Characterization of a Novel N4-Methylcytosine Restriction-Modification System in Deinococcus radiodurans
Abstract
1. Introduction
2. Results
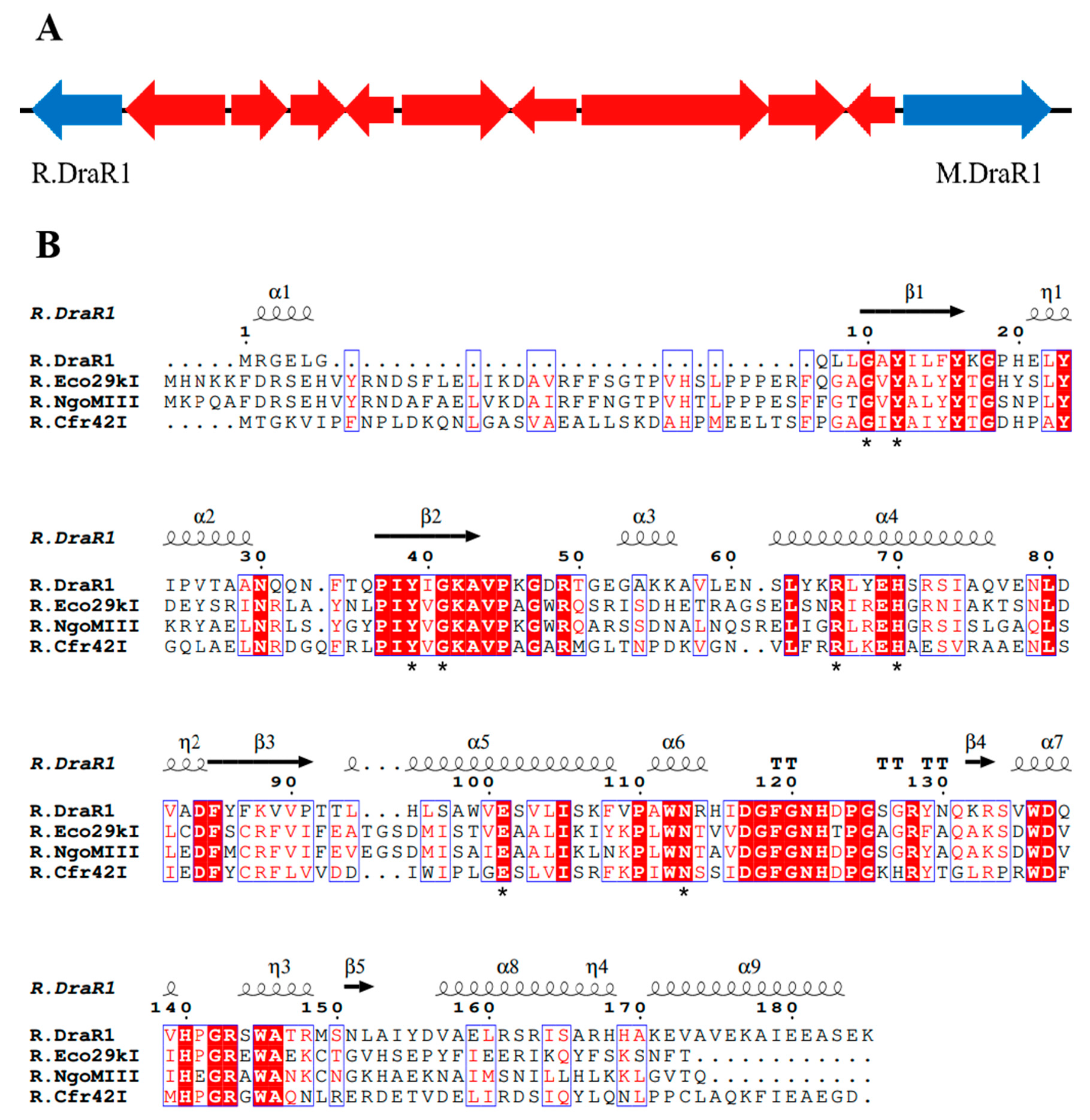
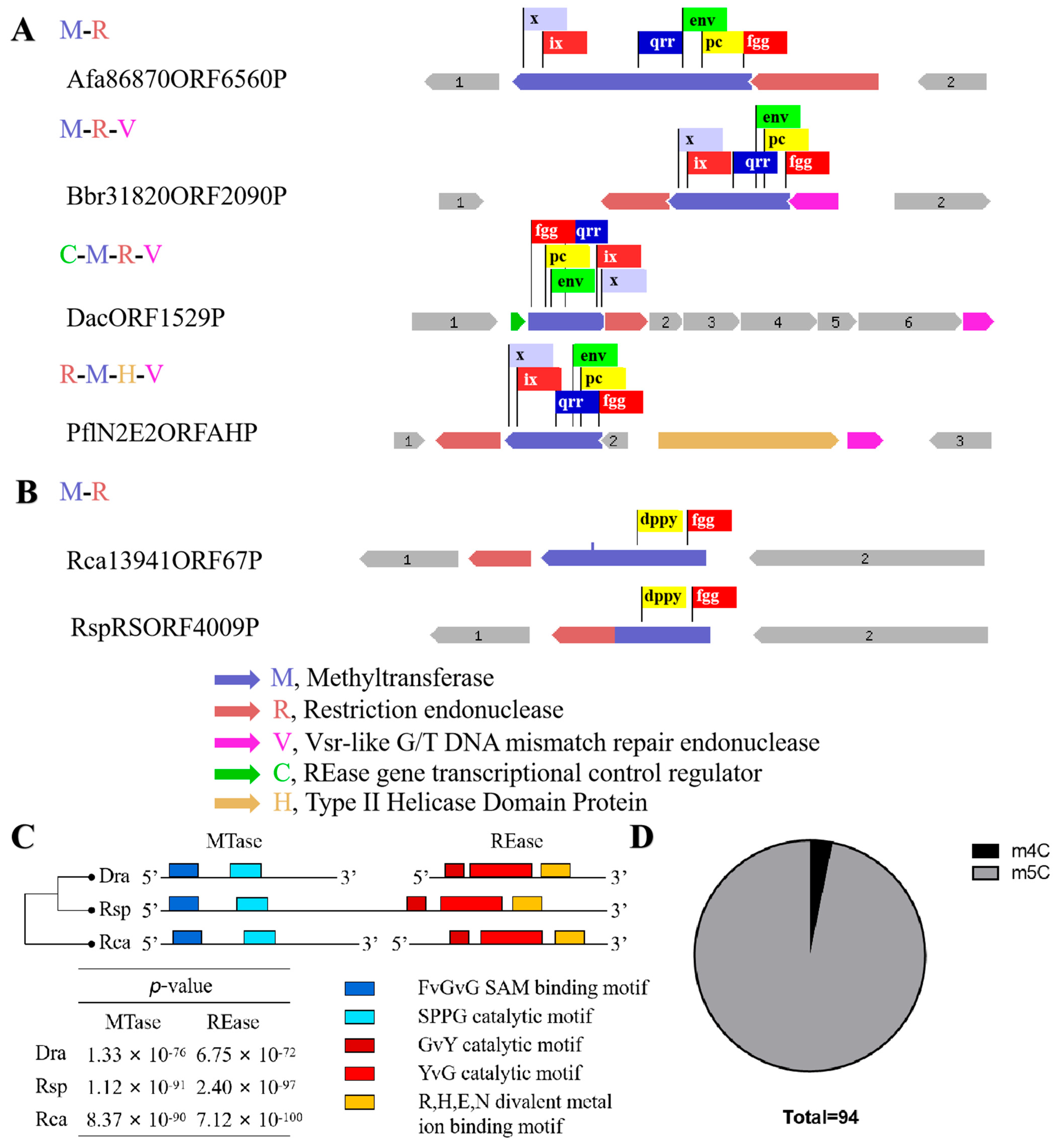
2.1. A m4C Restriction-Modification System DraI Existed in Deinococcus radiodurans
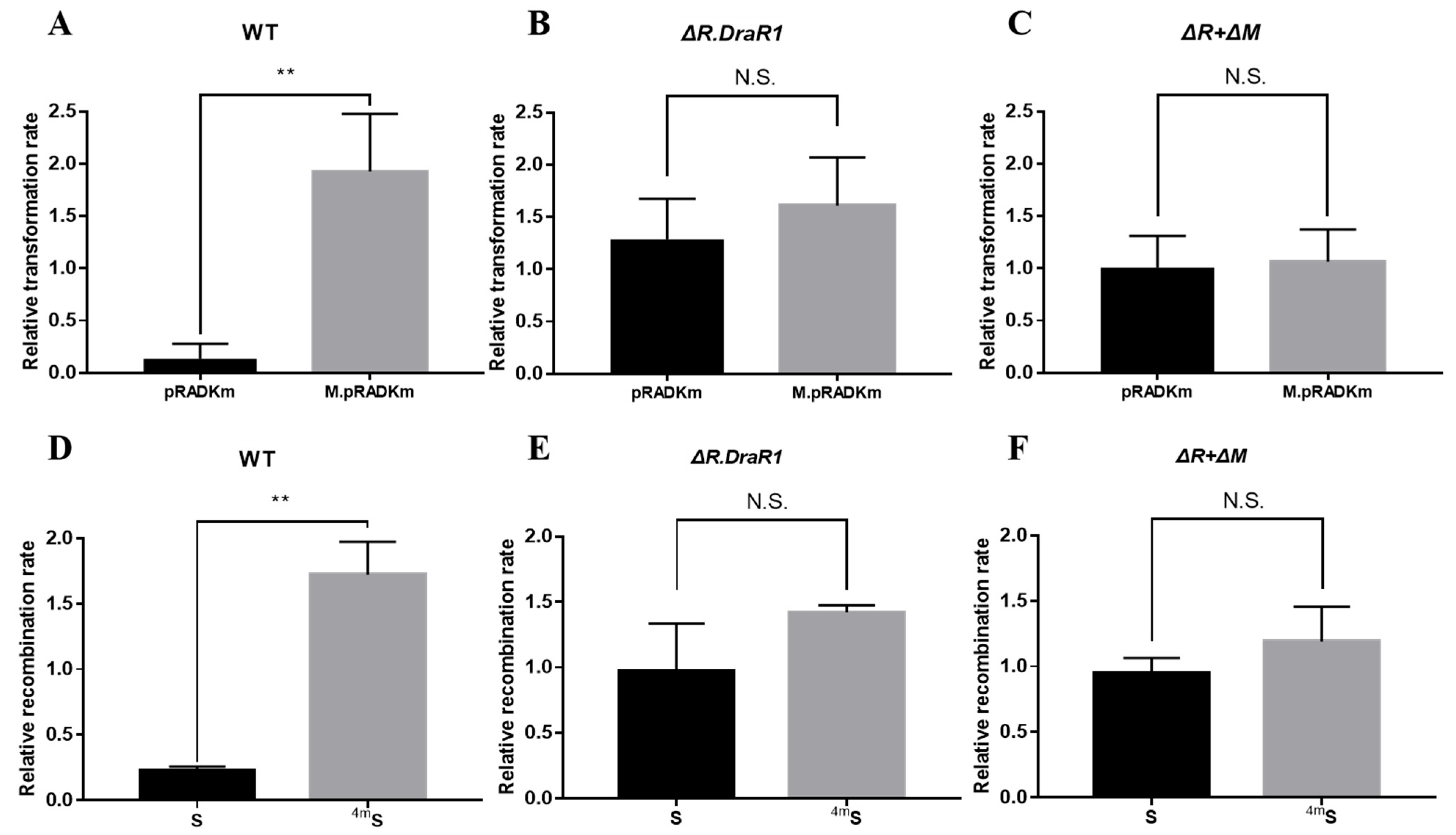
2.2. The m4C R-M System DraI Inhibits Foreign Plasmid and DNA Fragment Acquisition
2.3. R.DraR1 of the DraI System Could Specifically Cleave Unmethylated or m5C-Methylated 5′-CCGC/GG-3′ Sequences In Vitro
2.4. Imbalance in the DraI System Leads to a Significant Increase in Endogenous Cell Death
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. DNA Manipulation
4.3. Database Search for R-M Systems Which Recognize 5′-CCGCGG-3′ Sequence
4.4. Transformation and Recombination Efficiency Assays
4.5. Expression and Purification of R.DraR1
4.6. In Vitro M.DraR1 Protection and R.DraR1 Cleavage Assays
4.7. Fluorescence Microscopy and FACS Analysis
4.8. Over-Expression of R.DraR1 and β-Galactosidase Assay
4.9. RNA Isolation and Sequencing Analysis (RNA seq)
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, S.; Xu, T.; Taghizadeh, K.; Wishnok, J.S.; Zhou, X.; You, D.; Deng, Z.; Dedon, P.C. Phosphorothioation of DNA in bacteria by dnd genes. Nat. Chem. Biol. 2007, 3, 709–710. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, P.H.; Touchon, M.; Rocha, E.P.C. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014, 42, 10618–10631. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, J.J.; Kellner, S.M.; Yuan, Y.; Hutinet, G.; Thiaville, P.C.; Jumpathong, W.; Mohapatra, S.; Brochier-Armanet, C.; Letarov, A.V.; Hillebrand, R.; et al. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc. Natl. Acad. Sci. USA 2016, 113, E1452–E1459. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, T.; Sberro, H.; Weinstock, E.; Cohen, O.; Doron, S.; Charpak-Amikam, Y.; Afik, S.; Ofir, G.; Sorek, R. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2014, 34, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Ofir, G.; Melamed, S.; Sberro, H.; Mukamel, Z.; Silverman, S.; Yaakov, G.; Doron, S.; Sorek, R. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat. Microbiol. 2017, 3, 90–98. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Wolf, Y.I. Evolutionary Genomics of Defense Systems in Archaea and Bacteria. Annu. Rev. Microbiol. 2017, 71, 233–261. [Google Scholar] [CrossRef]
- Cohen, D.; Melamed, S.; Millman, A.; Shulman, G.; Oppenheimer-Shaanan, Y.; Kacen, A.; Doron, S.; Amitai, G.; Sorek, R. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 2019, 574, 691–695. [Google Scholar] [CrossRef]
- Bernheim, A.; Sorek, R. The pan-immune system of bacteria: Antiviral defence as a community resource. Nat. Rev. Microbiol. 2019, 18, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Tock, M.R.; Dryden, D.T.F. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005, 8, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Pingoud, A.; Jeltsch, A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001, 29, 3705–3727. [Google Scholar] [CrossRef]
- Gasiunas, G.; Sasnauskas, G.; Tamulaitis, G.; Urbanke, C.; Razaniene, D.; Siksnys, V. Tetrameric restriction enzymes: Expansion to the GIY-YIG nuclease family. Nucleic Acids Res. 2007, 36, 938–949. [Google Scholar] [CrossRef]
- Mak, A.N.-S.; Lambert, A.R.; Stoddard, B.L. Folding, DNA Recognition, and Function of GIY-YIG Endonucleases: Crystal Structures of R.Eco29kI. Structure 2010, 18, 1321–1331. [Google Scholar] [CrossRef]
- Adhikari, S.; Curtis, P.D.; Narberhaus, F. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol. Rev. 2016, 40, 575–591. [Google Scholar] [CrossRef]
- Casadesús, J.; Low, D. Epigenetic Gene Regulation in the Bacterial World. Microbiol. Mol. Biol. Rev. 2006, 70, 830–856. [Google Scholar] [CrossRef] [PubMed]
