Molecular Characterization, Expression, and Regulatory Signal Pathway Analysis of Inflammasome Component Apoptosis-Associated Speck-like Protein Containing a CARD Domain (ASC) in Large Yellow Croaker (Larimichthys crocea)
Abstract
1. Introduction
2. Results
2.1. Sequence Analysis of LcASC Gene
2.2. Tissue Distribution of LcASC Gene in Healthy Large Yellow Croaker
2.3. LcASC Gene Expression in Response to Immune Stimulation
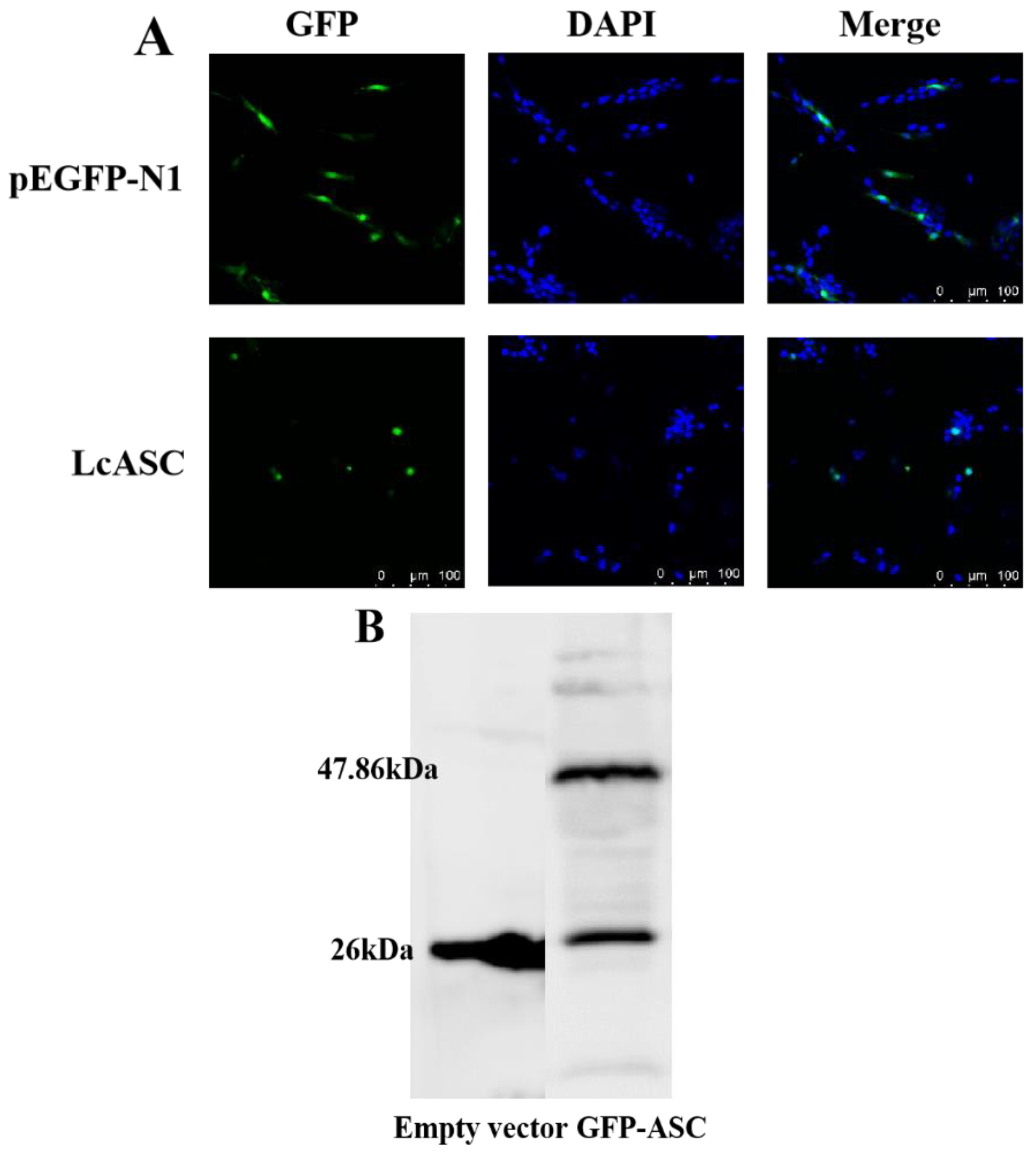
2.4. Subcellular Localization of LcASC Protein in Large Yellow Kidney Cells
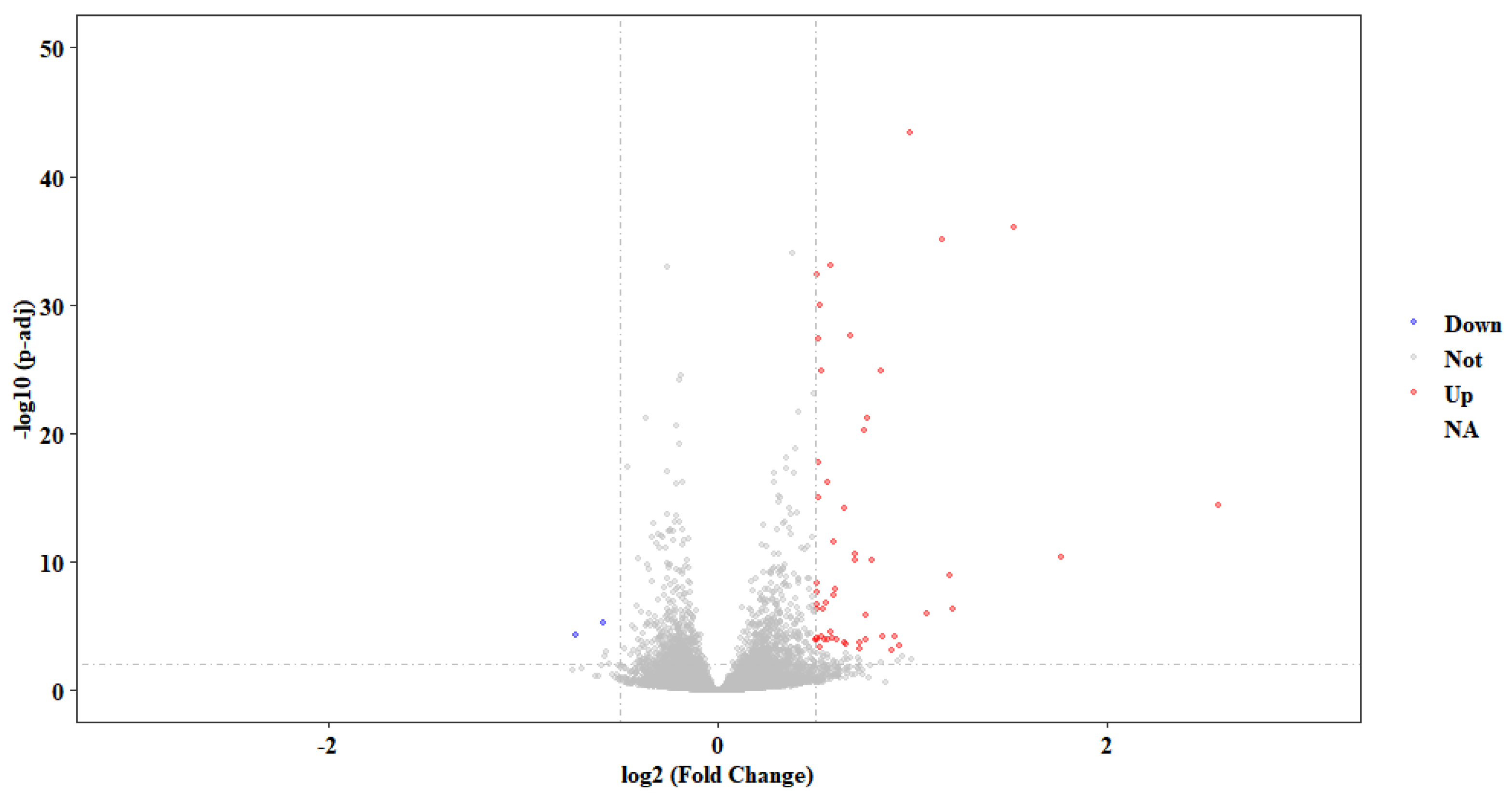
2.5. Transcriptome Analysis after LcASC Overexpression in HEK 293T Cells
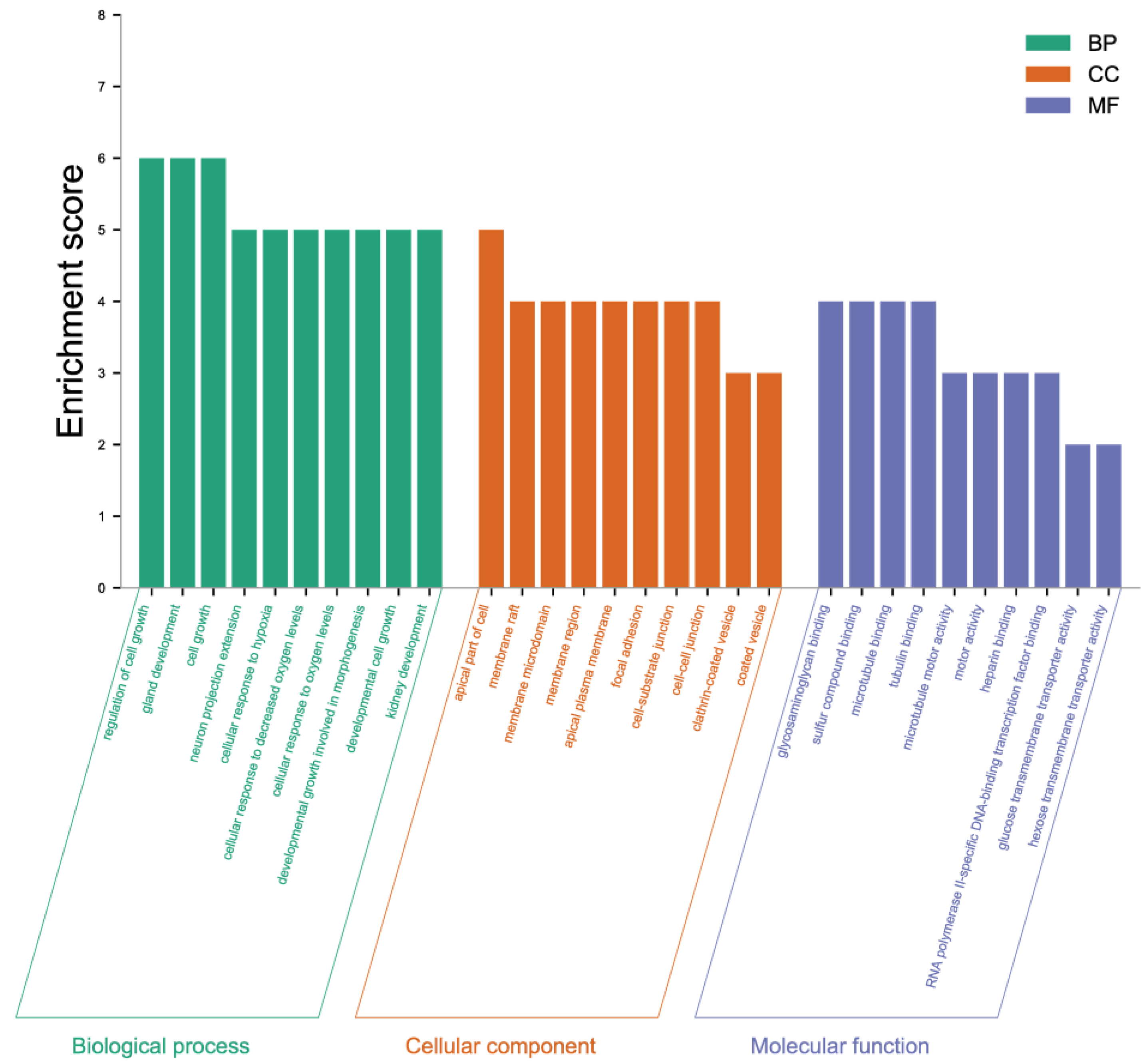
2.6. GO and KEGG Enrichment Analysis of DEGs
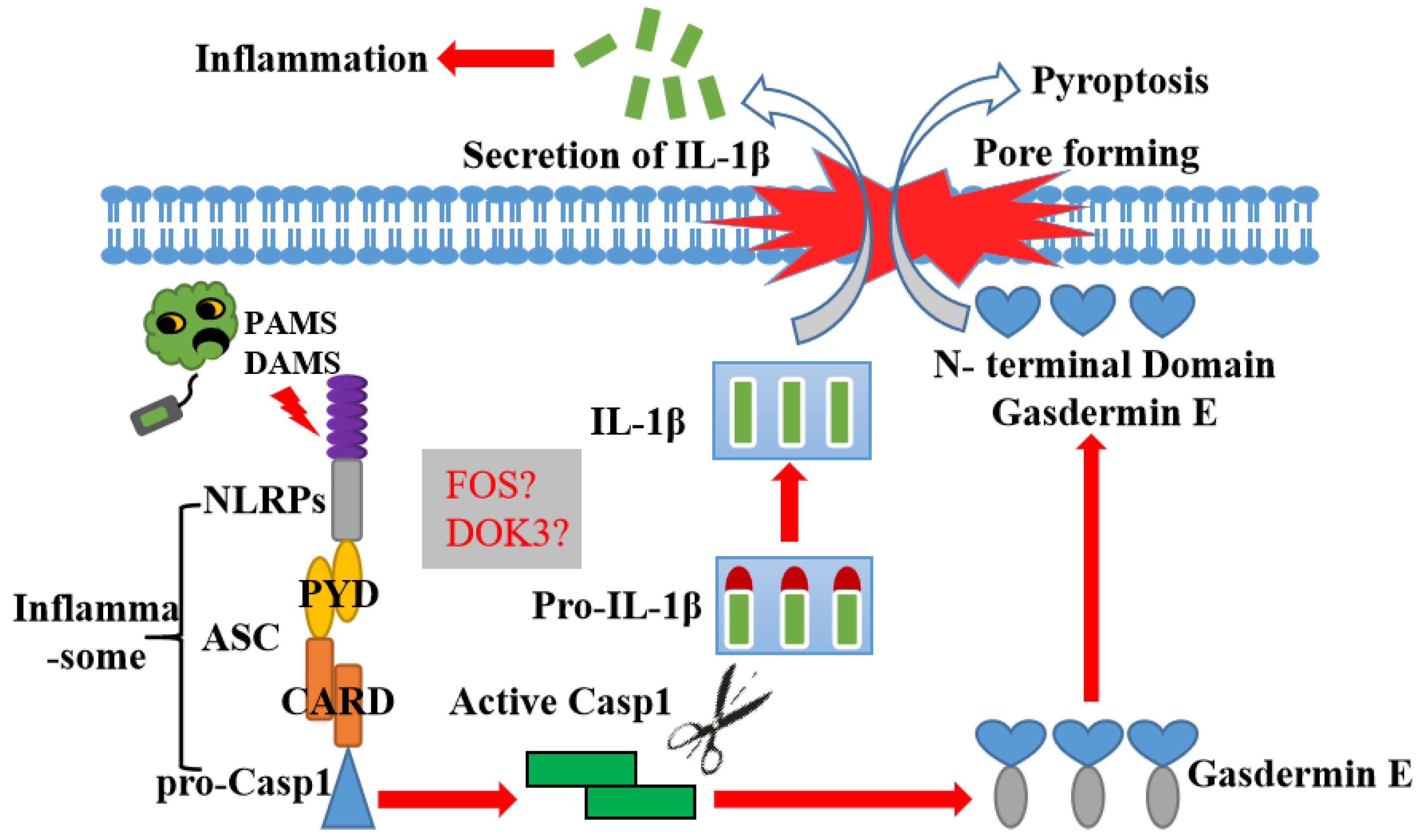
3. Discussion
4. Material and Methods
4.1. Ethical Statement
4.2. Fish and Sample
4.3. RNA Extraction and cDNA Synthesis
4.4. Identification of cDNA and Sequence Analysis
4.5. Tissue Distribution of LcASC and Expression Pattern Analysis
4.6. Subcellular Localization and Overexpression Analysis
4.7. Western Blot Analysis
4.8. Library Construction and Detection
4.9. Transcript Assembly and Differently Expressed Genes (DGEs) Analysis
4.10. Enrichment Analysis of DEGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mariathasan, S.; Monack, D.M. Inflammasome adaptors and sensors: Intracellular regulators of infection and inflammation. Nat. Rev. Immunol. 2007, 7, 31–40. [Google Scholar] [CrossRef]
- Li, J.-Y.; Gao, K.; Shao, T.; Fan, D.-D.; Hu, C.-B.; Sun, C.-C.; Dong, W.-R.; Lin, A.-F.; Xiang, L.-X.; Shao, J.-Z. Characterization of an NLRP1 Inflammasome from Zebrafish Reveals a Unique Sequential Activation Mechanism Underlying Inflammatory Caspases in Ancient Vertebrates. J. Immunol. 2018, 201, 1946–1966. [Google Scholar] [CrossRef]
