Specific Activation of Yamanaka Factors via HSF1 Signaling in the Early Stage of Zebrafish Optic Nerve Regeneration
Abstract
1. Introduction
2. Results
2.1. Rapid Increase of HSF1 Gene Expression in Zebrafish Retina after ONI
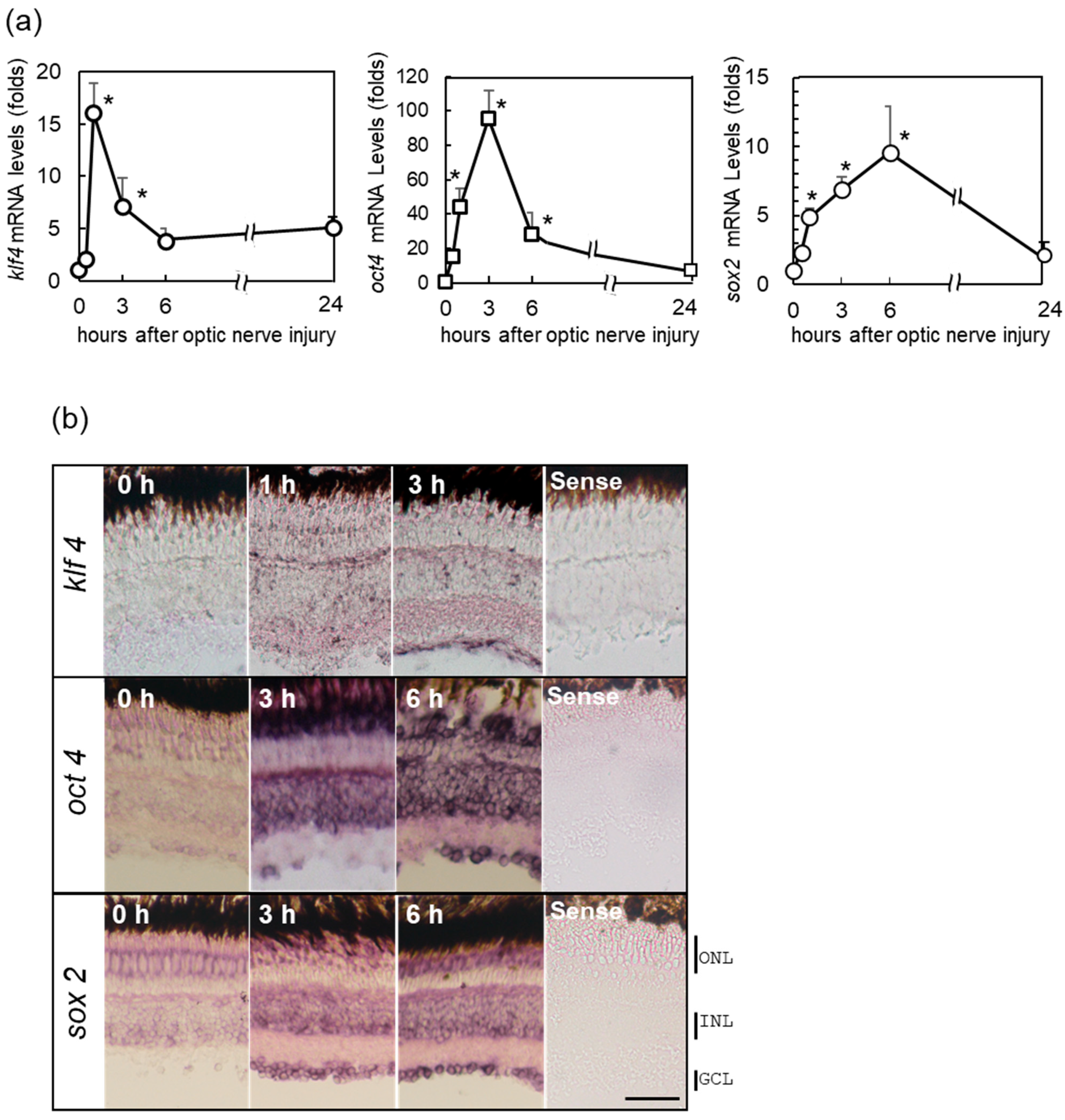
2.2. Increase in OSK Gene Expression in Zebrafish Retina after ONI
2.3. HSF1 Regulates Expression of OSK
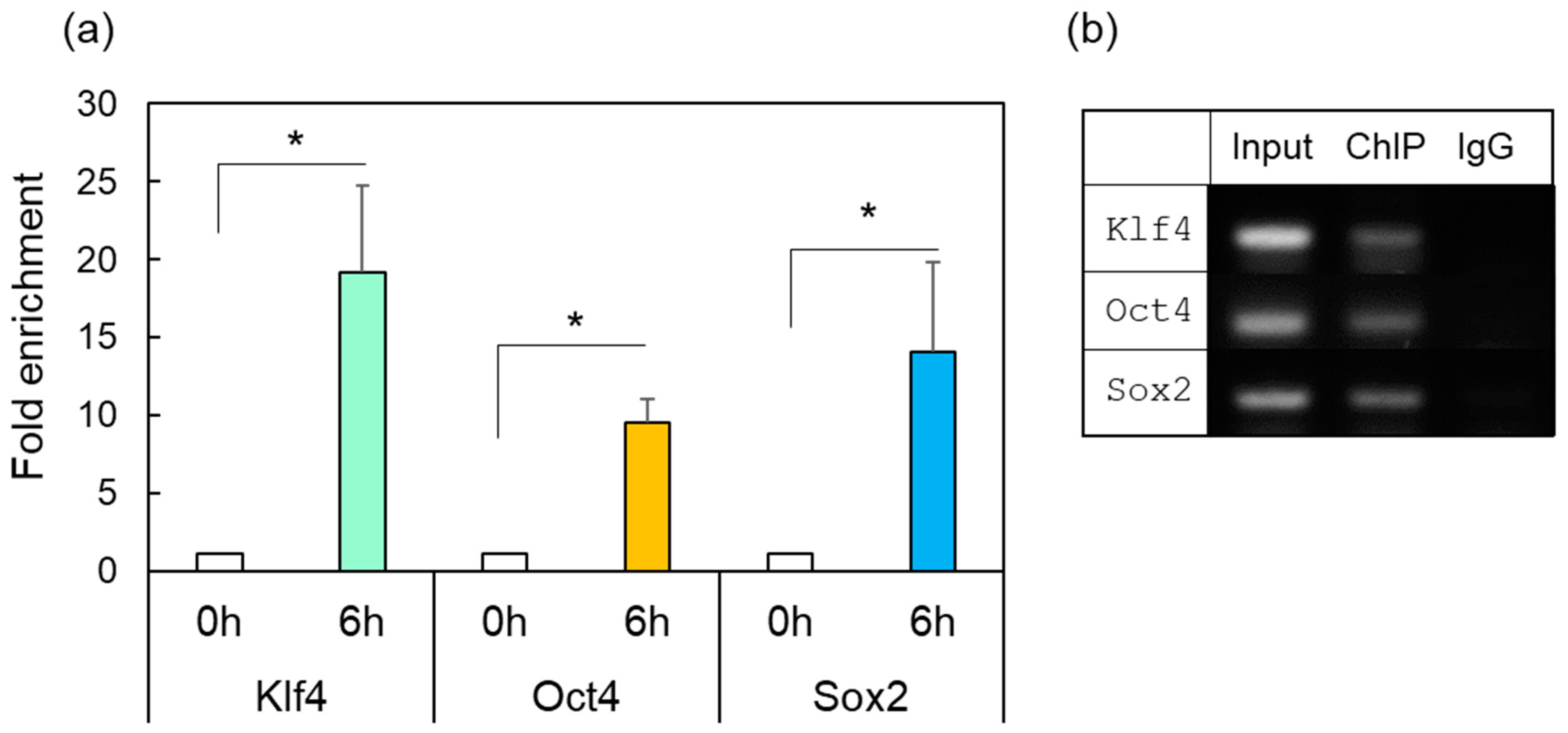
2.4. ChIP Assay of OSK in Response to HSF1
3. Discussion
3.1. Rapid Activation of OSK via HSF1 Signaling in the Zebrafish Retina after ONI
3.2. Role of Yamanaka Factors in the Injured Retina at the Early Stage of Optic Nerve Regeneration
4. Materials and Methods
4.1. Animals
4.2. Tissue Preparation
4.3. Total RNA Extraction and cDNA Synthesis
4.4. Quantitative Real-Time PCR
4.5. Immunohistochemistry
4.6. In Situ Hybridization
4.7. Chromatin Immunoprecipitation
4.8. Intraocular Injection of HSF1 Morpholino to Zebrafish Eye
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laha, B.; Stafford, B.K.; Huberman, A.D. Regenerating optic pathways from the eye to the brain. Science 2017, 356, 1031–1034. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.R.; Benowitz, L.I.; Goldberg, J.L.; He, Z. Axon Regeneration in the Mammalian Optic Nerve. Annu. Rev. Vis. Sci. 2020, 6, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Fague, L.; Liu, Y.A.; Marsh-Armstrong, N. The basic science of optic nerve regeneration. Ann. Transl. Med. 2021, 15, 1276. [Google Scholar] [CrossRef] [PubMed]
- Attardi, D.G.; Sperry, R.W. Preferential selection of central pathways by regenerating optic fibers. Exp. Neurol. 1963, 7, 46–64. [Google Scholar] [CrossRef]
- Sperry, R.W. Patterning of central synapses in regeneration of the optic nerve in teleosts. Physiol. Zool. 1948, 21, 351–361. [Google Scholar] [CrossRef]
- Becker, T.; Becker, C.G. Axonal regeneration in zebrafish. Curr. Opin. Neurobiol. 2014, 27, 186–191. [Google Scholar] [CrossRef]
- Lenkowski, J.R.; Raymond, P.A. Müller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish. Prog. Retin. Eye Res. 2014, 40, 94–123. [Google Scholar] [CrossRef]
- Goldman, D. Müller glial cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 2014, 15, 431–442. [Google Scholar] [CrossRef]
