miR-29a Is Downregulated in Progenies Derived from Chronically Stressed Males
Abstract
:1. Introduction
2. Results
2.1. RNAseq Analysis
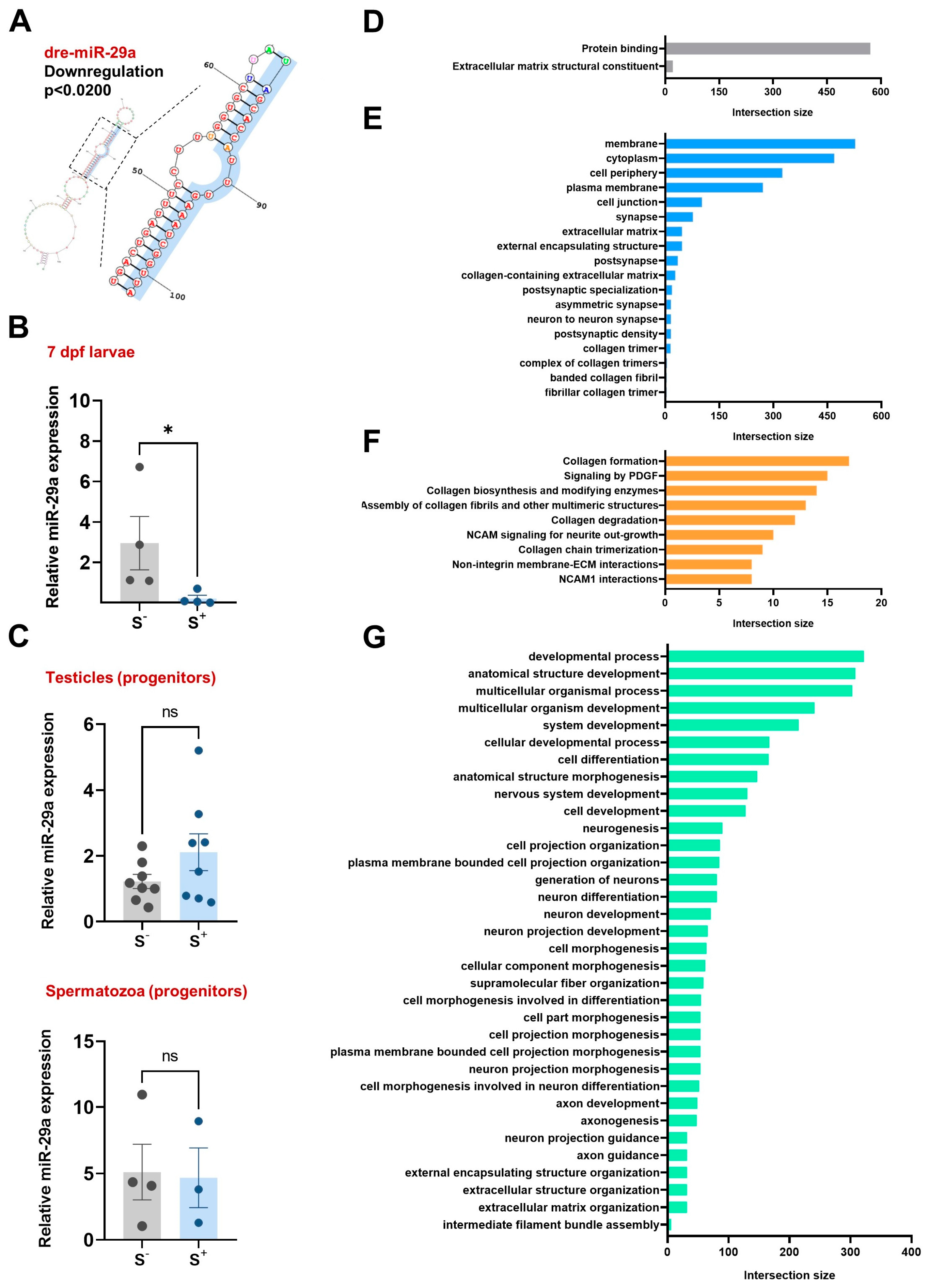
2.1.1. Differentially Expressed ncRNAs
2.1.2. RNAseq Validation
2.2. miR-29a Levels in Reproductive Cells and Tissues
2.3. Exploration of miR-29a Targets
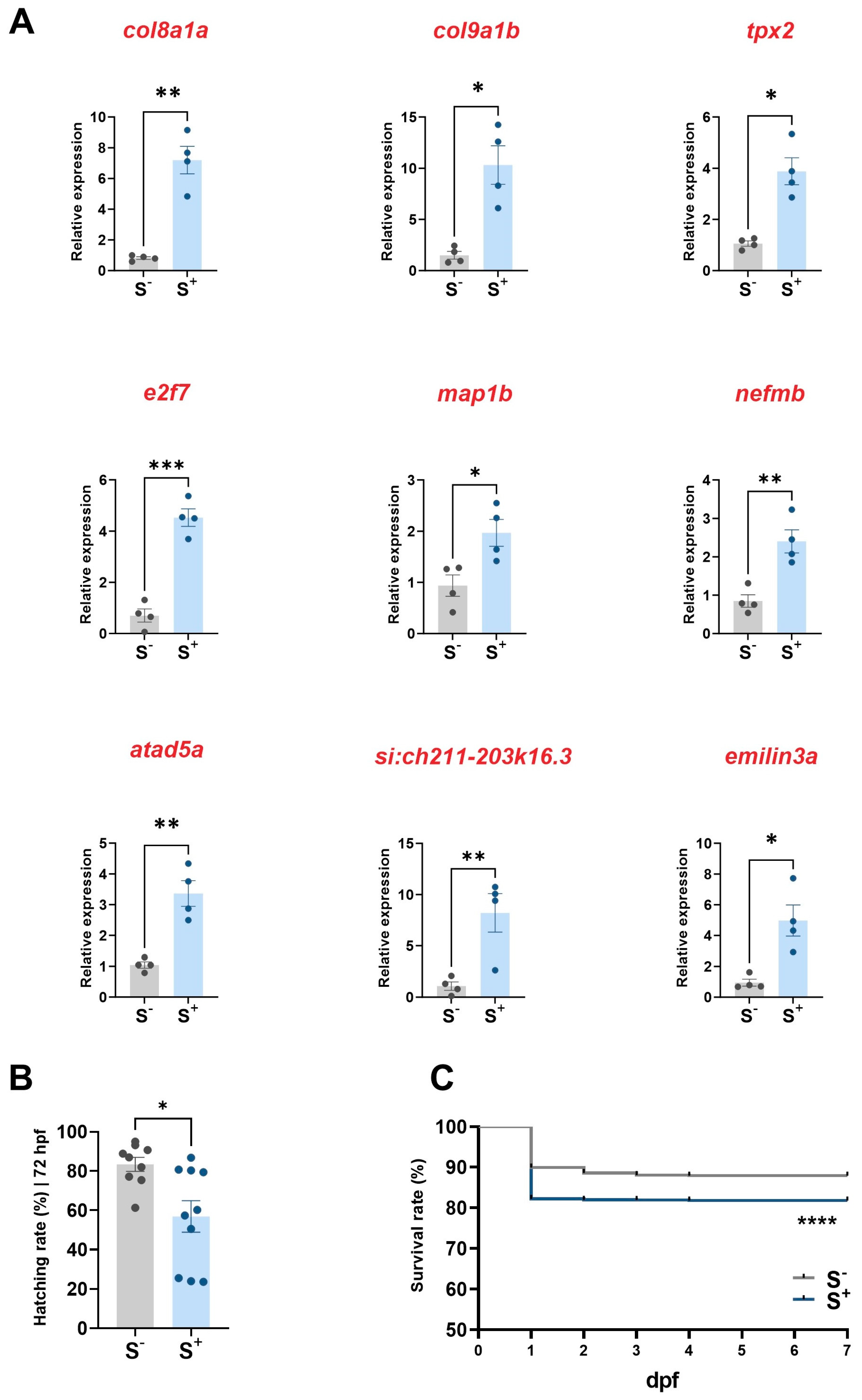
2.4. Expression of miR-29a Targets in Larvae
2.5. Progenies Development
2.5.1. Hatching Rate
2.5.2. Survival
2.6. Brain and Behavioral Analyses
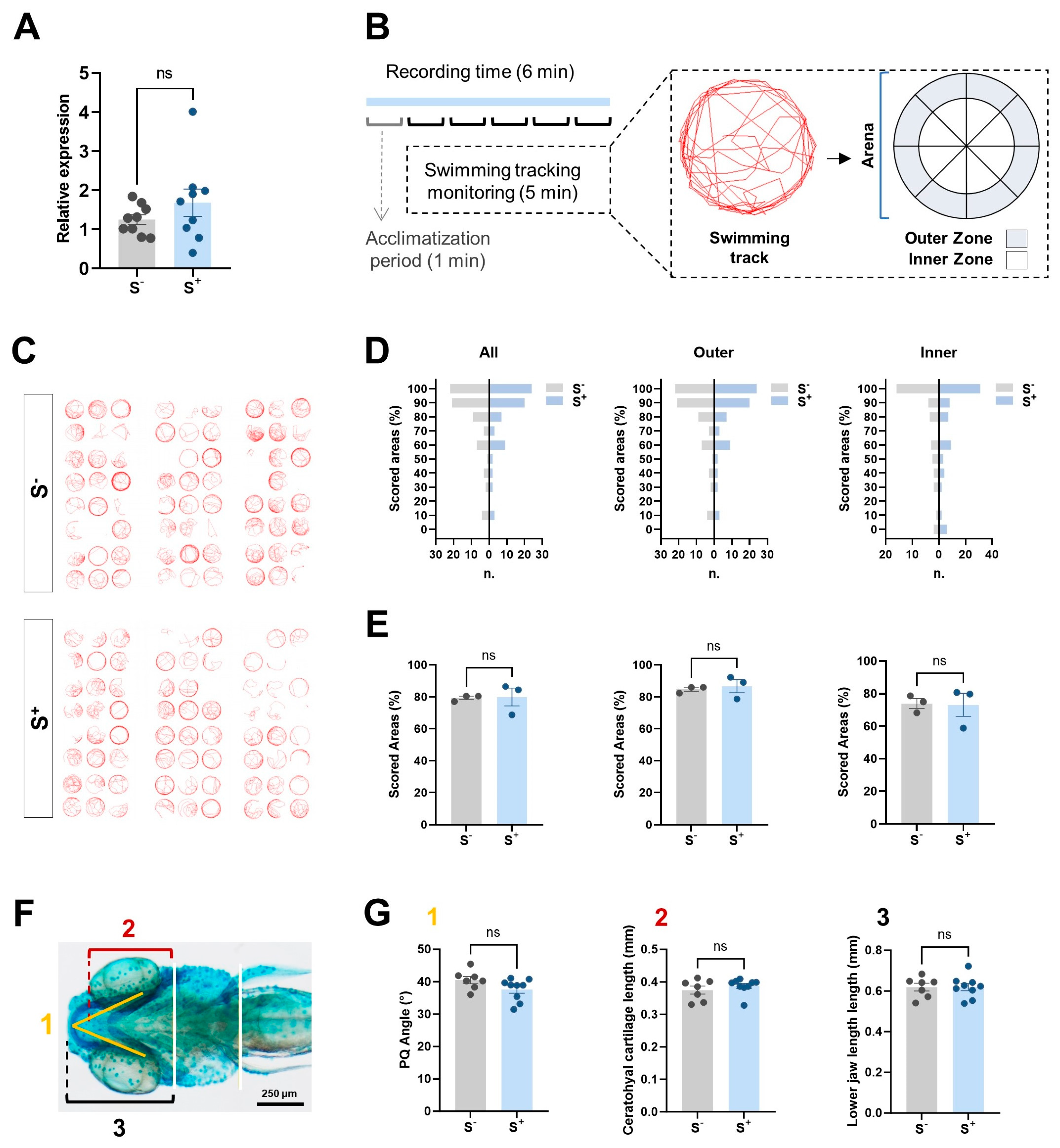
2.6.1. miR-29a Levels in Parental Brain Samples
2.6.2. Larvae and Adult Behavior Analysis
2.7. Cartilage Development
3. Discussion
4. Materials and Methods
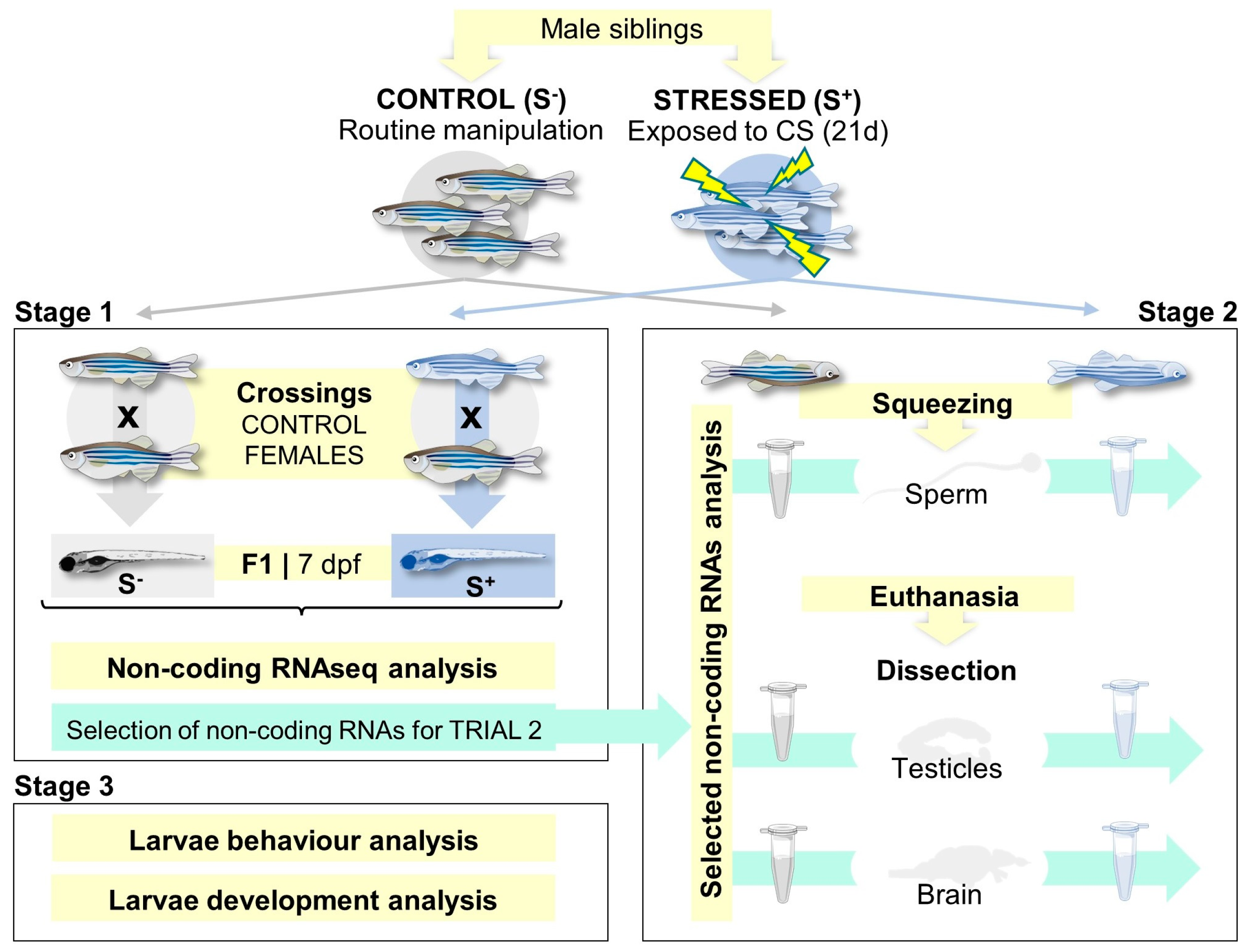
4.1. Animal Maintenance and Experimental Design
4.2. RNA Isolation
4.3. Small RNA-Seq Library Preparation and Sequencing
4.4. Small RNA-Seq Data Processing and Data Analysis
4.5. Retrotranscription
4.6. Target Prediction of miR-29a and Functional Enrichment Analysis
4.7. Primers Design
4.8. qPCR Analysis
4.8.1. qPCR Analysis for miRNAs
4.8.2. qPCR Analysis for mRNAs
4.9. Progeny Evaluation
4.10. Larvae and Adult Behavior Analyses
4.11. Alcian Blue Cartilage Staining
4.12. Statistics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Schreck, C.B.; Tort, L. The Concept of Stress in Fish. Fish Physiol. 2016, 35, 1–34. [Google Scholar] [CrossRef]