- Mainz, R.E.; Albers, S.; Haque, M.; Sonntag, R.; Treichel, N.S.; Clavel, T.; Latz, E.; Schneider, K.M.; Trautwein, C.; Otto, T. NLRP6 Inflammasome Modulates Disease Progression in a Chronic-Plus-Binge Mouse Model of Alcoholic Liver Disease. Cells 2022, 11, 182. [Google Scholar] [CrossRef]
- Tuladhar, S.; Kanneganti, T.-D. NLRP12 in innate immunity and inflammation. Mol. Asp. Med. 2020, 76, 100887. [Google Scholar] [CrossRef]
- Davis, B.K.; Wen, H.; Ting, J.P.-Y. The Inflammasome NLRs in Immunity, Inflammation, and Associated Diseases. Annu. Rev. Immunol. 2011, 29, 707–735. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Sarkar, A.; Walle, L.V.; Vitari, A.C.; Amer, A.O.; Wewers, M.D.; Tracey, K.J.; Kanneganti, T.-D.; Dixit, V.M. Inflammasome-Dependent Release of the Alarmin HMGB1 in Endotoxemia. J. Immunol. 2010, 185, 4385–4392. [Google Scholar] [CrossRef]
- Franchi, L.; Eigenbrod, T.; Núñez, G. Cutting Edge: TNF-α Mediates Sensitization to ATP and Silica via the NLRP3 Inflammasome in the Absence of Microbial Stimulation. J. Immunol. 2009, 183, 792–796. [Google Scholar] [CrossRef]
- Qiu, Y.; Huang, Y.; Chen, M.; Yang, Y.; Li, X.; Zhang, W. Mitochondrial DNA in NLRP3 inflammasome activation. Int. Immunopharmacol. 2022, 108, 108719. [Google Scholar] [CrossRef]
- Masumoto, J.; Taniguchi, S.; Ayukawa, K.; Sarvotham, H.; Kishino, T.; Niikawa, N.; Hidaka, E.; Katsuyama, T.; Higuchi, T.; Sagara, J. ASC, a Novel 22-kDa Protein, Aggregates during Apoptosis of Human Promyelocytic Leukemia HL-60 Cells. J. Biol. Chem. 1999, 274, 33835–33838. [Google Scholar] [CrossRef]
