MoaE Is Involved in Response to Oxidative Stress in Deinococcus radiodurans
Abstract
1. Introduction
2. Results
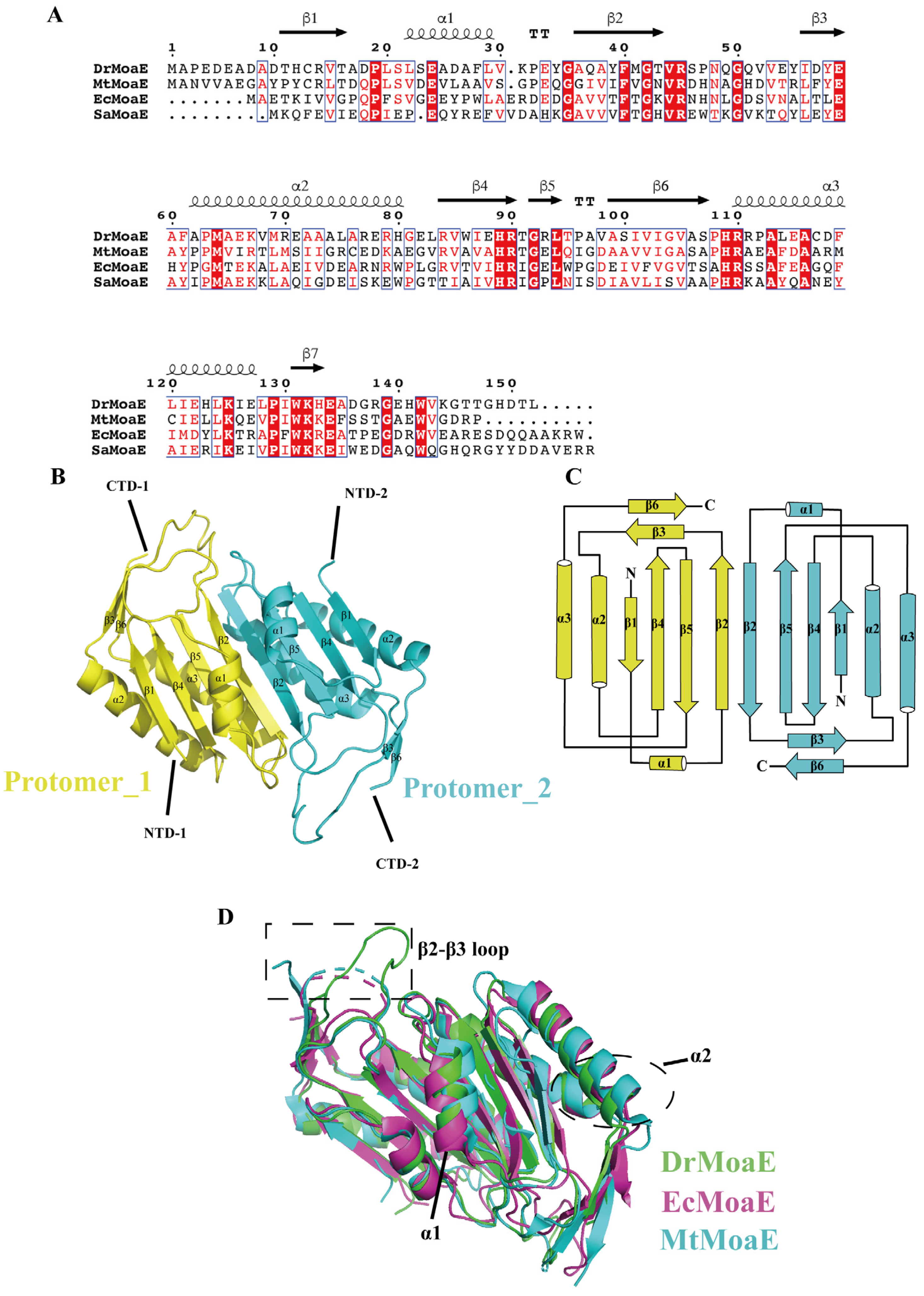
2.1. Overall Structure of DrMoaE
2.2. Arg110 Is a Key Residue for DrMoaE Dimerization
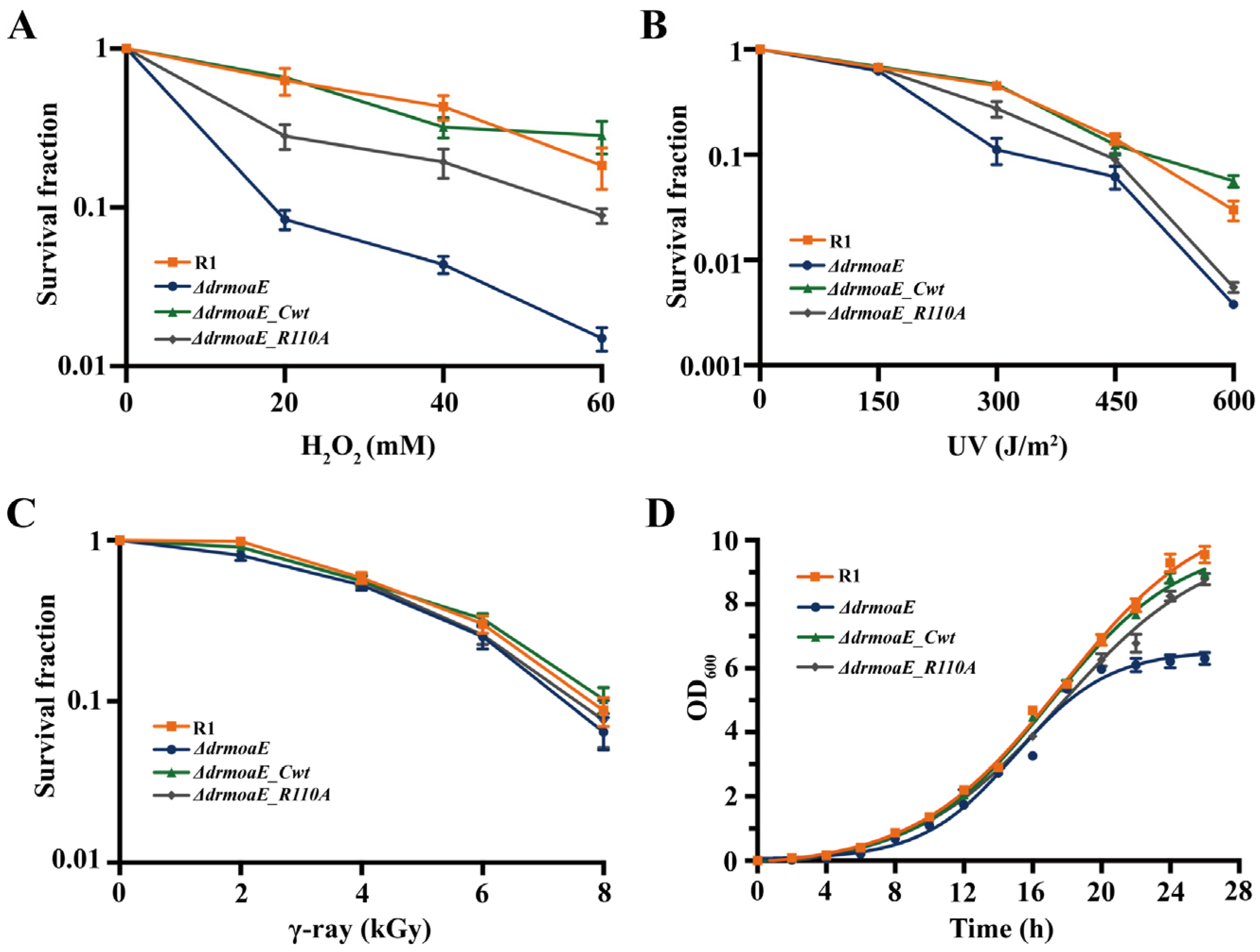
2.3. ΔdrmoaE Mutant Strain Is Sensitive to Damage Stress
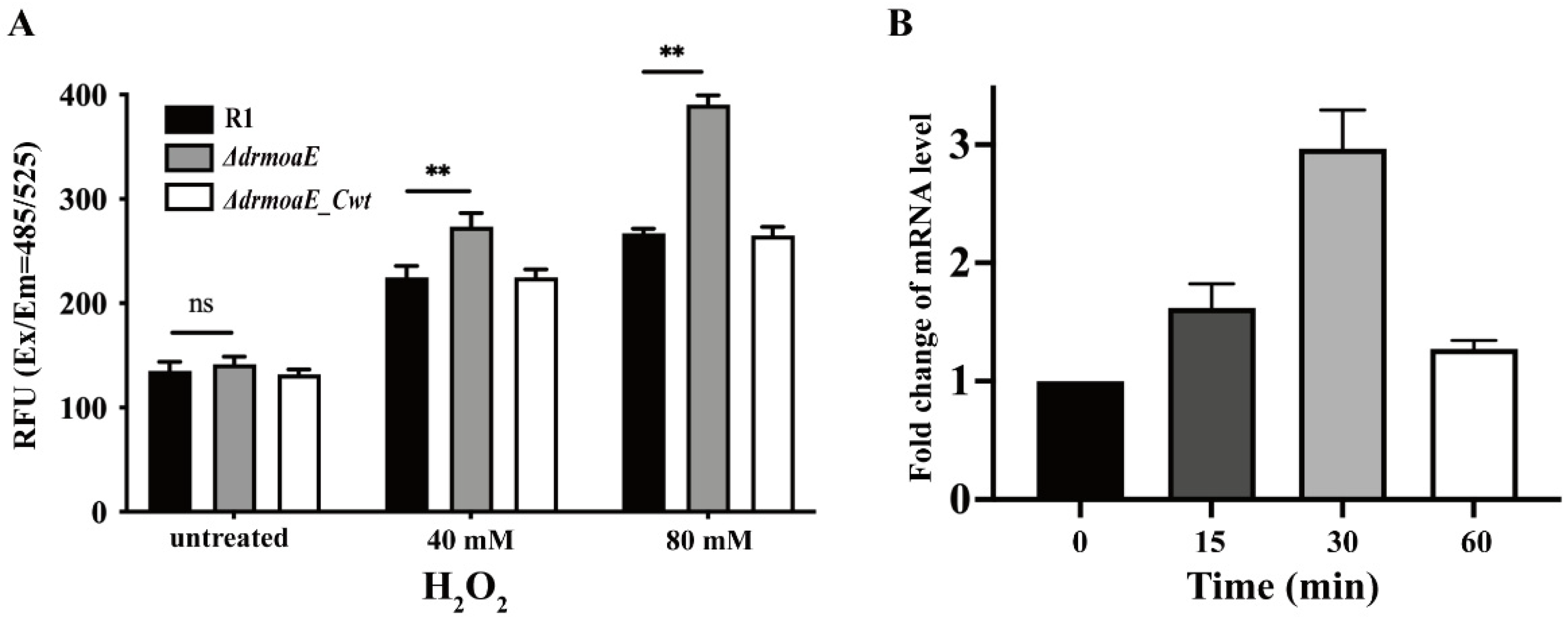
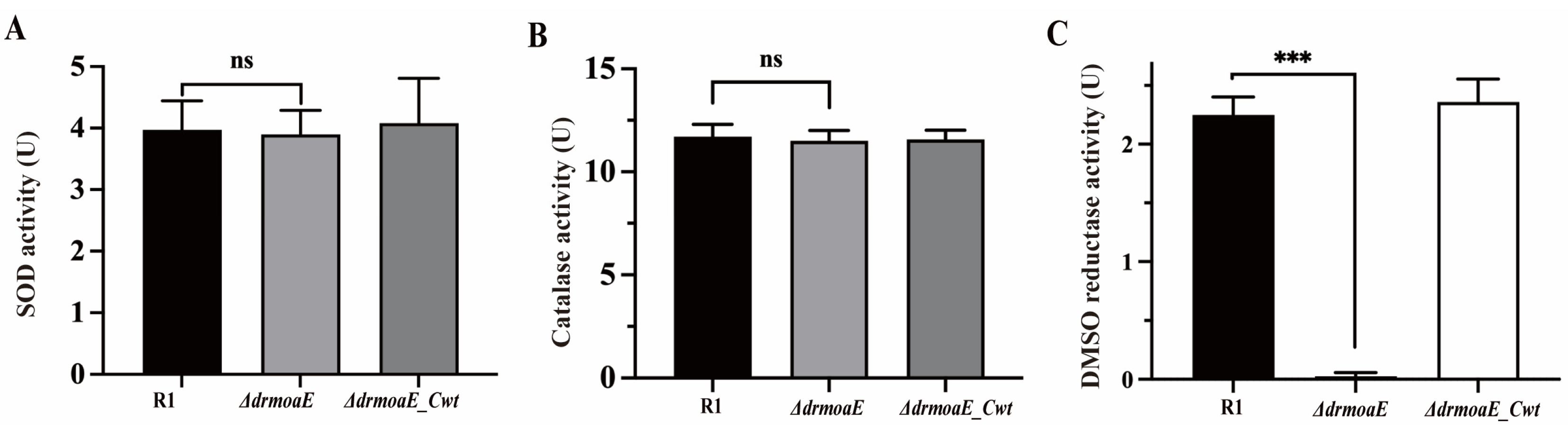
2.4. DrmoaE Contributes to the Antioxidant Process
2.5. DMSO Reductase Activity Is Decreased in the ΔdrmoaE Mutant Strain
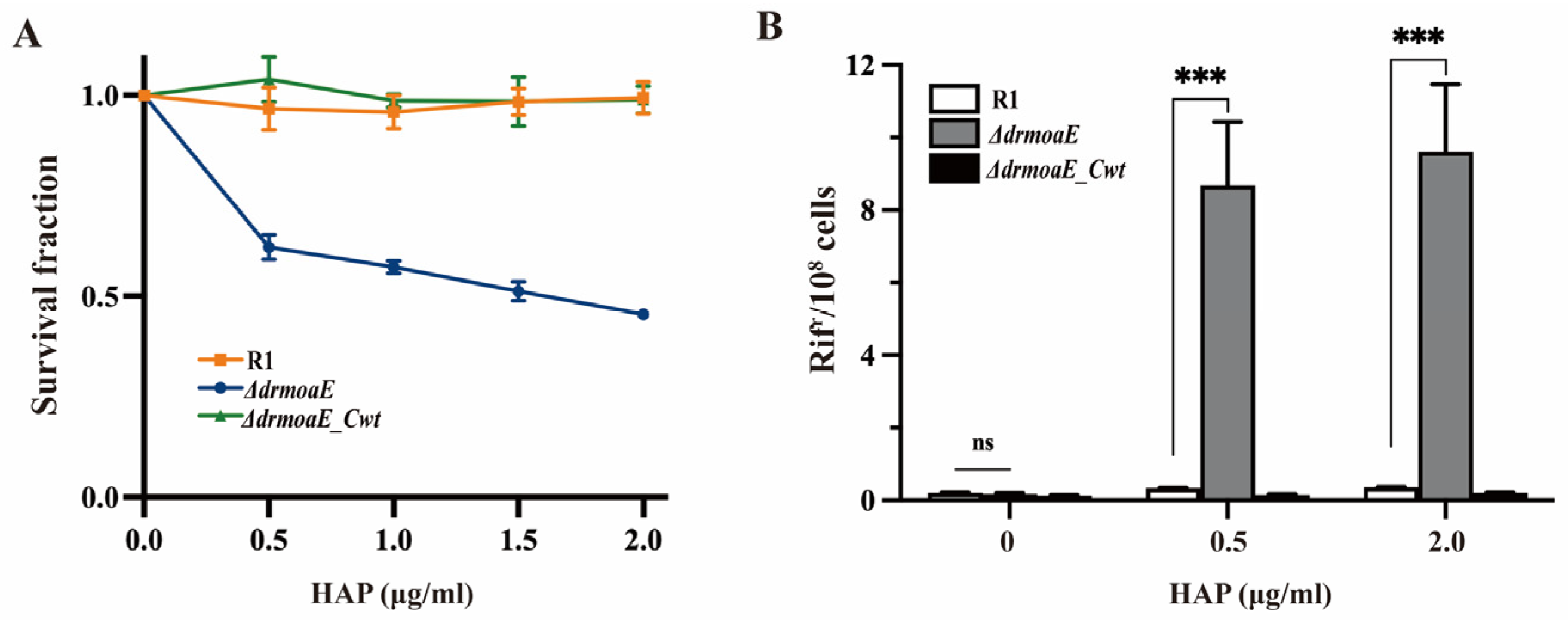
2.6. DrMoaE Responds to Base Analog Damage
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Expression and Purification of Proteins
4.3. Crystallization, Data Collection, and Structure Determination
4.4. Construction of Mutant and Complementary Strains
4.5. Phenotypic and Growth Curve Analysis
4.6. Detection of Intracellular ROS Content
4.7. Real-Time Quantitative PCR (RT-qPCR)
4.8. Analysis of Superoxide Dismutase (SOD) Activity
4.9. Analysis of Catalase Activity
4.10. Analysis of DMSO Reductase Activity
4.11. 6-Hydroxyaminopurine (HAP)-Mediated Growth Inhibition
4.12. Mutant Frequency Determinations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Gladyshev, V.N. Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 2008, 379, 881–899. [Google Scholar] [CrossRef] [PubMed]
- Leimkuhler, S. The biosynthesis of the molybdenum cofactors in Escherichia coli. Environ. Microbiol. 2020, 22, 2007–2026. [Google Scholar] [CrossRef] [PubMed]