- Timothy, C.; Yvonne, T.; Adam, S.; William, K.A.; Anna, P.; Brian, S.; Catherine, A.B. DNA Methylation by Restriction Modification Systems Affects the Global Transcriptome Profile in Borrelia burgdorferi. J. Bacteriol. 2018, 200, e00395-18. [Google Scholar] [CrossRef]
- Kumar, S.; Karmakar, B.C.; Nagarajan, D.; Mukhopadhyay, A.K.; Morgan, R.D.; Rao, D.N. N4-cytosine DNA methylation regulates transcription and pathogenesis in Helicobacter pylori. Nucleic Acids Res. 2018, 46, 3429–3445. [Google Scholar] [CrossRef]
- Sánchez-Romero, M.A.; Cota, I.; Casadesús, J. DNA methylation in bacteria: From the methyl group to the methylome. Curr. Opin. Microbiol. 2015, 25, 9–16. [Google Scholar] [CrossRef]
- Clark, T.A.; Murray, I.A.; Morgan, R.D.; Kislyuk, A.O.; Spittle, K.E.; Boitano, M.; Fomenkov, A.; Roberts, R.J.; Korlach, J. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012, 40, e29. [Google Scholar] [CrossRef]
- Fang, G.; Munera, D.; Friedman, D.I.; Mandlik, A.; Chao, M.C.; Banerjee, O.; Feng, Z.; Losic, B.; Mahajan, M.C.; Jabado, O.J.; et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 2012, 30, 1232–1239. [Google Scholar] [CrossRef]
- Rand, A.C.; Jain, M.; Eizenga, J.M.; Musselman-Brown, A.; Olsen, H.E.; Akeson, M.; Paten, B. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods 2017, 14, 411–413. [Google Scholar] [CrossRef]
- Cox, M.M.; Battista, J.R. Deinococcus radiodurans—The consummate survivor. Nat. Rev. Microbiol. 2005, 3, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cai, J.; Lu, H.; Mao, S.; Dai, S.; Hu, J.; Wang, L.; Hua, X.; Xu, H.; Tian, B.; et al. N4-Cytosine DNA Methylation Is Involved in the Maintenance of Genomic Stability in Deinococcus radiodurans. Front. Microbiol. 2019, 10, 1905. [Google Scholar] [CrossRef] [PubMed]
- Herbert, W.B. DNA restriction and modification mechanisms in bacteria. Annu. Rev. Microbiol. 1971, 25, 153–176. [Google Scholar] [CrossRef]
- Simmons, L.A.; Matthews, L.A.; Randall, J.R.; Schroeder, J.W.; Stevens, A.G.; van Gijtenbeek, L.A.; Nye, T.M. Methyltransferase DnmA is responsible for genome-wide N6-methyladenosine modifications at non-palindromic recognition sites in Bacillus subtilis. Nucleic Acids Res. 2020, 48, 5332–5348. [Google Scholar] [CrossRef]