- Kumar, M.; Roe, K.; Orillo, B.; Muruve, D.A.; Nerurkar, V.R.; Gale, M., Jr.; Verma, S. Correction for Kumar et al., “Inflammasome Adaptor Protein Apoptosis-Associated Speck-Like Protein Containing CARD (ASC) Is Critical for the Immune Response and Survival in West Nile Virus Encephalitis”. J. Virol. 2018, 92, e02176-17. [Google Scholar] [CrossRef]
- Pedra, J.H.; Sutterwala, F.S.; Sukumaran, B.; Ogura, Y.; Qian, F.; Montgomery, R.R.; Flavell, R.A.; Fikrig, E. ASC/PYCARD and caspase-1 regulate the IL-18/IFN-gamma axis during Anaplasma phagocytophilum infection. J. Immunol. 2007, 179, 4783–4791. [Google Scholar] [CrossRef]
- TeKippe, E.M.; Allen, I.C.; Hulseberg, P.D.; Sullivan, J.T.; McCann, J.R.; Sandor, M.; Braunstein, M.; Ting, J.P.-Y. Granuloma Formation and Host Defense in Chronic Mycobacterium tuberculosis Infection Requires PYCARD/ASC but Not NLRP3 or Caspase-1. PLoS ONE 2010, 5, e12320. [Google Scholar] [CrossRef]
- Ellebedy, A.H.; Lupfer, C.; Ghoneim, H.E.; DeBeauchamp, J.; Kanneganti, T.-D.; Webby, R. Inflammasome-independent role of the apoptosis-associated speck-like protein containing CARD (ASC) in the adjuvant effect of MF59. Proc. Natl. Acad. Sci. USA 2011, 108, 2927–2932. [Google Scholar] [CrossRef]
- Benoit, B.N.; Kobayashi, M.; Kawakubo, M.; Takeoka, M.; Sano, K.; Zou, J.; Itano, N.; Tsutsui, H.; Noda, T.; Fukuda, M.; et al. Role of ASC in the Mouse Model of Helicobacter pylori Infection. J. Histochem. Cytochem. 2009, 57, 327–338. [Google Scholar] [CrossRef]
- Abdelaziz, D.H.A.; Gavrilin, M.A.; Akhter, A.; Caution, K.; Kotrange, S.; Abu Khweek, A.; Abdulrahman, B.A.; Hassan, Z.A.; El-Sharkawi, F.Z.; Bedi, S.S.; et al. Asc-Dependent and Independent Mechanisms Contribute to Restriction of Legionella Pneumophila Infection in Murine Macrophages. Front. Microbiol. 2011, 2, 18. [Google Scholar] [CrossRef]
