Comparative Expression Profiling Reveals the Regulatory Effects of Dietary Mannan Oligosaccharides on the Intestinal Immune Response of Juvenile Megalobrama amblycephala against Aeromonas hydrophila Infection
Abstract
1. Introduction
2. Results
2.1. Sequencing Quality, Reads Mapping and Annotation
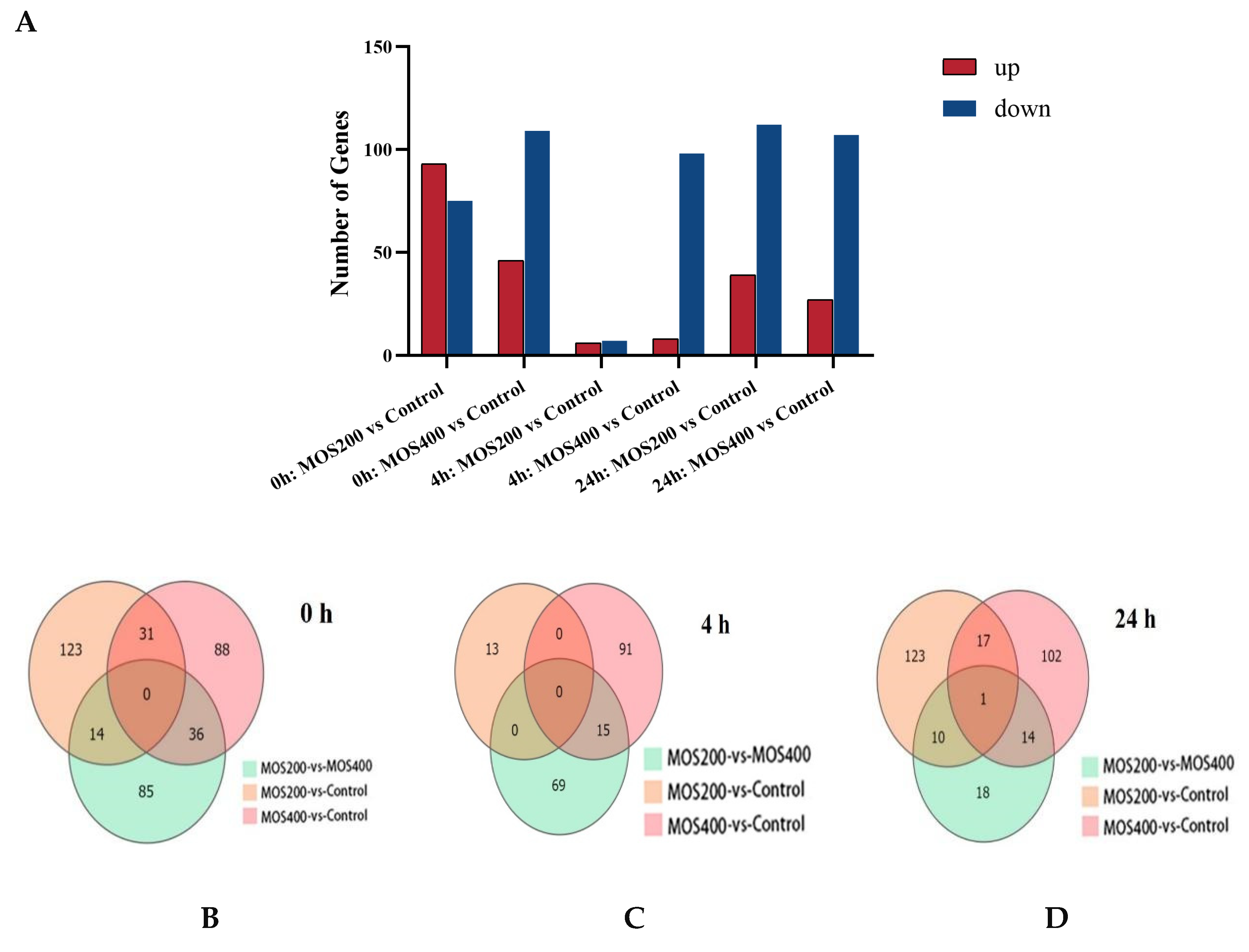
2.2. Identification and Statistics of Differentially Expressed Genes (DEGs)
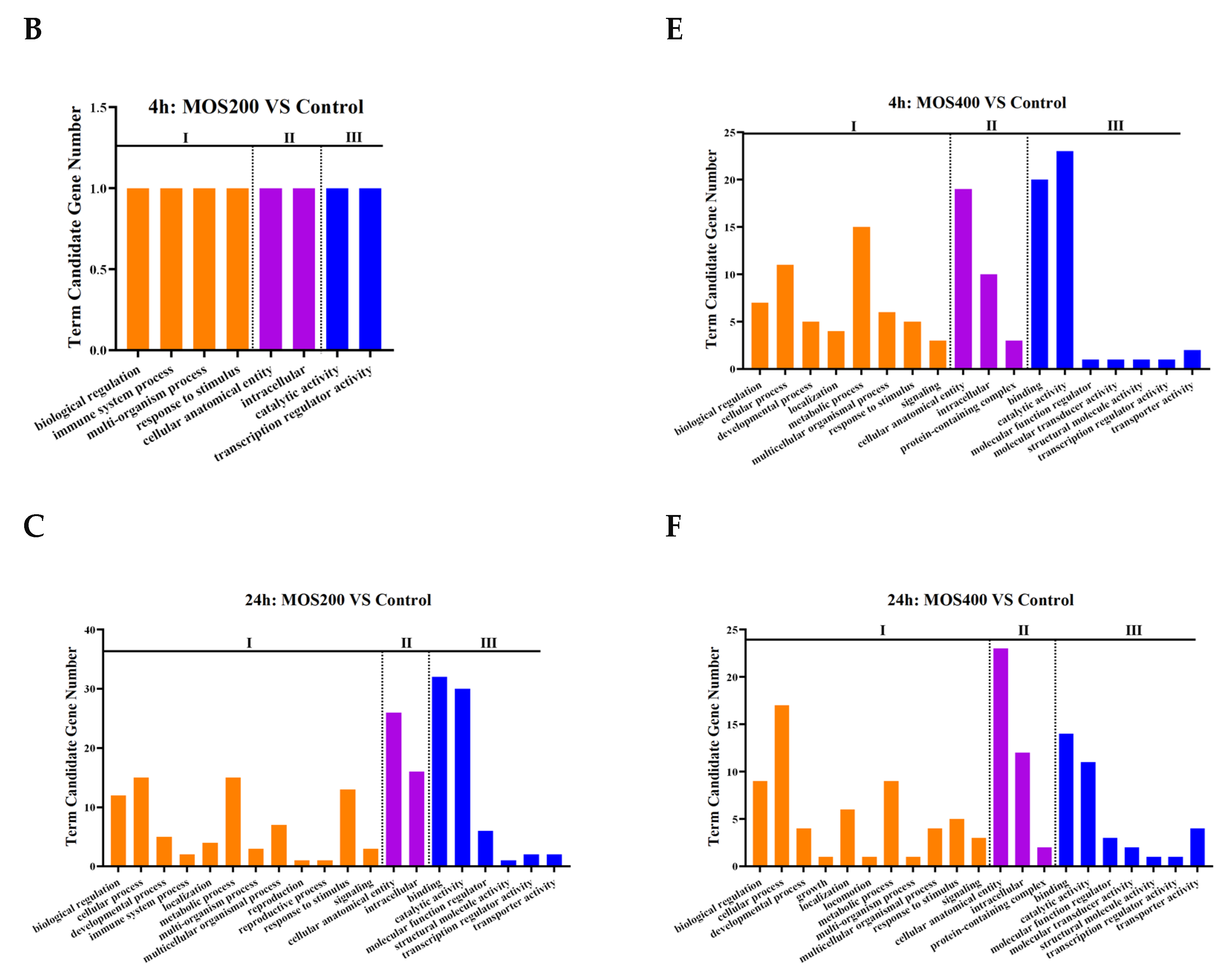
2.3. Functional Annotation of DEGs
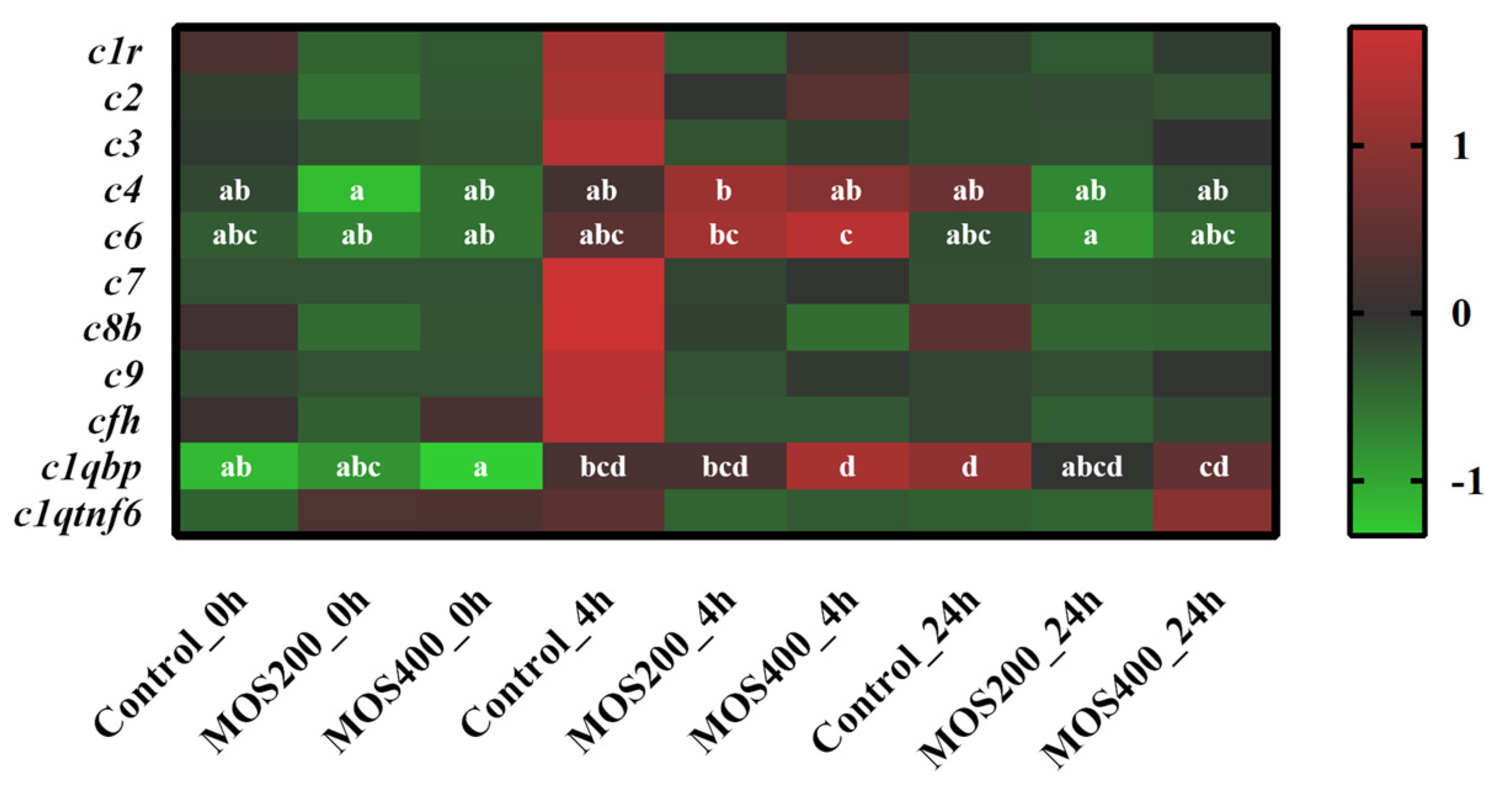
2.4. Regulation of MOS on Complement-Related Genes
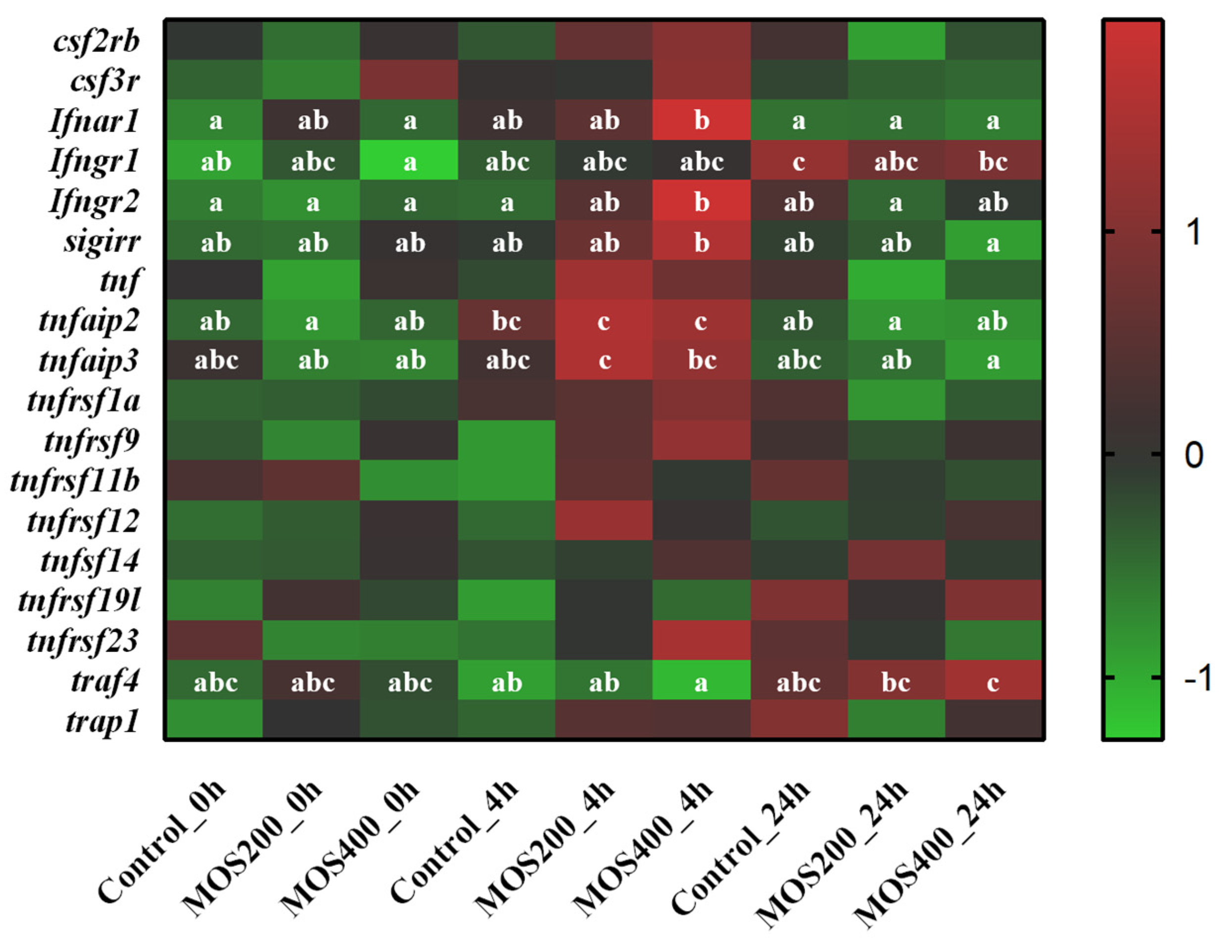
2.5. Activation of Inflammation-Related Cytokines by MOS
2.6. Transcriptional Factors Activated by MOS
2.7. Apoptosis-Related Genes Regulated by MOS
2.8. Identification of Tight Junction-Related Genes
2.9. qRT-PCR Verification
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Experiment Diets and Feeding Trial
4.3. Experimental Infections and Samples Collection
4.4. Total RNA Isolation, Illumina Sequencing and cDNA Preparation
4.5. Sequencing Quality Control and Reads Mapping
4.6. Differential Expression Analysis
4.7. qRT-PCR Assay
4.8. Western Blot
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, T.; Sun, R.; Zhang, Y.; Zhang, C.; Wang, Y.; Wang, Z.; Du, Y. Recent research and application prospect of functional oligosaccharides on Intestinal disease treatment. Molecules 2022, 27, 7622. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, X.; Bai, Y.; Zhou, X.; Chen, L.; Qiu, C.; Lu, C.; Jin, Z.; Long, J.; Xie, Z. Natural antimicrobial oligosaccharides in the food industry. Int. J. Food. Microbiol. 2022, 386, 110021. [Google Scholar] [CrossRef]
- Zhang, N.; Jin, M.; Wang, K.; Zhang, Z.; Shah, N.; Wei, H. Functional oligosaccharide fermentation in the gut: Improving intestinal health and its determinant factors-A review. Carbohyd. Polym. 2021, 284, 119043. [Google Scholar] [CrossRef] [PubMed]
- Dimitroglou, A.; Davies, S.; Sweetman, J.; Divanach, P.; Chatzifotis, S. Dietary supplementation of mannan oligosaccharide on white sea bream (Diplodus sargus L.) larvae: Effects on development, gut morphology and salinity tolerance. Aquac. Res. 2010, 41, 245–251. [Google Scholar] [CrossRef]
- Wang, T.; Wu, H.; Li, W.; Xu, R.; Qiao, F.; Du, Z.; Zhang, M. Effects of dietary mannan oligosaccharides (MOS) supplementation on metabolism, inflammatory response and gut microbiota of juvenile Nile tilapia (Oreochromis niloticus) fed with high carbohydrate diet. Fish Shellfish Immun. 2022, 130, 550–559. [Google Scholar] [CrossRef]
- Doan, H.; Hoseinifar, S.; Faggio, C.; Chitmanat, C.; Mai, N.; Jaturasitha, S.; Ringq, E. Effects of corncob derived xylooligosaccharide on innate immune response, disease resistance, and growth performance in Nile Tilapia (Oreochromis Niloticus) fingerlings. Aquaculture. 2018, 495, 786–793. [Google Scholar] [CrossRef]