- Gutzke, G.; Fischer, B.; Mendel, R.R.; Schwarz, G. Thiocarboxylation of molybdopterin synthase provides evidence for the mechanism of dithiolene formation in metal-binding pterins. J. Biol. Chem. 2001, 276, 36268–36274. [Google Scholar] [CrossRef] [PubMed]
- Daniels, J.N.; Wuebbens, M.M.; Rajagopalan, K.V.; Schindelin, H. Crystal structure of a molybdopterin synthase-precursor Z complex: Insight into its sulfur transfer mechanism and its role in molybdenum cofactor deficiency. Biochemistry 2008, 47, 3315. [Google Scholar] [CrossRef]
- Hasona, A.; Self, W.T.; Ray, R.M.; Shanmugam, K.T. Molybdate-dependent transcription of hyc and nar operons of Escherichia coli requires MoeA protein and ModE-molybdate. FEMS Microbiol. Lett. 1998, 169, 111–116. [Google Scholar] [CrossRef]
- Neumann, M.; Mittelstadt, G.; Seduk, F.; Iobbi-Nivol, C.; Leimkuhler, S. MocA Is a Specific Cytidylyltransferase Involved in Molybdopterin Cytosine Dinucleotide Biosynthesis in Escherichia coli. J. Biol. Chem. 2009, 284, 21891–21898. [Google Scholar] [CrossRef]
- Pitterle, D.M.; Rajagopalan, K.V. The biosynthesis of molybdopterin in Escherichia coli. Purification and characterization of the converting factor. J. Biol. Chem. 1993, 268, 13499–13505. [Google Scholar] [CrossRef] [PubMed]
- Wuebbens, M.M.; Rajagopalan, K.V. Investigation of the early steps of molybdopterin biosynthesis in Escherichia coli through the use of in vivo labeling studies. J. Biol. Chem. 1995, 270, 1082–1087. [Google Scholar] [CrossRef]
- Wuebbens, M.M.; Rajagopalan, K.V. Mechanistic and mutational studies of Escherichia coli molybdopterin synthase clarify the final step of molybdopterin biosynthesis. J. Biol. Chem. 2003, 278, 14523–14532. [Google Scholar] [CrossRef]