- Chrousos, G.P. Stress and Disorders of the Stress System. Nat. Rev. Endocrinol. 2009, 5, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Barton, B.A. Stress in Fishes: A Diversity of Responses with Particular Reference to Changes in Circulating Corticosteroids. Integr. Comp. Biol. 2002, 42, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Ilacqua, A.; Izzo, G.; Emerenziani, G.P.; Baldari, C.; Aversa, A. Lifestyle and Fertility: The Influence of Stress and Quality of Life on Male Fertility. Reprod. Biol. Endocrinol. 2018, 16, 115. [Google Scholar] [CrossRef]
- Rooney, K.L.; Domar, A.D. The Relationship between Stress and Infertility. Dialog. Clin. Neurosci. 2018, 20, 41–47. [Google Scholar] [CrossRef]
- Nargund, V.H. Effects of Psychological Stress on Male Fertility. Nat. Rev. Urol. 2015, 12, 373–382. [Google Scholar] [CrossRef]
- Bowers, M.E.; Yehuda, R. Intergenerational Transmission of Stress in Humans. Neuropsychopharmacology 2016, 41, 232–244. [Google Scholar] [CrossRef]
- Brunton, P.J. Effects of Maternal Exposure to Social Stress during Pregnancy: Consequences for Mother and Offspring. Reproduction 2013, 146, R175–R189. [Google Scholar] [CrossRef]
- Bagade, T.; Thapaliya, K.; Breuer, E.; Kamath, R.; Li, Z.; Sullivan, E.; Majeed, T. Investigating the Association between Infertility and Psychological Distress Using Australian Longitudinal Study on Women’s Health (ALSWH). Sci. Rep. 2022, 12, 10808. [Google Scholar] [CrossRef]
- Rodgers, A.B.; Morgan, C.P.; Bronson, S.L.; Revello, S.; Bale, T.L. Paternal Stress Exposure Alters Sperm MicroRNA Content and Reprograms Offspring HPA Stress Axis Regulation. J. Neurosci. 2013, 33, 9003–9012. [Google Scholar] [CrossRef]