- Robinson, K.M.; Ramanan, K.; Clay, M.; McHugh, K.J.; Pilewski, M.J.; Nickolich, K.L.; Corey, C.; Shiva, S.; Wang, J.; Alcorn, J.F. The inflammasome potentiates influenza/Staphylococcus aureus superinfection in mice. JCI Insight 2018, 3, e97470. [Google Scholar] [CrossRef]
- Tsuchiya, K.; Hara, H.; Fang, R.; Hernandez-Cuellar, E.; Sakai, S.; Daim, S.; Chen, X.; Dewamitta, S.R.; Qu, H.; Mitsuyama, M.; et al. The adaptor ASC exacerbates lethal Listeria monocytogenes infection by mediating IL-18 production in an inflammasome-dependent and -independent manner. Eur. J. Immunol. 2014, 44, 3696–3707. [Google Scholar] [CrossRef]
- Gross, O.; Poeck, H.; Bscheider, M.; Dostert, C.; Hannesschläger, N.; Endres, S.; Hartmann, G.; Tardivel, A.; Schweighoffer, E.; Tybulewicz, V.; et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature 2009, 459, 433–436. [Google Scholar] [CrossRef]
- Sun, Y.; Abbondante, S.; Karmakar, M.; de Jesus, C.S.; Che, C.; Hise, A.G.; Pearlman, E. Neutrophil Caspase-11 Is Required for Cleavage of Caspase-1 and Secretion of IL-1beta in Aspergillus fumigatus Infection. J. Immunol. 2018, 201, 2767–2775. [Google Scholar] [CrossRef]
- Stehlik, C.; Fiorentino, L.; Dorfleutner, A.; Bruey, J.M.; Ariza, E.M.; Sagara, J.; Reed, J.C. The PAAD/PYRIN-family protein ASC is a dual regulator of a conserved step in NF-κB activation pathways. J. Exp. Med. 2002, 196, 1605–1615. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, J.; Lao, H.; Yin, Z.; He, W.; Weng, S.; Yu, X.; Chan, S.; He, J. Molecular cloning and expression analysis of the ASC gene from mandarin fish and its regulation of NF-κB activation. Dev. Comp. Immunol. 2008, 32, 391–399. [Google Scholar] [CrossRef]
