich, V.V.; Sigler, P.B. The 2.8 Å crystal structure of visual arrestin: A model for arrestin’s regulation. Cell 1999, 97, 257–269. [Google Scholar] [CrossRef]
- Imamoto, Y.; Tamura, C.; Kamikubo, H.; Kataoka, M. Concentration-dependent tetramerization of bovine visual arrestin. Biophys. J. 2003, 85, 1186–1195. [Google Scholar] [CrossRef]
- Hanson, S.M.; Vishnivetskiy, S.A.; Hubbell, W.L.; Gurevich, V.V. Opposing effects of inositol hexakisphosphate on rod arrestin and arrestin2 self-association. Biochemistry 2008, 47, 1070–1075. [Google Scholar] [CrossRef]
- Chen, Q.; Zhuo, Y.; Kim, M.; Hanson, S.M.; Francis, D.J.; Vishnivetskiy, S.A.; Altenbach, C.; Klug, C.S.; Hubbell, W.L.; Gurevich, V.V. Self-association of arrestin family members. Handb. Exp. Pharmacol. 2014, 219, 205–223. [Google Scholar] [CrossRef]
- Kang, Y.; Zhou, X.E.; Gao, X.; He, Y.; Liu, W.; Ishchenko, A.; Barty, A.; White, T.A.; Yefanov, O.; Han, G.W.; et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 2015, 523, 561–567. [Google Scholar] [CrossRef]
- Hamm, H.E.; Bownds, M.D. Protein complement of rod outer segments of frog retina. Biochemistry 1986, 25, 4512–4523. [Google Scholar] [CrossRef]
- Kawamura, S. Phototransduction, excitation and adaptation. In Neurobiology and Clinical Aspects of the Outer Retina; Djamangoz, M.B.A., Arche, S.N., Vallerga, S., Eds.; Chapman & Hall: London, UK, 1995; pp. 105–131. [Google Scholar]
- Bayburt, T.H.; Grinkova, Y.V.; Sligar, S.G. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002, 2, 853–856. [Google Scholar] [CrossRef]
- Schnablegger, H.; Singh, Y. The SAXS Guide, 3rd ed.; Anton Paar: Graz, Austria, 2013. [Google Scholar]
- Imamoto, Y.; Kojima, K.; Oka, T.; Maeda, R.; Shichida, Y. Helical rearrangement of photoactivated rhodopsin in monomeric and dimeric forms probed by high-angle X-ray scattering. Photochem. Photobiol. Sci. 2015, 14, 1965–1973. [Google Scholar] [CrossRef]
- Imamoto, Y.; Kojima, K.; Oka, T.; Maeda, R.; Shichida, Y. Conformational differences among metarhodopsin I, metarhodopsin II, and opsin probed by wide-angle X-ray scattering. J. Phys. Chem. B 2019, 123, 9134–9142. [Google Scholar] [CrossRef]
- Choe, H.W.; Kim, Y.J.; Park, J.H.; Morizumi, T.; Pai, E.F.; Krauss, N.; Hofmann, K.P.; Scheerer, P.; Ernst, O.P. Crystal structure of metarhodopsin II. Nature 2011, 471, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Kuybeda, O.; de Waal, P.W.; Mukherjee, S.; Van Eps, N.; Dutka, P.; Zhou, X.E.; Bartesaghi, A.; Erramilli, S.; Morizumi, T.; et al. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 2018, 558, 553–558. [Google Scholar] [CrossRef]
- Kelleher, D.J.; Johnson, G.L. Phosphorylation of rhodopsin by protein kinase C in vitro. J. Biol. Chem. 1986, 261, 4749–4757. [Google Scholar] [CrossRef]
- Malmerberg, E.; Bovee-Geurts, P.H.M.; Katona, G.; Deupi, X.; Arnlund, D.; Wickstrand, C.; Johansson, L.C.; Westenhoff, S.; Nazarenko, E.; Schertler, G.F.X.; et al. Conformational activation of visual rhodopsin in native disc membranes. Sci. Signal. 2015, 8, ra26. [Google Scholar] [CrossRef] [PubMed]