- Gültepe, N.; Salnur, S.; Hoşsu, B.; Hisar, O. Dietary supplementation with Mannanoligosaccharides (MOS) from Bio-Mos enhances growth parameters and digestive capacity of gilthead sea bream (Sparus aurata). Aquacult. Nutr. 2011, 17, 482–487. [Google Scholar] [CrossRef]
- Salem, M.; Gaber, M.; Zaki, M.; Nour, A. Effects of dietary mannan oligosaccharides on growth, body composition and intestine of the sea bass. Aquac. Res. 2015, 47, 3516–3525. [Google Scholar] [CrossRef]
- Ali, S.; Ambasankar, K.; Praveena, E.; Nandakumar, S.; Syamadayal, J. Effect of dietary mannan oligosaccharide on growth, body composition, haematology and biochemical parameters of Asian seabass (Lates calcarifer). Aquac. Res. 2017, 48, 899–908. [Google Scholar] [CrossRef]
- Akter, M.; Sutriana, A.; Talpur, A.; Hashim, R. Dietary supplementation with mannan oligosaccharide influences growth, digestive enzymes, gut morphology, and microbiota in juvenile striped catfish, Pangasianodon hypophthalmus. Aquacult. Int. 2016, 24, 127–144. [Google Scholar] [CrossRef]
- Ding, Z.; Wang, X.; Liu, Y.; Zheng, Y.; Li, H.; Zhang, M.; He, Y.; Cheng, H.; Xu, J.; Chen, X.; et al. Dietary Mannan oligosaccharides enhance the non-specific Immunity, Intestinal health, and resistance capacity of juvenile blunt snout bream (Megalobrama amblycephala) against Aeromonas hydrophila. Front. Immunol. 2022, 13, 863657. [Google Scholar] [CrossRef] [PubMed]
- Welker, T.; Lim, C.; Yildirim-Aksoy, M.; Shelby, R.; Klesius, P. Immune response and resistance to stress and Edwardsiella ictaluri challenge in channel catfish, Ictalurus punctatus, fed diets containing commercial whole-cell yeast or yeast subcomponents. J. World Aquacult. Soc. 2010, 38, 24–35. [Google Scholar] [CrossRef]
- Lokesh, J.; Fernandes, J.; Korsnes, K.; Bergh, Q.; Brinchmann, M.; Kiron, V. Transcriptional regulation of cytokines in the intestine of Atlantic cod fed yeast derived mannan oligosaccharide or β-glucan and challenged with Vibrio anguillarum. Fish Shellfish Immun. 2012, 33, 626–631. [Google Scholar] [CrossRef]
- Lee, S.; Katya, K.; Hamidoghli, A.; Hong, J.; Kim, D.; Bai, S. Synergistic effects of dietary supplementation of Bacillus subtilis WB60 and mannanoligosaccharide (MOS) on growth performance, immunity and disease resistance in Japanese eel, Anguilla japonica. Fish Shellfish Immun. 2018, 83, 283–291. [Google Scholar] [CrossRef]
- Shiflhtt, D.; Clayburgh, D.; Koutsouris, A.; Turner, J.; Hecht, G. Enteropathgenic E. coli disrupts tight junction barier function and structure in vivo. Lab. Invest. 2005, 85, 1308–1324. [Google Scholar] [CrossRef] [PubMed]
- Torrecillas, S.; Makol, A.; Caballero, M.; Montero, D.; Dhanasiri, A.; Sweetman, J.; Izquierdo, M. Effects on mortality and stress response in European sea bass, Dicentrarchus labrax (L.), fed mannan oligosaccharides (MOS) after Vibrio anguillarum exposure. J. Fish Dis. 2012, 35, 591–602. [Google Scholar] [CrossRef]