- Zheng, X.; Li, Z.; Wang, G.; Wang, H.; Zhou, Y.; Zhao, X.; Cheng, C.Y.; Qiao, Y.; Sun, F. Sperm Epigenetic Alterations Contribute to Inter- and Transgenerational Effects of Paternal Exposure to Long-Term Psychological Stress via Evading Offspring Embryonic Reprogramming. Cell Discov. 2021, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Gapp, K.; Soldado-Magraner, S.; Alvarez-Sánchez, M.; Bohacek, J.; Vernaz, G.; Shu, H.; Franklin, T.B.; Wolfer, D.; Mansuy, I.M. Early Life Stress in Fathers Improves Behavioural Flexibility in Their Offspring. Nat. Commun. 2014, 5, 5466. [Google Scholar] [CrossRef] [PubMed]
- Valcarce, D.G.; Riesco, M.F.; Cuesta-Martín, L.; Esteve-Codina, A.; Martínez-Vázquez, J.M.; Robles, V. Stress Decreases Spermatozoa Quality and Induces Molecular Alterations in Zebrafish Progeny. BMC Biol. 2023, 21, 70. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Agarwal, A.; Rohra, V.K.; Assidi, M.; Abu-Elmagd, M.; Turki, R.F. Effects of Increased Paternal Age on Sperm Quality, Reproductive Outcome and Associated Epigenetic Risks to Offspring. Reprod. Biol. Endocrinol. 2015, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U. Paternal Contributions to Offspring Health: Role of Sperm Small Rnas in Intergenerational Transmission of Epigenetic Information. Front. Cell Dev. Biol. 2019, 7, 215. [Google Scholar] [CrossRef]
- Perez, M.F.; Lehner, B. Intergenerational and Transgenerational Epigenetic Inheritance in Animals. Nat. Cell Biol. 2019, 21, 143–151. [Google Scholar] [CrossRef]
- Sarangdhar, M.A.; Chaubey, D.; Srikakulam, N.; Pillai, B. Parentally Inherited Long Non-Coding RNA Cyrano Is Involved in Zebrafish Neurodevelopment. Nucleic Acids Res. 2018, 46, 9726–9735. [Google Scholar] [CrossRef]