- Marques, I.J.; Lupi, E.; Mercader, N. Model systems for regeneration: Zebrafish. Development 2019, 146, dev167692. [Google Scholar] [CrossRef]
- Sugitani, K.; Matsukawa, T.; Maeda, A.; Kato, S. Upregulation of transglutaminase in the goldfish retina during optic nerve regeneration. In Retinal Degenerative Diseases. Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 2006; Volume 572, pp. 525–530. [Google Scholar] [CrossRef]
- Kaneda, M.; Nagashima, M.; Nunome, T.; Muramatsu, T.; Yamada, Y.; Kubo, M.; Muramoto, K.; Matsukawa, T.; Koriyama, Y.; Sugitani, K.; et al. Changes of phospho-growth associated protein 43 (phospho-GAP43) in zebrafish retina after optic nerve injury: A long term observation. Neurosci. Res. 2008, 61, 281–288. [Google Scholar] [CrossRef]
- Sugitani, K.; Koriyama, Y.; Ogai, K.; Furukawa, A.; Kato, S. Alternative Splicing for Activation of Coagulation Factor XIII-A in the Fish Retina After Optic Nerve Injury. Adv. Exp. Med. Biol. 2018, 1074, 387–393. [Google Scholar]
- Sugitani, K.; Matsukawa, T.; Koriyama, Y.; Shintani, T.; Nakamura, T.; Noda, M.; Kato, S. Upregulation of retinal transglutaminase during the axonal elongation stage of goldfish optic nerve regeneration. Neuroscience 2006, 142, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Koriyama, Y.; Yasuda, R.; Homma, K.; Mawatari, K.; Nagashima, M.; Sugitani, K.; Matsukawa, T.; Kato, S. Nitric oxide-cGMP signaling regulates axonal elongation during optic nerve regeneration in the goldfish in vitro and in vivo. J. Neurochem. 2009, 110, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Sugitani, K.; Ogai, K.; Hitomi, K.; Nakamura-Yonehara, K.; Shintani, T.; Noda, M.; Koriyama, Y.; Tanii, H.; Matsukawa, T.; Kato, S. A distinct effect of transient and sustained upregulation of cellular factor XIII in the goldfish retina and optic nerve on optic nerve regeneration. Neurochem. Int. 2012, 61, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Devadas, M.; Okada, K.; Shimada, Y.; Ohkawa, M.; Muramoto, K.; Takizawa, N.; Matsukawa, T. Fast and slow recovery phases of goldfish behavior after transection of the optic nerve revealed by a