- Narrandes, N.C.; Machowski, E.E.; Mizrahi, V.; Kana, B.D. Cleavage of the moaX-encoded fused molybdopterin synthase from Mycobacterium tuberculosis is necessary for activity. BMC Microbiol. 2015, 15, 22. [Google Scholar] [CrossRef]
- Williams, M.; Mizrahi, V.; Kana, B.D. Molybdenum cofactor: A key component of Mycobacterium tuberculosis pathogenesis? Crit. Rev. Microbiol. 2014, 40, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Tang, H.; Wu, G.; Xu, P. Functional Identification of a Novel Gene, moaE, for 3-Succinoylpyridine Degradation in Pseudomonas putida S16. Sci. Rep. 2015, 5, 13464. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.W.; Nordan, H.C.; Cain, R.F.; Parrish, G.; Duggan, D. Studies on a Radio-Resistant Micrococcus. 1. Isolation, Morphology, Cultural Characteristics, and Resistance to Gamma Radiation. Food Technol.-Chic. 1956, 10, 575–578. [Google Scholar]
- Slade, D.; Radman, M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef] [PubMed]
- Julie, M.; Zimmerman, J.R.B. A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol. 2005, 5, 17. [Google Scholar]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Kiang, J.G.; Fukumoto, R.; Lee, D.Y.; Wehr, N.B.; Viteri, G.A.; Berlett, B.S.; Levine, R.L. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS ONE 2010, 5, e12570. [Google Scholar] [CrossRef]
- Anjem, A.; Varghese, S.; Imlay, J.A. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 2009, 72, 844–858. [Google Scholar] [CrossRef]
- Tanaka, A.; Hirano, H.H.; Kikuchi, M.; Kitayama, S.; Watanabe, H. Changes in cellular proteins of Deinococcus radiodurans following gamma-irradiation. Radiat. Environ. Bioph. 1996, 35, 95–99. [Google Scholar] [CrossRef]