- Ibryashkina, E.M.; Sasnauskas, G.; Solonin, A.S.; Zakharova, M.V.; Siksnys, V. Oligomeric Structure Diversity within the GIY-YIG Nuclease Family. J. Mol. Biol. 2009, 387, 10–16. [Google Scholar] [CrossRef]
- Lim, S.; Jung, J.-H.; Blanchard, L.; de Groot, A. Conservation and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus species. FEMS Microbiol. Rev. 2018, 43, 19–52. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE: A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2023, 51, D629–D630. [Google Scholar] [CrossRef]
- Klimasauskas, S.; Timinskas, A.; Menkevicius, S.; Butkienè, D.; Butkus, V.; Janulaitis, A. Sequence motifs characteristic of DNA[cytosine-N4]methyltransferases: Similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989, 17, 9823–9832. [Google Scholar] [CrossRef]
- Fox, K.R.; Allinson, S.L.; Sahagun-Krause, H.; Brown, T. Recognition of GT mismatches by Vsr mismatch endonuclease. Nucleic Acids Res. 2000, 28, 2535–2540. [Google Scholar] [CrossRef]
- McGeehan, J.E.; Papapanagiotou, I.; Streeter, S.D.; Kneale, G.G. Cooperative Binding of the C.AhdI Controller Protein to the C/R Promoter and its Role in Endonuclease Gene Expression. J. Mol. Biol. 2006, 358, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Chengjie, C.; Hao, C.; Yi, Z.; Hannah, R.T.; Margaret, H.F.; Yehua, H.; Rui, X. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Loenen, W.A.M.; Dryden, D.T.F.; Raleigh, E.A.; Wilson, G.G. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2013, 42, 20–44. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.N.; Dryden, D.T.F.; Bheemanaik, S. Type III restriction-modification enzymes: A historical perspective. Nucleic Acids Res. 2014, 42, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Truglio, J.J.; Rhau, B.; Croteau, D.L.; Wang, L.; Skorvaga, M.; Karakas, E.; DellaVecchia, M.J.; Wang, H.; Van Houten, B.; Kisker, C. Structural insights into the first incision reaction during nucleotide excision repair. EMBO J. 2005, 24, 885–894. [Google Scholar] [CrossRef] [PubMed]