- Li, S.; Chen, X.; Peng, W.; Hao, G.; Geng, X.; Zhan, W.; Sun, J. Cloning and characterization of apoptosis-associated speck-like protein containing a CARD domain (ASC) gene from Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2016, 54, 294–301. [Google Scholar] [CrossRef]
- Xie, J.; Belosevic, M. Functional characterization of apoptosis-associated speck-like protein (ASC) of the goldfish (Carassius auratus L.). Dev. Comp. Immunol. 2016, 65, 201–210. [Google Scholar] [CrossRef]
- Li, Y.; Huang, Y.; Cao, X.; Yin, X.; Jin, X.; Liu, S.; Jiang, J.; Jiang, W.; Xiao, T.; Zhou, R.; et al. Functional and structural characterization of zebrafish ASC. FEBS J. 2018, 285, 2691–2707. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.; Li, C.; Zhang, Y.; Wang, L.; Wei, J.; Qin, Q. Characterization of orange-spotted grouper (Epinephelus coioides) ASC and caspase-1 involved in extracellular ATP-mediated immune signaling in fish. Fish Shellfish Immunol. 2019, 97, 58–71. [Google Scholar] [CrossRef]
- Wang, W.; Tan, J.; Wang, Z.; Zhang, Y.; Liu, Q.; Yang, D. Characterization of the inflammasome component SmASC in turbot (Scophthalmus maximus). Fish Shellfish Immunol. 2020, 100, 324–333. [Google Scholar] [CrossRef]
- Morimoto, N.; Okamura, Y.; Maekawa, S.; Wang, H.-C.; Aoki, T.; Kono, T.; Sakai, M.; Hikima, J.-I. ASC-deficiency impairs host defense against Aeromonas hydrophila infection in Japanese medaka, Oryzias latipes. Fish Shellfish Immunol. 2020, 105, 427–437. [Google Scholar] [CrossRef]
- Morimoto, N.; Okamura, Y.; Kono, T.; Sakai, M.; Hikima, J.-I. Characterization and expression analysis of tandemly-replicated asc genes in the Japanese medaka, Oryzias latipes. Dev. Comp. Immunol. 2021, 115, 103894. [Google Scholar] [CrossRef]