- Beyriere, F.; Sommer, M.E.; Szczepek, M.; Bartl, F.J.; Hofmann, K.P.; Heck, M.; Ritter, E. Formation and decay of the arrestin·rhodopsin complex in native disc membranes. J. Biol. Chem. 2015, 290, 12919–12928. [Google Scholar] [CrossRef] [PubMed]
- Svergun, D.I.; Baberato, C.; Koch, M.H.J. CRYSOL-a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
- Okada, T.; Sugihara, M.; Bondar, A.N.; Elstner, M.; Entel, P.; Buss, V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 2004, 342, 571–583. [Google Scholar] [CrossRef]
- Tsukamoto, H.; Sinha, A.; DeWitt, M.; Farrens, D.L. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J. Mol. Biol. 2010, 399, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Grinkova, Y.V.; Lazarides, A.A.; Sligar, S.G. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 2004, 126, 3477–3487. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Grinkova, Y.V.; Sligar, S.G. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch. Biochem. Biophys. 2006, 450, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Bayburt, T.H.; Leitz, A.J.; Xie, G.; Oprian, D.D.; Sligar, S.G. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J. Biol. Chem. 2007, 282, 14875–14881. [Google Scholar] [CrossRef] [PubMed]
- Szundi, I.; Funatogawa, C.; Guo, Y.; Yan, E.C.Y.; Kliger, D.S. Protein sequence and membrane lipid roles in the activation kinetics of bovine and human rhodopsins. Biophys. J. 2017, 113, 1934–1944. [Google Scholar] [CrossRef] [PubMed]
- Strissel, K.J.; Sokolov, M.; Trieu, L.H.; Arshavsky, V.Y. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J. Neurosci. 2006, 26, 1146–1153. [Google Scholar] [CrossRef]
- Mendez, A.; Lem, J.; Simon, M.; Chen, J. Light-dependent translocation of arrestin in the absence of rhodopsin phosphorylation and transducin signaling. J. Neurosci. 2003, 23, 3124–3129. [Google Scholar] [CrossRef]
- Nair, K.S.; Hanson, S.M.; Mendez, A.; Gurevich, E.V.; Kennedy, M.J.; Shestopalov, V.I.; Vishnivetskiy, S.A.; Chen, J.; Hurley, J.B.; Gurevich, V.V.; et al. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron 2005, 46, 555–567. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, J.F.; Matsumoto, B.; Whelan, J.P.; Cao, W. Cytoskeleton participation in subcellular trafficking of signal transduction proteins in rod photoreceptor cells. J. Neurosci. Res. 2002, 67, 290–297. [Google Scholar] [CrossRef]
- Shukla, A.K.; Manglik, A.; Kruse, A.C.; Xiao, K.; Reis, R.I.; Tseng, W.C.; Staus, D.P.; Hilger, D.; Uysal, S.; Huang, L.Y.; et al. Structure of active b-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 2013, 497, 137–141. [Google Scholar] [CrossRef]
- Kim, Y.J.; Hofmann, K.P.; Ernst, O.P.; Scheerer, P.; Choe, H.W.; Sommer, M.E. Crystal structure of pre-activated arrestin p44. Nature 2013, 497, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K.; Westfield, G.H.; Xiao, K.; Reis, R.I.; Huang, L.Y.; Tripathi-Shukla, P.; Qian, J.; Li, S.; Blanc, A.; Oleskie, A.N.; et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 2014, 512, 218–222. [Google Scholar] [CrossRef]