- Nagamalleswari, E.; Rao, S.; Vasu, K.; Nagaraja, V. Restriction endonuclease triggered bacterial apoptosis as a mechanism for long time survival. Nucleic Acids Res. 2017, 45, 8423–8434. [Google Scholar] [CrossRef] [PubMed]
- Beaulaurier, J.; Schadt, E.E.; Fang, G. Deciphering bacterial epigenomes using modern sequencing technologies. Nat. Rev. Genet. 2018, 20, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Gaidamakova, E.K.; Matrosova, V.Y.; Bennett, B.; Daly, M.J.; Hoffman, B.M. Responses of Mn2+ speciation in Deinococcus radiodurans and Escherichia coli to γ-radiation by advanced paramagnetic resonance methods. Proc. Natl. Acad. Sci. USA 2013, 110, 5945–5950. [Google Scholar] [CrossRef]
- Kobayashi, I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001, 15, 3742–3756. [Google Scholar] [CrossRef]
- Asakura, Y.; Kobayashi, I. From damaged genome to cell surface: Transcriptome changes during bacterial cell death triggered by loss of a restriction–modification gene complex. Nucleic Acids Res. 2009, 37, 3021–3031. [Google Scholar] [CrossRef]
- von Kugelgen, A.; van Dorst, S.; Vikram, A.; Tanmay, A.M.B. A multidomain connector links the outer membrane and cell wall in phylogenetically deep-branching bacteria. Proc. Natl. Acad. Sci. USA 2022, 119, e2203156119. [Google Scholar] [CrossRef]
- Erental, A.; Kalderon, Z.; Saada, A.; Smith, Y.; Engelberg-Kulka, H.; Rosa, P. Apoptosis-Like Death, an Extreme SOS Response in Escherichia coli. mBio 2014, 5, e01426-14. [Google Scholar] [CrossRef]
- Tanaka, M.; Earl, A.M.; Howell, H.A.; Park, M.-J.; Eisen, J.A.; Peterson, S.N.; Battista, J.R. Analysis of Deinococcus radiodurans’s Transcriptional Response to Ionizing Radiation and Desiccation Reveals Novel Proteins That Contribute to Extreme Radioresistance. Genetics 2004, 168, 21–33. [Google Scholar] [CrossRef]