- Riesco, M.F.; Valcarce, D.G.; Martínez-Vázquez, J.M.; Robles, V. Effect of Low Sperm Quality on Progeny: A Study on Zebrafish as Model Species. Sci. Rep. 2019, 9, 11192. [Google Scholar] [CrossRef]
- Salas-Huetos, A.; James, E.R.; Aston, K.I.; Carrell, D.T.; Jenkins, T.G.; Yeste, M. The Role of MiRNAs in Male Human Reproduction: A Systematic Review. Andrology 2020, 8, 7–26. [Google Scholar] [CrossRef]
- Khawar, M.B.; Mehmood, R.; Roohi, N. MicroRNAs: Recent Insights towards Their Role in Male Infertility and Reproductive Cancers. Bosn. J. Basic Med Sci. 2019, 19, 31. [Google Scholar] [CrossRef]
- Chen, S.R.; Liu, Y.X. Regulation of Spermatogonial Stem Cell Self-Renewal and Spermatocyte Meiosis by Sertoli Cell Signaling. Reproduction 2015, 149, R159–R167. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Li, Z.; Lin, H.; Gu, L.; Lin, Q.; Lu, Z.; Tzeng, C.M. Integrated MiRNA and MRNA Expression Profiling to Identify MRNA Targets of Dysregulated MiRNAs in Non-Obstructive Azoospermia. Sci. Rep. 2015, 5, 7922. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, X.; Mata, A.; Bassas, L.; Larriba, S. Altered MiRNA Signature of Developing Germ-Cells in Infertile Patients Relates to the Severity of Spermatogenic Failure and Persists in Spermatozoa. Sci. Rep. 2015, 5, 17991. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shi, J.; Hernandez, R.; Li, X.; Konchadi, P.; Miyake, Y.; Chen, Q.; Zhou, T.; Zhou, C. Paternal Phthalate Exposure-Elicited Offspring Metabolic Disorders Are Associated with Altered Sperm Small RNAs in Mice. Environ. Int. 2023, 172, 107769. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the Sequence and Temporal Expression of Let-7Heterochronic Regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Altuvia, Y.; Landgraf, P.; Lithwick, G.; Elefant, N.; Pfeffer, S.; Aravin, A.; Brownstein, M.J.; Tuschl, T.; Margalit, H. Clustering and Conservation Patterns of Human MicroRNAs. Nucleic Acids Res. 2005, 33, 2697–2706. [Google Scholar] [CrossRef]
- Piferrer, F.; Ribas, L. The Use of the Zebrafish as a Model in Fish Aquaculture Research. Fish Physiol. 2020, 38, 273–313. [Google Scholar] [CrossRef]
- Laan, M.; Richmond, H.; He, C.; Campbell, R.K. Zebrafish as a Model for Vertebrate Reproduction: Characterization of the First Functional Zebrafish (Danio rerio) Gonadotropin Receptor. Gen. Comp. Endocrinol. 2002, 125, 349–364. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The Zebrafish Reference Genome Sequence and Its Relationship to the Human Genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Jørgensen, L. von G. Zebrafish as a Model for Fish Diseases in Aquaculture. Pathogens 2020, 9, 609. [Google Scholar] [CrossRef]
- Hoo, J.Y.; Kumari, Y.; Shaikh, M.F.; Hue, S.M.; Goh, B.H. Zebrafish: A Versatile Animal Model for Fertility Research. BioMed Res. Int. 2016, 2016, 9732780. [Google Scholar] [CrossRef] [PubMed]
- Piato, A.L.; Capiotti, K.M.; Tamborski, A.R.; Oses, J.P.; Barcellos, L.J.G.; Bogo, M.R.; Lara, D.R.; Vianna, M.R.; Bonan, C.D. Unpredictable Chronic Stress Model in Zebrafish (Danio Rerio): Behavioral and Physiological Responses. Prog. Neuropsychopharmacol. Biol. Psychiatry 2011, 35, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Russell, G.; Lightman, S. The Human Stress Response. Nat. Rev. Endocrinol. 2019, 15, 525–534. [Google Scholar] [CrossRef]