- Feuerstein, S.E.; Pulvermüller, A.; Hartmann, R.; Granzin, J.; Stoldt, M.; Henklein, P.; Ernst, O.P.; Heck, M.; Willbold, D.; Koenig, B.W. Helix formation in arrestin accompanies recognition of photoactivated rhodopsin. Biochemistry 2009, 48, 10733–10742. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, V.V.; Benovic, J.L. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J. Biol. Chem. 1993, 268, 11628–11638. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.; Imamoto, Y.; Maeda, R.; Yamashita, T.; Shichida, Y. Rod visual pigment optimizes active state to achieve efficient G protein activation as compared with cone visual pigments. J. Biol. Chem. 2014, 289, 5061–5073. [Google Scholar] [CrossRef]
- Buczyłko, J.; Palczewski, K. Purification of arrestin from bovine retinas. Methods Neurosci. 1993, 15, 226–236. [Google Scholar]
- Puig, J.; Arendt, A.; Tomson, F.L.; Abdulaeva, G.; Miller, R.; Hargrave, P.A.; McDowell, J.H. Synthetic phosphopeptide from rhodopsin sequence induces retinal arrestin binding to photoactivated unphosphorylated rhodopsin. FEBS Lett. 1995, 362, 185–188. [Google Scholar] [CrossRef]
- Palczewski, K.; Buczyłko, J.; Kaplan, M.W.; Polans, A.S.; Crabb, J.W. Mechanism of rhodopsin kinase activation. J. Biol. Chem. 1991, 266, 12949–12955. [Google Scholar] [CrossRef]
- Lee, K.A.; Craven, K.B.; Niemi, G.A.; Hurley, J.B. Mass spectrometric analysis of the kinetics of in vivo rhodopsin phosphorylation. Protein Sci. 2002, 11, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Glatter, O.; Kratky, O. Small Angle X-ray Scattering; Academic Press: New York, NY, USA, 1982. [Google Scholar]
- Shen, Y.C.; Sasaki, T.; Matsuyama, T.; Yamashita, T.; Shichida, Y.; Okitsu, T.; Yamano, Y.; Wada, A.; Ishizuka, T.; Yawo, H.; et al. Red-tuning of the channelrhodopsin spectrum using long conjugated retinal analogues. Biochemistry 2018, 57, 5544–5556. [Google Scholar] [CrossRef] [PubMed]




| (µM) 1 | (µM) | |
|---|---|---|
| rRh/ND Dark | 41.5 | 211 |
| rRh/ND Light | 41.5 | 167 |
| P-rRh/ND Dark | 41.5 | 109 |
| P-rRh/ND Light | 41.5 | 7.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imamoto, Y.; Kojima, K.; Maeda, R.; Shichida, Y.; Oka, T. Role of Monomer/Tetramer Equilibrium of Rod Visual Arrestin in the Interaction with Phosphorylated Rhodopsin. Int. J. Mol. Sci. 2023, 24, 4963. https://doi.org/10.3390/ijms24054963
Imamoto Y, Kojima K, Maeda R, Shichida Y, Oka T. Role of Monomer/Tetramer Equilibrium of Rod Visual Arrestin in the Interaction with Phosphorylated Rhodopsin. International Journal of Molecular Sciences. 2023; 24(5):4963. https://doi.org/10.3390/ijms24054963
Chicago/Turabian StyleImamoto, Yasushi, Keiichi Kojima, Ryo Maeda, Yoshinori Shichida, and Toshihiko Oka. 2023. "Role of Monomer/Tetramer Equilibrium of Rod Visual Arrestin in the Interaction with Phosphorylated Rhodopsin" International Journal of Molecular Sciences 24, no. 5: 4963. https://doi.org/10.3390/ijms24054963
APA StyleImamoto, Y., Kojima, K., Maeda, R., Shichida, Y., & Oka, T. (2023). Role of Monomer/Tetramer Equilibrium of Rod Visual Arrestin in the Interaction with Phosphorylated Rhodopsin. International Journal of Molecular Sciences, 24(5), 4963. https://doi.org/10.3390/ijms24054963