- Dai, S.; Jin, Y.; Li, T.; Weng, Y.; Xu, X.; Zhang, G.; Li, J.; Pang, R.; Tian, B.; Hua, Y. DR1440 is a potential iron efflux protein involved in maintenance of iron homeostasis and resistance of Deinococcus radiodurans to oxidative stress. PLoS ONE 2018, 13, e0202287. [Google Scholar] [CrossRef]
- Cheng, K.; Chen, X.; Xu, G.; Wang, L.; Xu, H.; Yang, S.; Zhao, Y.; Hua, Y.; Metcalf, W.W. Biochemical and Functional Characterization of the NurA-HerA Complex from Deinococcus radiodurans. J. Bacteriol. 2015, 197, 2048–2061. [Google Scholar] [CrossRef]
- Han, W.; Zhou, C.; Cheng, J.; Pan, M.; Hua, Y.; Zhao, Y. Characterization and role of a 2′,3′-cyclic phosphodiesterase from Deinococcus radiodurans. Biotechnol. Lett. 2017, 39, 1211–1217. [Google Scholar] [CrossRef]
- François, L.; Geneviève, C.; Suzanne, S.; Adriana, B. Vectors for regulated gene expression in the radioresistant bacterium Deinococcus radiodurans. Gene 2004, 336, 25–35. [Google Scholar] [CrossRef]
- Bonacossa de Almeida, C.; Coste, G.; Sommer, S.; Bailone, A. Quantification of RecA protein in Deinococcus radiodurans reveals involvement of RecA, but not LexA, in its regulation. Mol. Genet. Genom. 2002, 268, 28–41. [Google Scholar] [CrossRef]



| Gene ID | Function Annotation | ΔR.DraR1 vs. R1.DEG | ΔM.DraR1 vs. R1.DEG | ||
|---|---|---|---|---|---|
| log2FC | p-Value | log2FC | p-Value | ||
| DNA damage response protein | |||||
| DR_0423 | Single-stranded DNA-binding protein, ddrA | −1.86 | 6.56 × 10−6 | 6.61 | 1.59 × 10−56 |
| DR_0070 | Single-stranded DNA-binding protein, DdrB | −1.47 | 1.56 × 10−4 | 4.66 | 5.89 × 10−27 |
| DR_0003 | DNA damage response protein C, ddrC | −2.39 | 3.07 × 10−9 | 4.36 | 9.44 × 10−19 |
| DR_0326 | DNA damage response protein D, ddrD | −1.39 | 1.15 × 10−3 | 5.89 | 2.11 × 10−26 |
| DR_B0100 | RNA ligase domain-containing protein, DdrP | −1.08 | 2.34 × 10−3 | 3.14 | 3.73 × 10−14 |
| DR_2574 | Transcriptional repressor of the RDR regulon, DdrO | −1.28 | 4.14 × 10−4 | 3.57 | 3.47 × 10−27 |