computer image processing system. Neuroscience 1999, 93, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Ogai, K.; Hisano, S.; Mawatari, K.; Sugitani, K.; Koriyama, Y.; Nakashima, H.; Kato, S. Upregulation of anti-apoptotic factors in upper motor neurons after spinal cord injury in adult zebrafish. Neurochem. Int. 2012, 61, 1202–1211. [Google Scholar] [CrossRef]
- Koriyama, Y.; Homma, K.; Sugitani, K.; Higuchi, Y.; Matsukawa, T.; Murayama, D.; Kato, S. Upregulation of IGF-I in the goldfish retinal ganglion cells during the early stage of optic nerve regeneration. Neurochem. Int. 2007, 50, 749–756. [Google Scholar] [CrossRef]
- Matsukawa, T.; Sugitani, K.; Mawatari, K.; Koriyama, Y.; Liu, Z.; Tanaka, M.; Kato, S. Role of purpurin as a retinol-binding protein in goldfish retina during the early stage of optic nerve regeneration: Its priming action on neurite outgrowth. J. Neurosci. 2004, 24, 8346–8353. [Google Scholar] [CrossRef]
- Sugitani, K.; Koriyama, Y.; Ogai, K.; Wakasugi, K.; Kato, S. A Possible Role of Neuroglobin in the Retina After Optic Nerve Injury: A Comparative Study of Zebrafish and Mouse Retina. Adv. Exp. Med. Biol. 2016, 854, 671–675. [Google Scholar] [PubMed]
- Sugitani, K.; Koriyama, Y.; Sera, M.; Arai, K.; Ogai, K.; Wakasugi, K. A novel function of neuroglobin for neuroregeneration in mice after optic nerve injury. Biochem. Biophys. Res. Commun. 2017, 493, 1254–1259. [Google Scholar] [CrossRef]
- Nagashima, M.; Fujikawa, C.; Mawatari, K.; Mori, Y.; Kato, S. HSP70, the earliest-induced gene in the zebrafish retina during optic nerve regeneration: Its role in cell survival. Neurochem. Int. 2011, 58, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, C.; Nagashima, M.; Mawatari, K.; Kato, S. HSP 70 gene expression in the zebrafish retina after optic nerve injury: A comparative study under heat shock stresses. Adv. Exp. Med. Biol. 2012, 723, 663–668. [Google Scholar] [PubMed]
- Kato, S.; Matsukawa, T.; Koriyama, Y.; Sugitani, K.; Ogai, K. A molecular mechanism of optic nerve regeneration in fish: The retinoid signaling pathway. Prog. Retin. Eye Res. 2013, 37, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, J.W.; Zhao, X.; De Cecco, M.; Peterson, A.L.; Pagliaroli, L.; Manivannan, J.; Hubbard, G.B.; Ikeno, Y.; Zhang, Y.; Feng, B.; et al. Reduced expression of MYC increases longevity and enhances healthspan. Cell 2015, 160, 477–488. [Google Scholar] [CrossRef]
- Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Okita, K.; Mochiduki, Y.; Takizawa, N.; Yamanaka, S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008, 26, 101–106. [Google Scholar] [CrossRef]
- Wernig, M.; Meissner, A.; Cassady, J.P.; Jaenisch, R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2008, 2, 10–12. [Google Scholar] [CrossRef]
- Pirkkala, L.; Nykänen, P.; Sistonen, L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001, 15, 1118–1131. [Google Scholar] [CrossRef]
- Hu, Z.; Killion, P.J.; Iyer, V.R. Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 2007, 39, 683–687. [Google Scholar] [CrossRef]
- Akerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, M.; Nakai, A. The heat shock factor family and adaptation to proteotoxic stress. FEBS J. 2010, 277, 4112–4125. [Google Scholar] [CrossRef] [PubMed]
- Anckar, J.; Sistonen, L. Regulation of HSF1 function in the heat stress response: Implications in aging and disease. Annu. Rev. Biochem. 2011, 80, 1089–1115. [Google Scholar] [CrossRef]
- Vihervaara, A.; Sistonen, L. HSF1 at a glance. J. Cell Sci. 2014, 127, 261–266. [Google Scholar] [CrossRef]
- Liu, W.; Xia, F.; Ha, Y.; Zhu, S.; Li., Y.; Folorunso, O.; Pashaei-Marandi, A.; Lin, P.Y.; Tilton, R.G.; Pierce, A.P.; et al. Neuroprotective Effects of HSF1 in Retinal Ischemia-Reperfusion Injury. Invest. Ophthalmol. Vis. Sci. 2019, 60, 965–977. [Google Scholar] [CrossRef]
- Episkopou, V. SOX2 functions in adult neural stem cells. Trends Neurosci. 2005, 28, 219–221. [Google Scholar] [CrossRef] [PubMed]
- Maucksch, C.; Jones, K.S.; Connor, B. Concise review: The involvement of SOX2 in direct reprogramming of induced neural stem/precursor cells. Stem Cells Transl. Med. 2013, 2, 579–583. [Google Scholar] [CrossRef] [PubMed]
- Amador-Arjona, A.; Cimadamore, F.; Huang, C.T.; Wright, R.; Lewis, S.; Gage, F.H.; Terskikh, A.V. SOX2 primes the epigenetic landscape in neural precursors enabling proper gene activation during hippocampal neurogenesis. Proc. Natl. Acad. Sci. USA 2015, 112, 1936–1945. [Google Scholar] [CrossRef]