- de Abreu, M.S.; Demin, K.A.; Giacomini, A.C.V.V.; Amstislavskaya, T.G.; Strekalova, T.; Maslov, G.O.; Kositsin, Y.; Petersen, E.V.; Kalueff, A.V. Understanding How Stress Responses and Stress-Related Behaviors Have Evolved in Zebrafish and Mammals. Neurobiol. Stress 2021, 15, 100405. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Novo, A.; Pérez-Garnelo, S.S.; Villagrá, A.; Pérez-Villalobos, N.; Astiz, S. The Effect of Stress on Reproduction and Reproductive Technologies in Beef Cattle—A Review. Animals 2020, 10, 2096. [Google Scholar] [CrossRef]
- Pourhosein Sarameh, S.; Falahatkar, B.; Takami, G.A.; Efatpanah, I. Effects of Different Photoperiods and Handling Stress on Spawning and Reproductive Performance of Pikeperch Sander Lucioperca. Anim. Reprod. Sci. 2012, 132, 213–222. [Google Scholar] [CrossRef]
- Ibarra-Zatarain, Z.; Martín, I.; Rasines, I.; Fatsini, E.; Rey, S.; Chereguini, O.; Duncan, N. Exploring the Relationship between Stress Coping Styles and Sex, Origin and Reproductive Success, in Senegalese Sole (Solea Senegalensis) Breeders in Captivity. Physiol. Behav. 2020, 220, 112868. [Google Scholar] [CrossRef]
- Morandini, L.; Ramallo, M.R.; Moreira, R.G.; Höcht, C.; Somoza, G.M.; Silva, A.; Pandolfi, M. Serotonergic Outcome, Stress and Sexual Steroid Hormones, and Growth in a South American Cichlid Fish Fed with an L-Tryptophan Enriched Diet. Gen. Comp. Endocrinol. 2015, 223, 27–37. [Google Scholar] [CrossRef]