- Zhang, Z.X.; Wang, Z.Y.; Fang, M.; Ye, K.; Tang, X.; Zhang, D.L. Genome-wide association analysis on host resistance against the rotten body disease in a naturally infected population of large yellow croaker Larimichthys crocea. Aquaculture 2022, 548 Pt 1, 737615. [Google Scholar] [CrossRef]
- Tang, X.; Wang, Z.; Jiang, D.; Chen, M.; Zhang, D. Expression profile, subcellular localization of MARCH4 and transcriptome analysis of its potential regulatory signaling pathway in large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol. 2022, 130, 273–282. [Google Scholar] [CrossRef]
- Zhang, X.; Mu, Y.; Mu, P.; Ao, J.; Chen, X. Transcriptome Analysis Reveals Comprehensive Insights into the Early Immune Response of Large Yellow Croaker (Larimichthys crocea) Induced by Trivalent Bacterial Vaccine. PLoS ONE 2017, 12, e0170958. [Google Scholar] [CrossRef]
- Guo, H.; Callaway, J.B.; Ting, J.P.-Y. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 2015, 21, 677–687. [Google Scholar] [CrossRef]
- Chavarria-Smith, J.; Vance, R.E. The NLRP1 inflammasomes. Immunol. Rev. 2015, 265, 22–34. [Google Scholar] [CrossRef]
- Shiohara, M.; Taniguchi, S.; Masumoto, J.; Yasui, K.; Koike, K.; Komiyama, A.; Sagara, J. ASC, which is composed of a PYD and a CARD, is up-regulated by inflammation and apoptosis in human neutrophils. Biochem. Biophys. Res. Commun. 2002, 293, 1314–1318. [Google Scholar] [CrossRef]
- Liepinsh, E.; Barbals, R.; Dahl, E.; Sharipo, A.; Staub, E.; Otting, G. The Death-domain Fold of the ASC PYRIN Domain, Presenting a Basis for PYRIN/PYRIN Recognition. J. Mol. Biol. 2003, 332, 1155–1163. [Google Scholar] [CrossRef]
- Shaulian, E.; Karin, M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002, 4, E131–E136. [Google Scholar] [CrossRef]