- Northmore, D.P. Quantitative electrophysiological studies of regenerating visuotopic maps in goldfish-II. Delayed recovery of sensitivity to small light flashes. Neuroscience 1989, 32, 749–757. [Google Scholar] [CrossRef]
- Murray, M.; Grafstein, B. Changes in the morphology and amino acid incorporation of regenerating goldfish optic neurons. Exp. Neurol. 1969, 23, 544–560. [Google Scholar] [CrossRef]
- Devadas, M.; Sugawara, K.; Shimada, Y.; Sugitani, K.; Liu, Z.W.; Matsukawa, T.; Kato, S. Slow recovery of goldfish retinal ganglion cells’ soma size during regeneration. Neurosci. Res. 2000, 37, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Brommer, B.; Tian, X.; Krishnan, A.; Meer, M.; Wang, C.; Vera, D.L.; Zeng, Q.; Yu, D.; Bonkowski, M.S.; et al. Reprogramming to recover youthful epigenetic information and restore vision. Nature 2020, 88, 124–129. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugitani, K.; Mokuya, T.; Homma, S.; Maeda, M.; Konno, A.; Ogai, K. Specific Activation of Yamanaka Factors via HSF1 Signaling in the Early Stage of Zebrafish Optic Nerve Regeneration. Int. J. Mol. Sci. 2023, 24, 3253. https://doi.org/10.3390/ijms24043253
Sugitani K, Mokuya T, Homma S, Maeda M, Konno A, Ogai K. Specific Activation of Yamanaka Factors via HSF1 Signaling in the Early Stage of Zebrafish Optic Nerve Regeneration. International Journal of Molecular Sciences. 2023; 24(4):3253. https://doi.org/10.3390/ijms24043253
Chicago/Turabian StyleSugitani, Kayo, Takumi Mokuya, Shuichi Homma, Minami Maeda, Ayano Konno, and Kazuhiro Ogai. 2023. "Specific Activation of Yamanaka Factors via HSF1 Signaling in the Early Stage of Zebrafish Optic Nerve Regeneration" International Journal of Molecular Sciences 24, no. 4: 3253. https://doi.org/10.3390/ijms24043253
APA StyleSugitani, K., Mokuya, T., Homma, S., Maeda, M., Konno, A., & Ogai, K. (2023). Specific Activation of Yamanaka Factors via HSF1 Signaling in the Early Stage of Zebrafish Optic Nerve Regeneration. International Journal of Molecular Sciences, 24(4), 3253. https://doi.org/10.3390/ijms24043253