- White, O. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 1999, 286, 1571–1577. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, J.; Omelchenko, M.V.; Beliaev, A.S.; Venkateswaran, A.; Stair, J.; Wu, L.; Thompson, D.K.; Xu, D.; Rogozin, I.B.; et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. USA 2003, 100, 4191–4196. [Google Scholar] [CrossRef]
- Yang, Y.M.; Won, Y.B.; Ji, C.J.; Kim, J.H.; Ryu, S.H.; Ok, Y.H.; Lee, J.W. Cleavage of molybdopterin synthase MoaD-MoaE linear fusion by JAMM/MPN(+) domain containing metalloprotease DR0402 from Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 2018, 502, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.L.; Pan, C.M.; Zhao, Y.; Xu, H.; Tian, B.; Wang, L.Y.; Hua, Y.J. DRJAMM Is Involved in the Oxidative Resistance in Deinococcus radiodurans. Front. Microbiol. 2022, 12, 756867. [Google Scholar] [CrossRef]
- Rudolph, M.J.; Wuebbens, M.M.; Turque, O.; Rajagopalan, K.V.; Schindelin, H. Structural studies of molybdopterin synthase provide insights into its catalytic mechanism. J. Biol. Chem. 2003, 278, 14514–14522. [Google Scholar] [CrossRef] [PubMed]
- Miralles-Robledillo, J.M.; Torregrosa-Crespo, J.; Martinez-Espinosa, R.M.; Pire, C. DMSO Reductase Family: Phylogenetics and Applications of Extremophiles. Int. J. Mol. Sci. 2019, 20, 3349. [Google Scholar] [CrossRef] [PubMed]
- Kozmin, S.G.; Pavlov, Y.I.; Dunn, R.L.; Schaaper, R.M. Hypersensitivity of Escherichia coli Delta(uvrB-bio) mutants to 6-hydroxylaminopurine and other base analogs is due to a defect in molybdenum cofactor biosynthesis. J. Bacteriol. 2000, 182, 3361–3367. [Google Scholar] [CrossRef] [PubMed]