- Hop, H.T.; Arayan, L.T.; Huynh, T.H.; Reyes, A.W.B.; Vu, S.H.; Min, W.; Lee, H.J.; Rhee, M.H.; Chang, H.H.; Kim, S. The Key Role of c-Fos for Immune Regulation and Bacterial Dissemination in Brucella Infected Macrophage. Front. Cell. Infect. Microbiol. 2018, 8, 287. [Google Scholar] [CrossRef]
- Robson, J.D.; Davidson, D.; Veillette, A. Inhibition of the Jun N-Terminal Protein Kinase Pathway by SHIP-1, a Lipid Phosphatase That Interacts with the Adaptor Molecule Dok-3. Mol. Cell. Biol. 2004, 24, 2332–2343. [Google Scholar] [CrossRef]
- Peng, Q.; O’Loughlin, J.L.; Humphrey, M.B. DOK3 Negatively Regulates LPS Responses and Endotoxin Tolerance. PLoS ONE 2012, 7, e39967. [Google Scholar] [CrossRef]
- Kim, S.S.; Lee, K.G.; Chin, C.S.; Ng, S.K.; Pereira, N.A.; Xu, S.; Lam, K.P. DOK3 is required for IFN-beta production by enabling TRAF3/TBK1 complex formation and IRF3 activation. J. Immunol. 2014, 193, 840–848. [Google Scholar] [CrossRef]
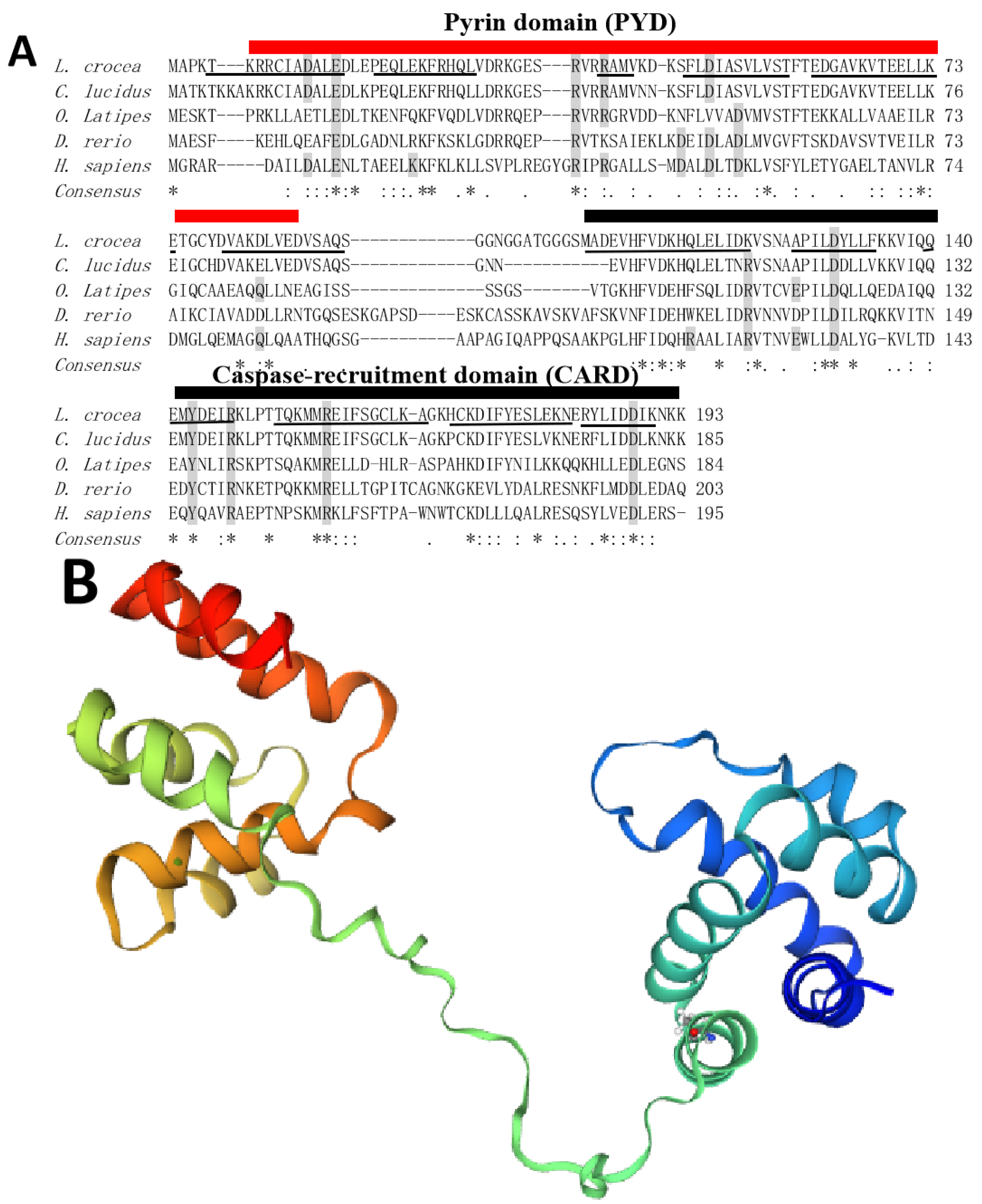
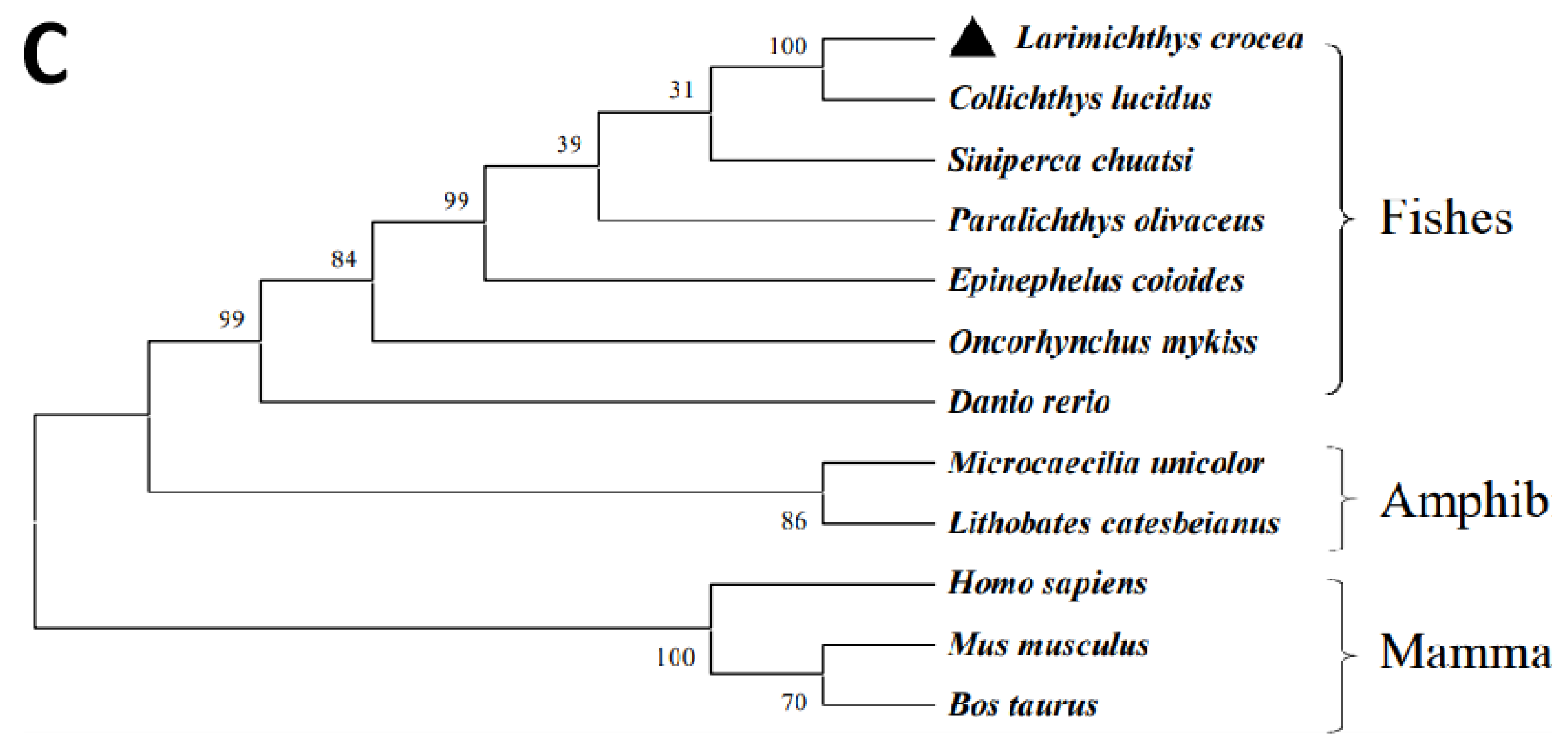
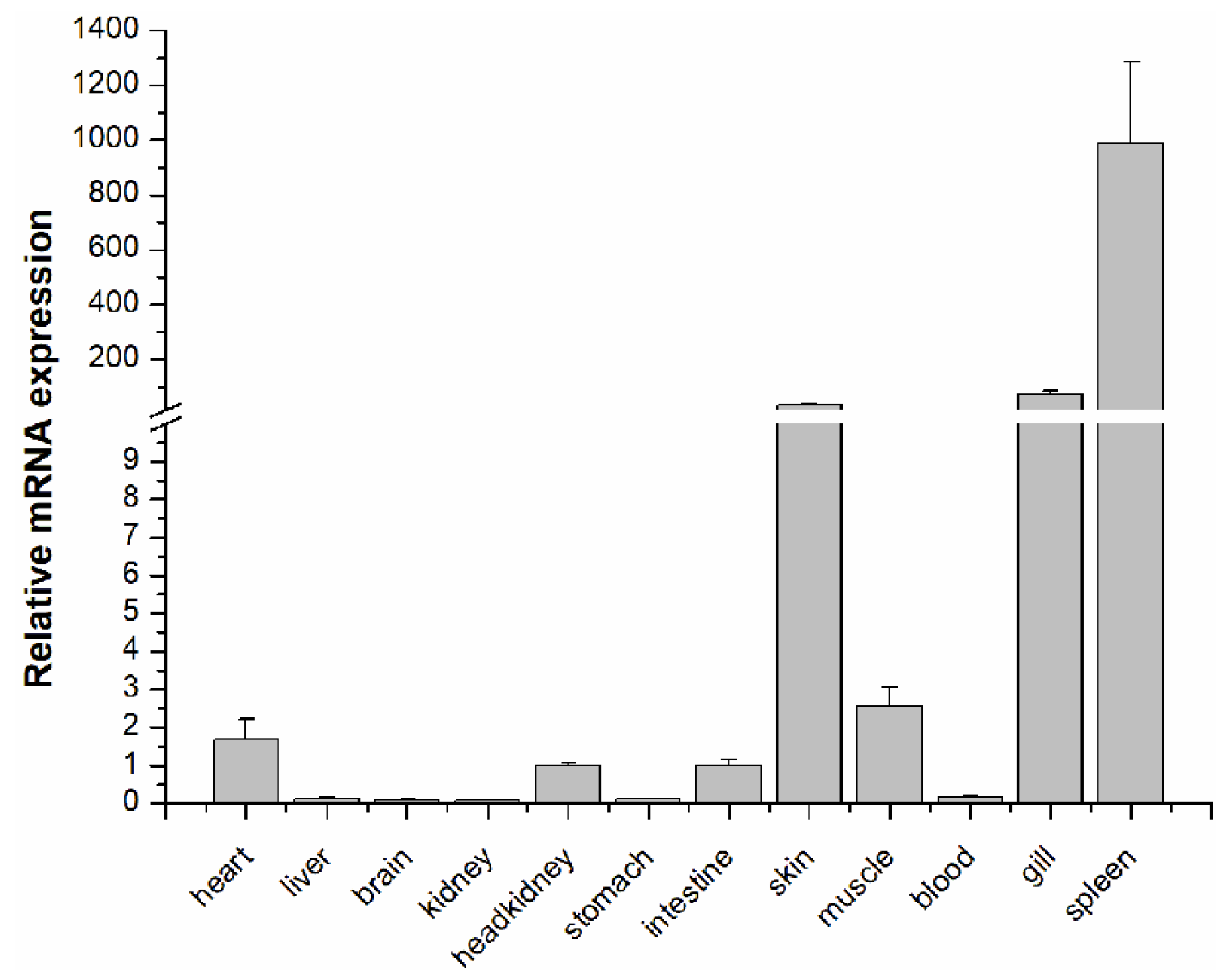
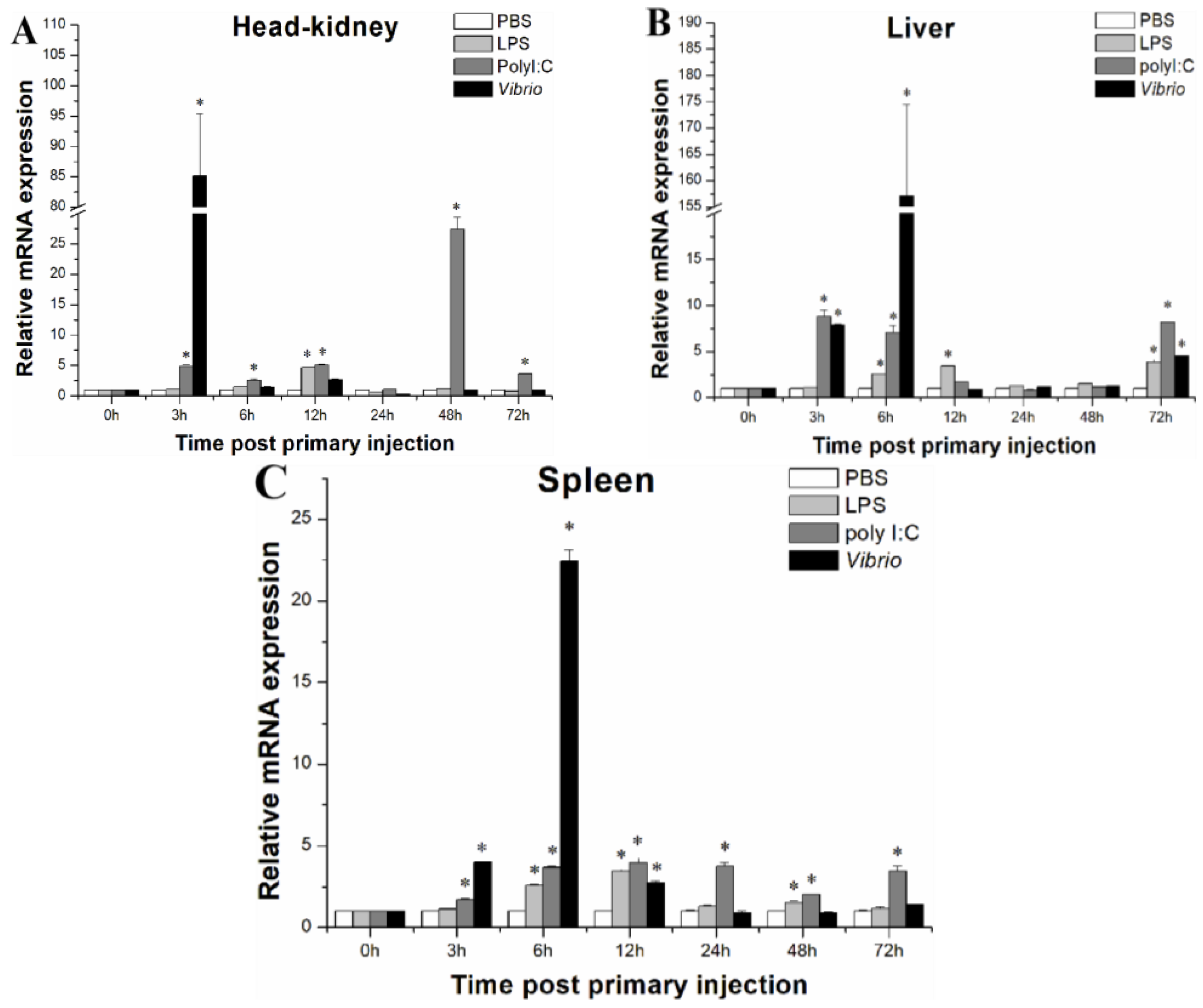

| GeneID | log2FoldChange | Function Annotation |
|---|---|---|
| KIAA0754 | 2.575797 | Related to autoimmune syndrome, Sjögren’s syndrome, metabolic syndrome, and inflammation |
| POMK | 1.761321 | Regulates dystroglycan function, as candidate gene for Walker-Warburg syndrome with meningoencephalocele and muscular dystrophy-dystroglycanopathy |
| NCR3LG1 | 1.522483 | Regulates tumor cell response |
| FOS | 1.21052 | Binds c-Jun protein to form transcription factor active protein1 |
| TSIX | 1.1931 | participates in the choice of the inactive X and in Xist regulation |
| SLC2A3 | 1.153975 | acts as a tumor promoter |