| DR_A0346 | DNA repair protein PprA | −1.52 | 1.53 × 10−3 | 3.13 | 6.37 × 10−12 |
| DR_C0012 | GerE family transcriptional regulator | 2.37 | 1.40 × 10−8 | 9.35 | 2.05 × 10−7 |
| Transporter | |||||
| DR_0203 | ABC transporter permease | −1.18 | 1.36 × 10−2 | 1.79 | 6.25 × 10−5 |
| DR_1036 | branched-chain amino acid ABC transporter permease | 1.05 | 2.38 × 10−3 | −0.96 | 1.61 × 10−2 |
| DR_1142 | protein transporter | −2.18 | 1.91 × 10−10 | 1.22 | 8.28 × 10−3 |
| DR_2612 | undecaprenyl phosphate transporter, DedA | −1.31 | 1.09 × 10−2 | 1.57 | 1.18 × 10−4 |
| DR_B0083 | Potassium-transporting ATPase subunit B, kdpB | −1.11 | 1.82 × 10−2 | 2.58 | 1.36 × 10−3 |
| DR_B0086 | Potassium-transporting ATPase subunit A, kdpA | −1.32 | 1.60 × 10−2 | 2.23 | 4.56 × 10−4 |
| DR_A0073 | cation-transporting P-type ATPase | −1.18 | 4.98 × 10−4 | 2.05 | 1.12 × 10−9 |
| Energy production | |||||
| DR_A0076 | AAA+ ATPase domain-containing protein | −1.72 | 4.25 × 10−3 | 3.48 | 4.34 × 10−5 |
| DR_C0016 | Phosphodiester glycosidase domain-containing protein | −5.12 | 7.99 × 10−25 | 4.44 | 2.27 × 10−6 |
| DR_C0009 | KAP NTPase domain-containing protein | 2.08 | 1.53 × 10−6 | 1.23 | 1.04 × 10−2 |
| DR_C0023 | N-terminal DNA-binding domain-containing protein, KfrA | −1.27 | 2.53 × 10−2 | 7.66 | 3.71 × 10−4 |
| Biosynthetic pathway | |||||
| DR_0133 | acetyl xylan esterase | −1.00 | 2.84 × 10−2 | 2.00 | 5.51 × 10−7 |
| DR_0422 | Trans-aconitate 2-methyltransferase, tam | −1.26 | 7.85 × 10−5 | 5.17 | 3.86 × 10−31 |
| DR_0598 | Peptidase M23 domain-containing protein, LytH | −1.05 | 5.65 × 10−3 | 3.26 | 5.56 × 10−3 |
| DR_0659 | frnE protein | −1.35 | 2.57 × 10−5 | 3.15 | 2.59 × 10−4 |
| DR_A0029 | 4-aminobutyrate aminotransferase, argD | −1.20 | 1.23 × 10−3 | −1.66 | 1.33 × 10−3 |
| DR_A0178 | xanthine dehydrogenase C-terminal subunit | 1.10 | 7.00 × 10−3 | −1.17 | 1.03 × 10−3 |
| DR_A0179 | xanthine dehydrogenase accessory protein XdhC | 1.39 | 4.09 × 10−5 | −1.15 | 4.02 × 10−3 |