- Teng, T.; Xi, B.; Xie, J.; Chen, K.; Xu, P.; Pan, L. Molecular cloning and expression analysis of Megalobrama amblycephala transferrin gene and effects of exposure to iron and infection with Aeromonas hydrophila. Fish Physiol. Biochem. 2017, 43, 987–997. [Google Scholar] [CrossRef]
- Hwang, J.; Kwon, M.; Jung, S.; Park, M.; Kim, D.; Cho, W.; Park, J.; Son, M. RNA-Seq transcriptome analysis of the olive flounder (Paralichthys olivaceus) kidney response to vaccination with heat-inactivated viral hemorrhagic septi-cemia virus. Fish. Shellfish. Immun. 2017, 62, 221–226. [Google Scholar] [CrossRef]
- Wu, Q.; Ning, X.; Jiang, S.; Sun, L. Transcriptome analysis reveals seven key immune pathways of Japanese flounder (Paralichthys olivaceus) involved in megalocytivirus infection. Fish. Shellfish. Immun. 2020, 103, 150–158. [Google Scholar] [CrossRef]
- Yi, S.; Gao, Z.; Zhao, H.; Zeng, C.; Luo, W.; Chen, B.; Wang, W. Identification and characterization of microRNAs involved in growth of blunt snout bream (Megalobrama amblycephala) by Solexa sequencing. BMC Genomics. 2013, 14, 754. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.; Blake, J.; Botstein, D.; Butler, H.; Cherry, J.; Davis, A.; Dolinski, K.; Dwight, S.; Eppig, J.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Wang, S.; Zhang, Y.; Huang, T.; Cai, Y. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PloS. One. 2017, 12, e0184129. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wang, X.; Liu, P.; Wang, X. Complement-related proteins in crustacean immunity. Dev. Comp. Immunol. 2022, 139, 104577. [Google Scholar] [CrossRef] [PubMed]
- Ricklin, D.; Hajishengallis, G.; Yang, K.; Lambris, J. Complement: A key system for immune surveillance and homeostasis. Nat. Immunol. 2010, 11, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, W.; Park, E.; Park, S. C1qTNF-related protein-6 increases the expression of interleukin-10 in macrophages. Mol. Cells. 2010, 30, 59–64. [Google Scholar] [CrossRef]
- Wong, G.; Krawczyk, S.; Kitidis-Mitrokostas, C.; Revett, T.; Gimeno, R.; Lodish, H. Molecular, biochemical and functional characterizations of C1q/TNF family members: Adipose-tissue-selective expression patterns, regulation by PPAR-γ agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem. J. 2008, 416, 161–177. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, H.; Li, H.; Wang, A. Transcriptome profiling of grass carp (Ctenopharyngodon idellus) infected with Aeromonas hydrophila. Fish Shellfish Immun. 2016, 51, 329–336. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, Y.; Han, Z.; Zheng, Y.; Wang, X.; Zhang, M.; Li, H.; Xu, J.; Chen, X.; Ding, Z.; et al. Comparative effects of dietary soybean oil and fish oil on the growth performance, fatty acid composition and lipid metabolic signaling of grass carp, Ctenopharyngodon idella. Aquacult. Rep. 2022, 22, 101002. [Google Scholar] [CrossRef]