- Lyu, G.; Guan, Y.; Zhang, C.; Zong, L.; Sun, L.; Huang, X.; Huang, L.; Zhang, L.; Tian, X.L.; Zhou, Z.; et al. TGF-β Signaling Alters H4K20me3 Status via MiR-29 and Contributes to Cellular Senescence and Cardiac Aging. Nat. Commun. 2018, 9, 2560. [Google Scholar] [CrossRef]
- Lin, H.Y.; Yang, Y.L.; Wang, P.W.; Wang, F.S.; Huang, Y.H. The Emerging Role of MicroRNAs in NAFLD: Highlight of MicroRNA-29a in Modulating Oxidative Stress, Inflammation, and Beyond. Cells 2020, 9, 1041. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, Y.; Shi, J.; Zhuang, H.; Ci, L.; Huang, Q.; Wan, Z.; Yang, H.; Zhang, M.; Tan, Y.; et al. MiR-29a/B1 Regulates the Luteinizing Hormone Secretion and Affects Mouse Ovulation. Front. Endocrinol. 2021, 12, 636220. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Zhang, Q.; Wang, D.D.; Yan, W.; Sha, H.H.; Zhao, J.H.; Yang, S.J.; Zhang, H.D.; Hou, J.C.; Xu, H.Z.; et al. MiR-29a: A Potential Therapeutic Target and Promising Biomarker in Tumors. Biosci. Rep. 2018, 38, 20171265. [Google Scholar] [CrossRef] [PubMed]
- Horita, M.; Farquharson, C.; Stephen, L.A. The Role of MiR-29 Family in Disease. J. Cell. Biochem. 2021, 122, 696–715. [Google Scholar] [CrossRef] [PubMed]
- Mitxelena, J.; Apraiz, A.; Vallejo-Rodriguez, J.; Malumbres, M.; Zubiaga, A.M. E2F7 Regulates Transcription and Maturation of Multiple MicroRNAs to Restrain Cell Proliferation. Nucleic Acids Res. 2016, 44, 5557–5570. [Google Scholar] [CrossRef]
- Lee, K.Y.; Fu, H.; Aladjem, M.I.; Myung, K. ATAD5 Regulates the Lifespan of DNA Replication Factories by Modulating PCNA Level on the Chromatin. J. Cell Biol. 2013, 200, 31–44. [Google Scholar] [CrossRef]