| LBH | 1.077569 | Related to cancer and Rheumatoid Arthritis |
| DOK3 | −0.58951 | Regulates B cell receptor, TLR3 and TLR4 signaling |
| RNA28SN5 | −0.72932 | uncharacterized |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, X.; Liu, X.; Wang, Z.; Chen, M.; Zhang, D. Molecular Characterization, Expression, and Regulatory Signal Pathway Analysis of Inflammasome Component Apoptosis-Associated Speck-like Protein Containing a CARD Domain (ASC) in Large Yellow Croaker (Larimichthys crocea). Int. J. Mol. Sci. 2023, 24, 2175. https://doi.org/10.3390/ijms24032175
Tang X, Liu X, Wang Z, Chen M, Zhang D. Molecular Characterization, Expression, and Regulatory Signal Pathway Analysis of Inflammasome Component Apoptosis-Associated Speck-like Protein Containing a CARD Domain (ASC) in Large Yellow Croaker (Larimichthys crocea). International Journal of Molecular Sciences. 2023; 24(3):2175. https://doi.org/10.3390/ijms24032175
Chicago/Turabian StyleTang, Xin, Xiande Liu, Zhiyong Wang, Meiling Chen, and Dongling Zhang. 2023. "Molecular Characterization, Expression, and Regulatory Signal Pathway Analysis of Inflammasome Component Apoptosis-Associated Speck-like Protein Containing a CARD Domain (ASC) in Large Yellow Croaker (Larimichthys crocea)" International Journal of Molecular Sciences 24, no. 3: 2175. https://doi.org/10.3390/ijms24032175
APA StyleTang, X., Liu, X., Wang, Z., Chen, M., & Zhang, D. (2023). Molecular Characterization, Expression, and Regulatory Signal Pathway Analysis of Inflammasome Component Apoptosis-Associated Speck-like Protein Containing a CARD Domain (ASC) in Large Yellow Croaker (Larimichthys crocea). International Journal of Molecular Sciences, 24(3), 2175. https://doi.org/10.3390/ijms24032175