| DR_A0180 | guanine deaminase, guaD | 1.14 | 3.86 × 10−4 | −1.12 | 3.02 × 10−3 |
| DR_B0060 | Spore coat protein U domain-containing protein | −2.40 | 9.31 × 10−6 | 6.46 | 8.75 × 10−6 |
| DR_C0013 | N-acetylmuramoyl-L-alanine amidase | −4.18 | 4.30 × 10−14 | 6.07 | 2.26 × 10−5 |
| Nucleoid-associated gene | |||||
| DR_0906 | DNA gyrase subunit B, GyrB | −1.07 | 1.61 × 10−2 | 0.89 | 5.65 × 10−3 |
| Exonuclease | |||||
| DR_C0006 | putative 3′-5′ ssDNA/RNA exonuclease, TatD | 1.56 | 1.69 × 10−5 | 1.42 | 5.19 × 10−3 |
| Natural competence regulated gene | |||||
| DR_2338 | cinA protein | −1.25 | 1.59 × 10−3 | 2.56 | 2.18 × 10−15 |
| Peptidase activity | |||||
| DR_A0028 | DUF4274 domain-containing protein | −1.36 | 5.85 × 10−3 | −1.77 | 4.53 × 10−4 |
| DR_A0283 | S8 serine protease | 1.23 | 1.39 × 10−2 | −0.96 | 1.06 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, C.; Wang, L.; Xu, H.; Zhao, Y.; Tian, B.; Hua, Y. Characterization of a Novel N4-Methylcytosine Restriction-Modification System in Deinococcus radiodurans. Int. J. Mol. Sci. 2024, 25, 1660. https://doi.org/10.3390/ijms25031660
Shi C, Wang L, Xu H, Zhao Y, Tian B, Hua Y. Characterization of a Novel N4-Methylcytosine Restriction-Modification System in Deinococcus radiodurans. International Journal of Molecular Sciences. 2024; 25(3):1660. https://doi.org/10.3390/ijms25031660
Chicago/Turabian StyleShi, Chenxiang, Liangyan Wang, Hong Xu, Ye Zhao, Bing Tian, and Yuejin Hua. 2024. "Characterization of a Novel N4-Methylcytosine Restriction-Modification System in Deinococcus radiodurans" International Journal of Molecular Sciences 25, no. 3: 1660. https://doi.org/10.3390/ijms25031660
APA StyleShi, C., Wang, L., Xu, H., Zhao, Y., Tian, B., & Hua, Y. (2024). Characterization of a Novel N4-Methylcytosine Restriction-Modification System in Deinococcus radiodurans. International Journal of Molecular Sciences, 25(3), 1660. https://doi.org/10.3390/ijms25031660