- Swartz, C.D.; Parks, N.; Umbach, D.M.; Ward, W.O.; Schaaper, R.M.; DeMarini, D.M. Enhanced mutagenesis of Salmonella tester strains due to deletion of genes other than uvrB. Environ. Mol. Mutagen 2007, 48, 694–705. [Google Scholar] [CrossRef]
- Barros, M.P.; Hollnagel, H.C.; Glavina, A.B.; Soares, C.O.; Ganini, D.; Dagenais-Bellefeuille, S.; Morse, D.; Colepicolo, P. Molybdate:sulfate ratio affects redox metabolism and viability of the dinoflagellate Lingulodinium polyedrum. Aquat. Toxicol. 2013, 142, 195–202. [Google Scholar] [CrossRef]
- Feig, D.I.; Sowers, L.C.; Loeb, L.A. Reverse Chemical Mutagenesis—Identification of the Mutagenic Lesions Resulting from Reactive Oxygen Species-Mediated Damage to DNA. Proc. Natl. Acad. Sci. USA 1994, 91, 6609–6613. [Google Scholar] [CrossRef]
- Fujikawa, K.; Kamiya, H.; Yakushiji, H.; Fujii, Y.; Nakabeppu, Y.; Kasai, H. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem. 1999, 274, 18201–18205. [Google Scholar] [CrossRef]
- Michaels, M.L.; Miller, J.H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 1992, 174, 6321–6325. [Google Scholar] [CrossRef]
- Kozmin, S.G.; Schaaper, R.M.; Shcherbakova, P.V.; Kulikov, V.N.; Noskov, V.N.; Guetsova, M.L.; Alenin, V.V.; Rogozin, I.B.; Makarova, K.S.; Pavlov, Y.I. Multiple antimutagenesis mechanisms affect mutagenic activity and specificity of the base analog 6-N-hydroxylaminopurine in bacteria and yeast. Mutat. Res. 1998, 402, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Simandan, T.; Sun, J.; Dix, T.A. Oxidation of DNA bases, deoxyribonucleosides and homopolymers by peroxyl radicals. Biochem. J. 1998, 335 Pt 2, 233–240. [Google Scholar] [CrossRef]