- Lu, Z.; Feng, L.; Jiang, W.; Wu, P.; Liu, Y.; Jiang, J.; Kuang, S.; Tang, L.; Li, S.; Liu, X.; et al. Mannan oligosaccharides application: Multipath restriction from Aeromonas hydrophila Infection in the skin barrier of grass carp (Ctenopharyngodon idella). Front. Immunol. 2021, 12, 4280. [Google Scholar] [CrossRef]
- Modanloo, M.; Soltanian, S.; Akhlaghi, M.; Hoseinifar, S. The effects of single or combined administration of galactooligosaccharide and Pediococcus acidilactici on cutaneous mucus immune parameters, humoral immune responses and immune related genes expression in common carp (Cyprinus Carpio) fingerlings. Fish Shellfish Immun. 2017, 70, 391–397. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Zhang, J.; Liu, R.; Zhao, H.; Shan, S.; Yang, G. Identification of three inflammatory Caspases in common carp (Cyprinus carpio L.) and its role in immune response against bacterial infection. Fish Shellfish Immun. 2022, 131, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. Mechanisms of caspase activation and Inhibition during apoptosis. Mol. Cell. 2002, 9, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Mittra, N.; Kumar, V.; Patel, D.; Singh, C. Inflammation and B-cell lymphoma-2 associated X protein regulate zinc-induced apoptotic degeneration of rat nigrostriatal dopaminergic neurons. Mol. neurobiol. 2016, 53, 5782–5795. [Google Scholar] [CrossRef] [PubMed]
- Hockenbery, D.; Nuñez, G.; Milliman, C.; Schreiber, R.; Korsmeyer, S. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature. 1990, 348, 334–336. [Google Scholar] [CrossRef]
- Meyer, T.; Schwesinger, C.; Denker, B. Zonula occludens-1 is a scaffolding protein for signaling molecules: Gα12 directly binds to the Src homology 3 domain and regulates paracellular permeability in epithelial cells. J. Biol. Chem. 2002, 277, 24855–24858. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Zhao, X.; Zhan, Q.; Cui, L.; Sun, Q.; Lin, L.; Wang, W.; Liu, H. Characterization and expression analysis of an intelectin gene from Megalobrama amblycephala with excellent bacterial binding and agglutination activity. Fish Shellfish Immun. 2017, 61, 100–110. [Google Scholar] [CrossRef]
- Cui, H.; Li, H.; Zhang, M.; Li, H.; Wang, X.; Wang, Z.; Zhai, W.; Chen, X.; Cheng, H.; Xu, J.; et al. Molecular characterization, expression, evolutionary selection, and biological activity analysis of CD68 gene from Megalobrama amblycephala. Int. J. Mol. Sci. 2022, 23, 13133. [Google Scholar] [CrossRef]
- Cock, P.; Fields, C.; Goto, N.; Heuer, M.; Rice, P. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic. Acids. Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Liu, H.; Chen, C.; Chen, Z.; Min, J.; Min, Y.; Min, J.; Min, X.; Min, H.; Ebersberger, I.; Ebersberger, M.; et al. The draft genome of blunt snout bream (Megalobrama amblycephala) reveals the development of intermuscular bone and adaptation to herbivorous diet. Gigascience. 2017, 6, 1–13. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S. HISAT: A fast spliced aligner with low memory requirements. Nat. methods. 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Lynch, G.; Guo, W.; Sarkar, S.; Finner, H. The control of the false discovery rate in fixed sequence multiple testing. Electron. J. Stat. 2017, 11, 4649–4673. [Google Scholar] [CrossRef]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Song, C.; Liu, B.; Xu, P.; Xie, J.; Ge, X.; Zhou, Q.; Sun, C.; Zhang, H.; Shan, F.; Yang, Z. Oxidized fish oil injury stress in Megalobrama amblycephala: Evaluated by growth, intestinal physiology, and transcriptome-based PI3K-Akt/NF-κB/TCR inflammatory signaling. Fish shellfish immun. 2018, 81, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, Y.; Li, H.; Wang, X.; Zhang, M.; Shen, X.; Cheng, H.; Xu, J.; Chen, X.; Wang, X.; et al. Novel insights into the immune regulatory effects of Megalobrama amblycephala intelectin on the phagocytosis and killing activity of macrophages. Mol. Immunol. 2021, 137, 145–154. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, X.; Chen, L.; Chen, N.; Li, Y.; Wang, W.; Wang, H. Sequence analysis and expression differentiation of chemokine receptor CXCR4b among three populations of Megalobrama amblycephala. Dev. Comp. Immunol. 2013, 40, 195–201. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cui, H.; Shen, X.; Zheng, Y.; Guo, P.; Gu, Z.; Gao, Y.; Zhao, X.; Cheng, H.; Xu, J.; Chen, X.; et al. Identification, expression patterns, evolutionary characteristics and recombinant protein activities analysis of CD209 gene from Megalobrama amblycephala. Fish Shellfish Immun. 2022, 126, 47–56. [Google Scholar] [CrossRef]


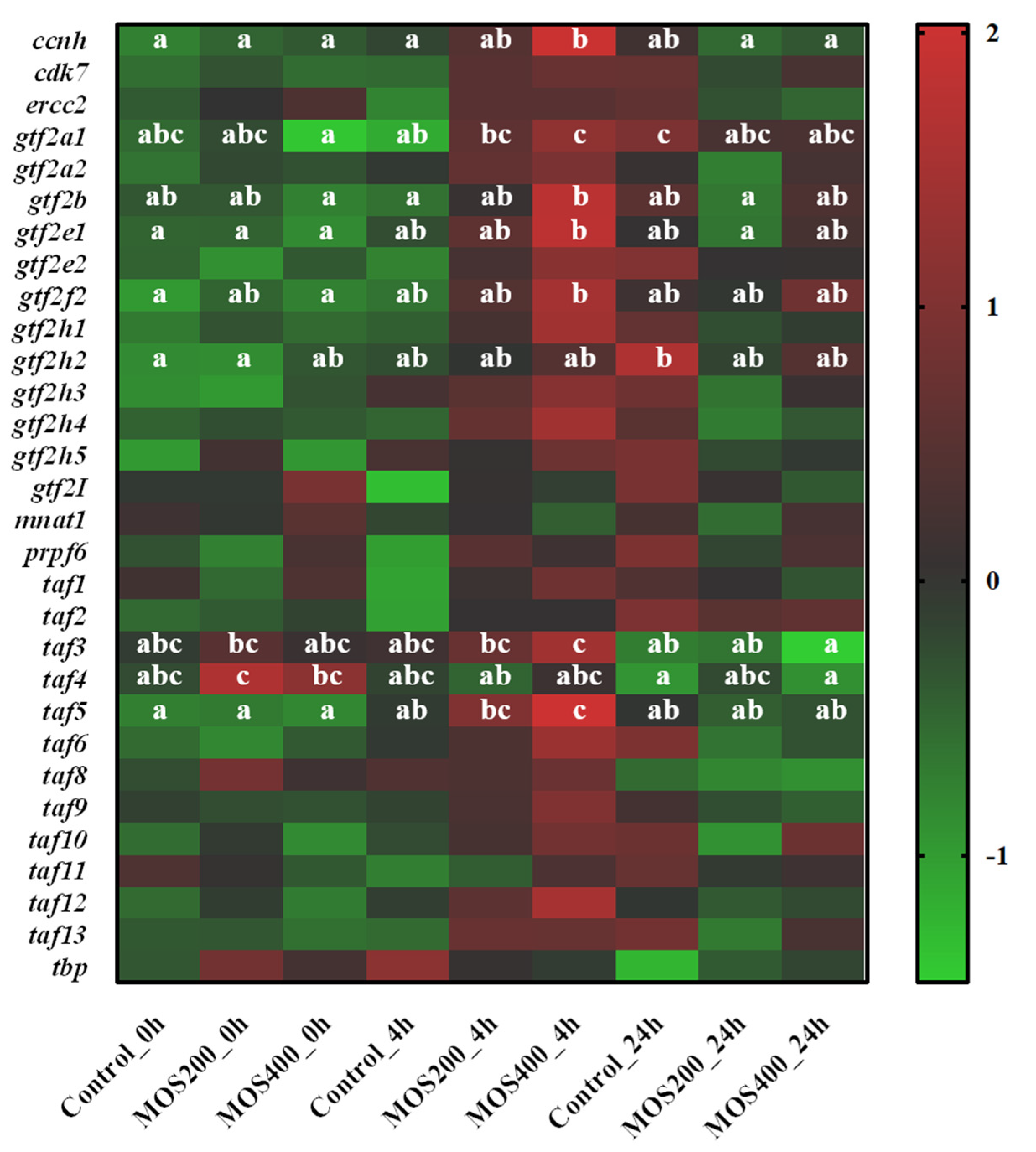

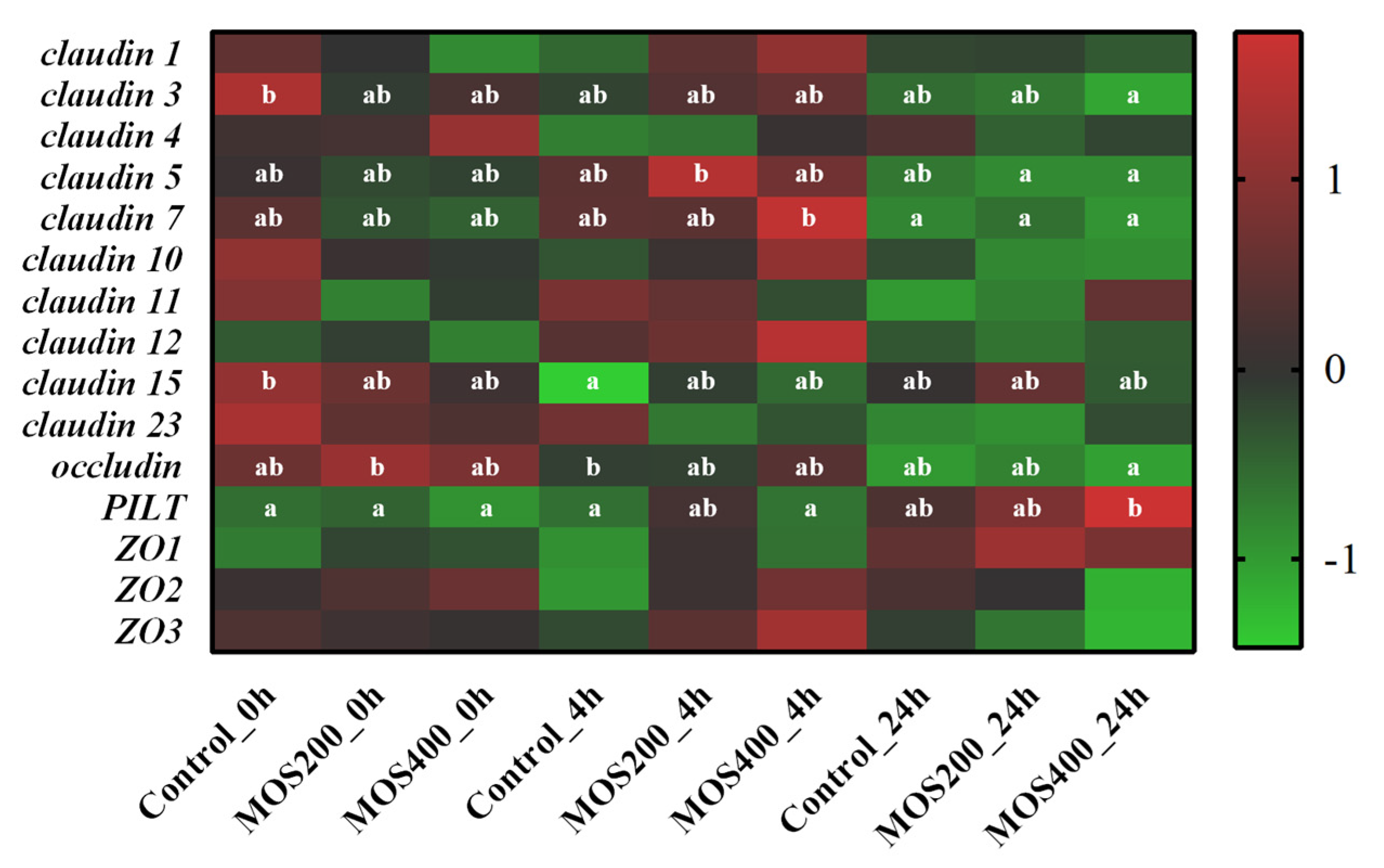
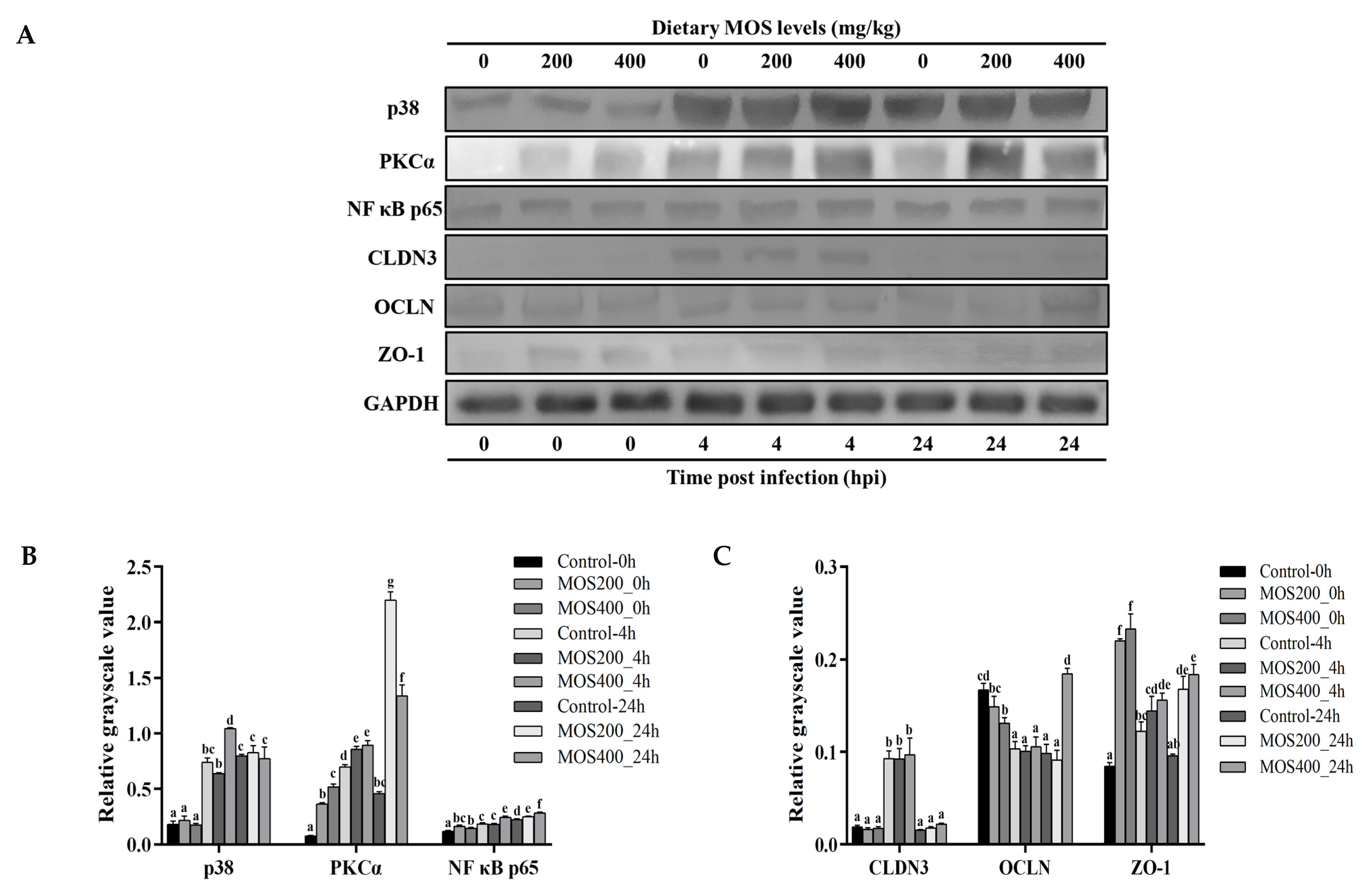
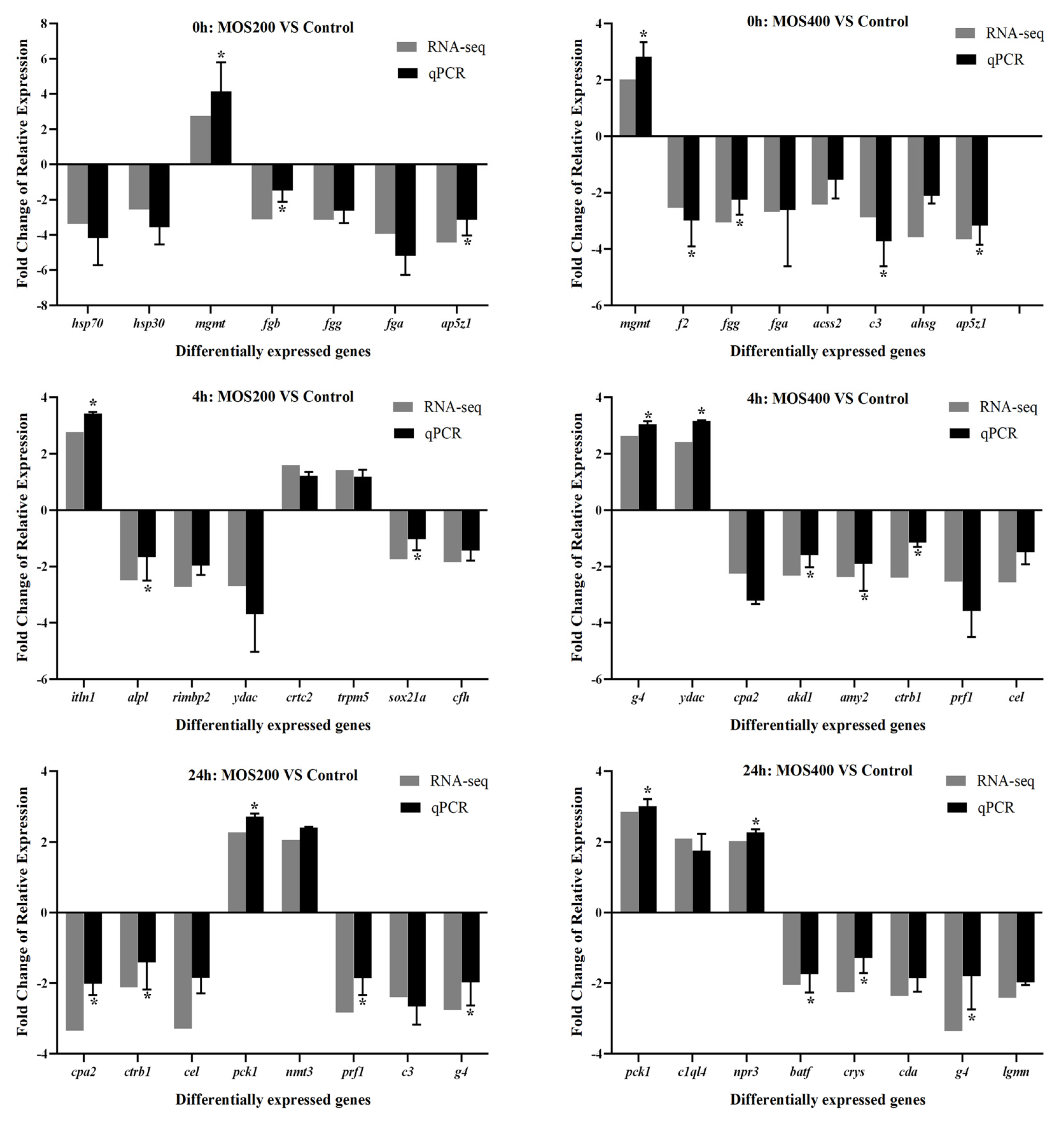
| Samples | Raw Reads (M) | Clean Reads (M) | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|
| Control_0_1 | 21.75 | 21.42 | 0.03 | 97.90 | 93.55 | 45.34 |
| Control_0_2 | 21.75 | 21.47 | 0.03 | 97.84 | 93.38 | 46.73 |
| Control_0_3 | 21.75 | 21.49 | 0.02 | 98.03 | 93.92 | 47.11 |
| Control_4_1 | 21.75 | 21.50 | 0.02 | 98.06 | 93.99 | 46.51 |
| Control_4_2 | 21.75 | 21.48 | 0.03 | 98.08 | 94.11 | 46.45 |
| Control_4_3 | 21.75 | 21.48 | 0.03 | 98.33 | 94.47 | 47.33 |
| Control_24_1 | 21.75 | 21.49 | 0.02 | 98.13 | 94.28 | 45.93 |
| Control_24_2 | 21.75 | 21.5 | 0.03 | 97.88 | 93.54 | 46.14 |
| Control_24_3 | 21.75 | 21.46 | 0.02 | 98.03 | 94.01 | 46.13 |
| MOS200_0_1 | 20.44 | 20.15 | 0.03 | 97.82 | 93.55 | 46.38 |
| MOS200_0_2 | 21.75 | 21.37 | 0.03 | 97.85 | 93.57 | 46.03 |
| MOS200_0_3 | 21.75 | 21.46 | 0.03 | 98.02 | 94.06 | 45.56 |
| MOS200_4_1 | 21.75 | 21.48 | 0.03 | 98.00 | 93.85 | 46.26 |
| MOS200_4_2 | 21.75 | 21.45 | 0.02 | 97.75 | 93.27 | 46.67 |
| MOS200_4_3 | 21.75 | 21.45 | 0.02 | 97.94 | 93.87 | 45.79 |
| MOS200_24_1 | 21.75 | 21.46 | 0.03 | 97.99 | 93.84 | 46.02 |