- Kozmin, S.G.; Schaaper, R.M. Molybdenum cofactor-dependent resistance to N-hydroxylated base analogs in Escherichia coli is independent of MobA function. Mutat. Res. 2007, 619, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Clement, B.; Kunze, T. The Reduction of 6-N-Hydroxylaminopurine to Adenine by Xanthine-Oxidase. Biochem. Pharmacol. 1992, 44, 1501–1509. [Google Scholar] [CrossRef] [PubMed]
- Kozmin, S.G.; Leroy, P.; Pavlov, Y.I.; Schaaper, R.M. YcbX and yiiM, two novel determinants for resistance of Escherichia coli to N-hydroxylated base analogues. Mol. Microbiol. 2008, 68, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Li, S.; Cai, J.; Lu, H.; Mao, S.; Dai, S.; Hu, J.; Wang, L.; Hua, X.; Xu, H.; Tian, B.; et al. N (4)-Cytosine DNA Methylation Is Involved in the Maintenance of Genomic Stability in Deinococcus radiodurans. Front Microbiol. 2019, 10, 1905. [Google Scholar] [CrossRef]
- Dai, J.; Gao, K.; Yao, T.; Lu, H.; Zhou, C.; Guo, M.; Dai, S.; Wang, L.; Xu, H.; Tian, B.; et al. Late embryogenesis abundant group3 protein (DrLEA3) is involved in antioxidation in the extremophilic bacterium Deinococcus radiodurans. Microbiol. Res. 2020, 240, 126559. [Google Scholar] [CrossRef]
- Dai, S.; Jin, Y.; Li, T.; Weng, Y.; Xu, X.; Zhang, G.; Li, J.; Pang, R.; Tian, B.; Hua, Y. DR1440 is a potential iron efflux protein involved in maintenance of iron homeostasis and resistance of Deinococcus radiodurans to oxidative stress. PLoS ONE 2018, 13, e0202287. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Chen, Z.; Hou, J.; Wang, Y.; Guo, M.; Cao, J.; Wang, L.; Xu, H.; Tian, B.; Zhao, Y. MazEF Toxin-Antitoxin System-Mediated DNA Damage Stress Response in Deinococcus radiodurans. Front Genet. 2021, 12, 632423. [Google Scholar] [CrossRef] [PubMed]
- Miranda, H.V.; Nembhard, N.; Su, D.; Hepowit, N.; Krause, D.J.; Pritz, J.R.; Phillips, C.; Soll, D.; Maupin-Furlow, J.A. E1-and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc. Natl. Acad. Sci. USA 2011, 108, 4417–4422. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, J.; Zhang, M.; Chen, Z.; Zhao, Y.; Xu, H.; Tian, B.; Wang, L.; Hua, Y. MoaE Is Involved in Response to Oxidative Stress in Deinococcus radiodurans. Int. J. Mol. Sci. 2023, 24, 2441. https://doi.org/10.3390/ijms24032441
Cai J, Zhang M, Chen Z, Zhao Y, Xu H, Tian B, Wang L, Hua Y. MoaE Is Involved in Response to Oxidative Stress in Deinococcus radiodurans. International Journal of Molecular Sciences. 2023; 24(3):2441. https://doi.org/10.3390/ijms24032441
Chicago/Turabian StyleCai, Jianling, Maoxu Zhang, Zijing Chen, Ye Zhao, Hong Xu, Bing Tian, Liangyan Wang, and Yuejin Hua. 2023. "MoaE Is Involved in Response to Oxidative Stress in Deinococcus radiodurans" International Journal of Molecular Sciences 24, no. 3: 2441. https://doi.org/10.3390/ijms24032441
APA StyleCai, J., Zhang, M., Chen, Z., Zhao, Y., Xu, H., Tian, B., Wang, L., & Hua, Y. (2023). MoaE Is Involved in Response to Oxidative Stress in Deinococcus radiodurans. International Journal of Molecular Sciences, 24(3), 2441. https://doi.org/10.3390/ijms24032441