| MOS200_24_2 | 21.75 | 21.47 | 0.03 | 98.26 | 94.6 | 45.80 |
| MOS200_24_3 | 21.75 | 21.46 | 0.03 | 98.20 | 94.38 | 45.85 |
| MOS400_0_1 | 20.35 | 20.07 | 0.03 | 97.76 | 93.43 | 46.45 |
| MOS400_0_2 | 21.75 | 21.44 | 0.03 | 98.06 | 94.10 | 45.73 |
| MOS400_0_3 | 21.75 | 21.48 | 0.03 | 98.07 | 94.13 | 45.67 |
| MOS400_4_1 | 21.75 | 21.43 | 0.03 | 97.97 | 93.85 | 46.33 |
| MOS400_4_2 | 21.75 | 21.45 | 0.02 | 97.81 | 93.53 | 46.14 |
| MOS400_4_3 | 21.75 | 21.48 | 0.03 | 98.01 | 93.99 | 46.21 |
| MOS400_24_1 | 21.75 | 21.48 | 0.03 | 98.06 | 94.04 | 46.47 |
| MOS400_24_2 | 21.75 | 21.46 | 0.04 | 98.02 | 93.86 | 46.29 |
| MOS400_24_3 | 21.75 | 21.48 | 0.03 | 97.90 | 93.64 | 45.84 |
| Groups | GO (%) | KEGG (%) | Up-Regulation (%) | Down-Regulation (%) | Total |
|---|---|---|---|---|---|
| 0 h MOS200 vs. Control | 53 (31.55) | 155 (92.26) | 93 (55.36) | 75 (44.64) | 168 |
| 0 h MOS400 vs. Control | 38 (24.52) | 150 (96.77) | 46 (29.68) | 109 (70.32) | 155 |
| 4 h MOS200 vs. Control | 3 (23.08) | 9 (69.23) | 6 (46.15) | 7 (53.85) | 13 |
| 4 h MOS400 vs. Control | 35 (33.02) | 101 (95.28) | 8 (7.55) | 98 (72.45) | 106 |
| 24 h MOS200 vs. Control | 54 (35.76) | 132 (87.42) | 39 (25.83) | 112 (74.17) | 151 |
| 24 h MOS400 vs. Control | 33 (24.63) | 126 (94.03) | 27 (20.15) | 107 (79.85) | 134 |
| Total | 176 | 570 | 198 | 434 | 622 |
| Ko ID | KEGG Terms | Number of DEGs | FDR ≤ 0.01 |
|---|---|---|---|
| 0 h MOS200 vs. Control | |||
| map04975 | Fat digestion and absorption | 10 | 5.01 × 10−10 |
| map04610 | Complement and coagulation cascades | 14 | 8.37 × 10−9 |
| map03320 | PPAR signaling pathway | 9 | 1.98 × 10−6 |
| map04977 | Vitamin digestion and absorption | 6 | 2.45 × 10−5 |
| map04979 | Cholesterol metabolism | 8 | 2.15 × 10−5 |
| map04152 | AMPK signaling pathway | 10 | 4.47 × 10−5 |
| map04920 | Adipocytokine signaling pathway | 6 | 1.31 × 10−3 |
| map01212 | Fatty acid metabolism | 5 | 2.16 × 10−3 |
| map00591 | Linoleic acid metabolism | 4 | 2.91 × 10−3 |
| map00061 | Fatty acid biosynthesis | 3 | 3.47 × 10−3 |
| 0 h MOS400 vs. Control | |||
| map00100 | Steroid biosynthesis | 8 | 3.00 × 10−11 |
| map04610 | Complement and coagulation cascades | 14 | 1.88 × 10−9 |
| map00900 | Terpenoid backbone biosynthesis | 6 | 2.10 × 10−7 |
| map04976 | Bile secretion | 7 | 8.21 × 10−5 |
| map03320 | PPAR signaling pathway | 6 | 5.40 × 10−4 |
| map04979 | Cholesterol metabolism | 6 | 6.16 × 10−4 |
| map00120 | Primary bile acid biosynthesis | 3 | 1.54 × 10−3 |
| map04115 | p53 signaling pathway | 5 | 5.83 × 10−3 |
| 4 h MOS200 vs. Control | |||
| map00730 | Thiamine metabolism | 1 | 5.42 × 10−3 |
| 4 h MOS400 vs. Control | |||
| map04972 | Pancreatic secretion | 14 | 1.17 × 10−12 |
| map04650 | Natural killer cell-mediated cytotoxicity | 13 | 1.47 × 10−9 |
| map04974 | Protein digestion and absorption | 12 | 1.52 × 10−8 |
| map00260 | Glycine, serine and threonine metabolism | 5 | 7.61 × 10−6 |
| map04210 | Apoptosis | 9 | 4.12 × 10−5 |
| map04973 | Carbohydrate digestion and absorption | 4 | 2.67 × 10−4 |
| map04979 | Cholesterol metabolism | 5 | 5.49 × 10−4 |
| map00100 | Steroid biosynthesis | 2 | 8.93 × 10−3 |
| map00620 | Pyruvate metabolism | 3 | 8.44 × 10−3 |
| 24 h MOS200 vs. Control | |||
| map04972 | Pancreatic secretion | 10 | 4.97 × 10−7 |
| map04650 | Natural killer cell-mediated cytotoxicity | 9 | 6.92 × 10−5 |
| map04974 | Protein digestion and absorption | 8 | 3.78 × 10−4 |
| map04068 | FoxO signaling pathway | 7 | 6.60 × 10−4 |
| map04152 | AMPK signaling pathway | 7 | 6.78 × 10−4 |
| map04210 | Apoptosis | 8 | 1.05 × 10−3 |
| map03320 | PPAR signaling pathway | 5 | 1.26 × 10−3 |
| map04066 | HIF-1 signaling pathway | 5 | 8.01 × 10−3 |
| 24 h MOS400 vs. Control | |||
| map04910 | Insulin signaling pathway | 6 | 9.62 × 10−3 |
| map04142 | Lysosome | 14 | 3.62 × 10−12 |
| map00511 | Other glycan degradation | 4 | 5.29 × 10−5 |
| map04612 | Antigen processing and presentation | 7 | 8.44 × 10−5 |
| map00603 | Glycosphingolipid biosynthesis—globo and isoglobo series | 3 | 2.71 × 10−4 |
| map04152 | AMPK signaling pathway | 5 | 6.07 × 10−3 |
| map00531 | Glycosaminoglycan degradation | 2 | 9.05 × 10−3 |
| map04145 | Phagosome | 7 | 7.95 × 10−3 |
| Target Genes | Primer Sequences (5′-3′) |
|---|---|
| ap5z1 | Forward: GCTGGATCATGTGAAGTCT Reverse: CACCTGGGTAGCAACTGG |
| acss2 | Forward: GACAGAGTCGCCATCTACCT Reverse: CAGAGCCTCAGACGCAAT |
| ahsg | Forward: ACACCCACGGTTACAAAT Reverse: AATGCCACAACAGACAGAC |
| alpl | Forward: ACTCCCACGCATTTACCT Reverse: GTCTGGACGCTTGTTGTTA |
| ak9 | Forward: ACACTGAAACCGACTGAA Reverse: CACTAAAGGCATCCATCT |
| amy2 | Forward: CCGACTACATGAACAAGC Reverse: CTCAGTCACCCTTCCAAT |
| batf | Forward: AAAAGAAATCGCTGAACTG Reverse: GAATCTGAGGTAAAGGGTG |
| c3 | Forward: ACAAGCCCATCTACACGC Reverse: ATCCCAGAACTGGCAACC |
| crtc2 | Forward: GGTTCTTGTCCCGCTCCC Reverse: TCTCCTCCGCCTGCCTCT |
| cfh | Forward: AAAGATAGACGGAAGTAGTGA Reverse: TTGAACCCTGGTGAACAT |
| cpa2 | Forward: CATTGCGGACTTCATCAC Reverse: AGGGCTTTGTTTGCTTGTG |
| ctrb1 | Forward: CACTTCTGCGGTGGCTCT Reverse: CAGGAGGATGTCGTTGTTGA |
| cel | Forward: GGAAAGGTTATCGTGGTG Reverse: GGAAGTTTACGCTGGCTC |
| c1q14 | Forward: TGTGGTCCAGAAGGTAGA Reverse: CTCAGTAGCCCTCAGTCG |
| cryS | Forward: CGAAACCAGGACAGAAGA Reverse: TTGTAGCAGAACGGAAGC |
| cda | Forward: TACTGTCCCTACAGCAAAT Reverse: TGGAGAAATAAACCGATC |
| fga | Forward: CAACAGGGTAAGGGCGAGAT Reverse: TCTGCCTCCGAGCCAACT |
| fgg | Forward: CGTGCCTTATGTGCTGAG Reverse: GTGCGAACTTGTCATTGTCT |
| fgb | Forward: TACATCCTAGCAGTTGGTCGTT Reverse: CTCCAGGTGTCCATTTGTCATT |
| f2 | Forward: CCTTCACGACCTCCTACT Reverse: ACCCTCCATATCTCCATCT |
| g4 | Forward: CTGGAGAATCCCATAAAC Reverse: CAGATACACCTGCTTCCT |
| hsp70 | Forward: ATGTGGCTCCTCTGTCCC Reverse: CAGCAGGTTGTTGTCTTTAGTC |
| hsp30 | Forward: GGACGACCCTTTCTTTGC Reverse: GCCGCTTGTTCTCATCTGTG |
| itln1 | Forward: TAGTGAGCAGGGCAGCAA Reverse: CAGGATGGAGGCAATGGT |
| lgmn | Forward: ACCTACCTGGGAGACTGG Reverse: CTTTAGAGTTACCCTGGAAT |
| mmgmt | Forward: GCGTGATGTGACTCTTTG Reverse: AACTCAGGGCTCATTTCT |
| nmt3 | Forward: GGATAACCAGCAGTACACCC Reverse: TCCCACAGCCGACATCTA |
| npr3 | Forward: GTCTACGGGAGCGAAGTG Reverse: TTTGGTGGACCTAAGTAATG |
| prf1 | Forward: CTGCTGTTCGGGTGTTTG Reverse: CTCCAGCTTCAGGTTATCATTC |
| pck1 | Forward: GTGCGCTCATCCCAACTC Reverse: ACACTCCGTGCGACCAAT |
| rimp2 | Forward: GTCACCAAACCCTGAAAC Reverse: CTGCTGGAACAGACGAAG |
| sox21a | Forward: GAAGCCAAACGACTTAGAGC Reverse: CGAGTAGCAGCAGCAACAG |
| trpm5 | Forward: TGGATTGCGGAGTGTTTC Reverse: TTGGCTTCCTGTGGTTGT |
| ydaC | Forward: CATACAACCCAACAACACG Reverse: CCTTCATGCGCTCACTAG |
| GAPDH | Forward: TGCCGGCATCTCCCTCAA Reverse: TCAGCAACACGGTGGCTGTAG |
| Antibodies | Host | Source | Catalog No. | Dilution for WB |
|---|---|---|---|---|
| p38 | Rabbit | Abcam | ab254122 | 1: 1000 |
| PKCα | Rabbit | GeneTex | GTX130453 | 1: 1000 |
| NF-κB p65 | Rabbit | Affinity | AF5006 | 1: 1000 |
| Claudin-3 | Rabbit | GeneTex | GTX135322 | 1: 1000 |
| Occludin | Rabbit | Self-made | - | 1: 500 |
| ZO-1 | Mouse | Thermo | 33-9100 | 1: 500 |
| GAPDH | Rabbit | GeneTex | GTX82899 | 1: 3000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Wang, X.; Li, H.; Liu, Y.; Zheng, Y.; Li, H.; Zhang, M.; Cheng, H.; Xu, J.; Chen, X.; et al. Comparative Expression Profiling Reveals the Regulatory Effects of Dietary Mannan Oligosaccharides on the Intestinal Immune Response of Juvenile Megalobrama amblycephala against Aeromonas hydrophila Infection. Int. J. Mol. Sci. 2023, 24, 2207. https://doi.org/10.3390/ijms24032207
Zhao X, Wang X, Li H, Liu Y, Zheng Y, Li H, Zhang M, Cheng H, Xu J, Chen X, et al. Comparative Expression Profiling Reveals the Regulatory Effects of Dietary Mannan Oligosaccharides on the Intestinal Immune Response of Juvenile Megalobrama amblycephala against Aeromonas hydrophila Infection. International Journal of Molecular Sciences. 2023; 24(3):2207. https://doi.org/10.3390/ijms24032207
Chicago/Turabian StyleZhao, Xiaoheng, Xu Wang, Hong Li, Yunlong Liu, Yancui Zheng, Hongping Li, Minying Zhang, Hanliang Cheng, Jianhe Xu, Xiangning Chen, and et al. 2023. "Comparative Expression Profiling Reveals the Regulatory Effects of Dietary Mannan Oligosaccharides on the Intestinal Immune Response of Juvenile Megalobrama amblycephala against Aeromonas hydrophila Infection" International Journal of Molecular Sciences 24, no. 3: 2207. https://doi.org/10.3390/ijms24032207
APA StyleZhao, X., Wang, X., Li, H., Liu, Y., Zheng, Y., Li, H., Zhang, M., Cheng, H., Xu, J., Chen, X., & Ding, Z. (2023). Comparative Expression Profiling Reveals the Regulatory Effects of Dietary Mannan Oligosaccharides on the Intestinal Immune Response of Juvenile Megalobrama amblycephala against Aeromonas hydrophila Infection. International Journal of Molecular Sciences, 24(3), 2207. https://doi.org/10.3390/ijms24032207