- Koike, Y.; Yin, C.; Sato, Y.; Nagano, Y.; Yamamoto, A.; Kitajima, T.; Shimura, T.; Kawamura, M.; Matsushita, K.; Okugawa, Y.; et al. TPX2 Is a Prognostic Marker and Promotes Cell Proliferation in Neuroblastoma. Oncol. Lett. 2022, 23, 1–9. [Google Scholar] [CrossRef]
- Halpain, S.; Dehmelt, L. The MAP1 Family of Microtubule-Associated Proteins. Genome Biol. 2006, 7, 1–7. [Google Scholar] [CrossRef]
- Hopfer, U.; Fukai, N.; Hopfer, H.; Wolf, G.; Joyce, N.; Li, E.; Olsen, B.R. Targeted Disruption of Col8a1 and Col8a2 Genes in Mice Leads to Anterior Segment Abnormalities in the Eye. FASEB J. 2005, 19, 1232–1244. [Google Scholar] [CrossRef]
- Gray, R.S.; Wilm, T.P.; Smith, J.; Bagnat, M.; Dale, R.M.; Topczewski, J.; Johnson, S.L.; Solnica-Krezel, L. Loss of Col8a1a Function during Zebrafish Embryogenesis Results in Congenital Vertebral Malformations. Dev. Biol. 2014, 386, 72–85. [Google Scholar] [CrossRef]
- Zhang, L.; Kendrick, C.; Jülich, D.; Holley, S.A. Cell Cycle Progression Is Required for Zebrafish Somite Morphogenesis but Not Segmentation Clock Function. Development 2008, 135, 2065. [Google Scholar] [CrossRef]
- Dey, A.; Flajšhans, M.; Pšenička, M.; Gazo, I. DNA Repair Genes Play a Variety of Roles in the Development of Fish Embryos. Front. Cell Dev. Biol. 2023, 11, 1119229. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zou, X.; Fu, M.; Bai, X.; Zhao, Y.; Chen, X.; Wang, X.; Wang, P.; Huang, S. Mild Cold Stress Specifically Disturbs Clustering Movement of DFCs and Sequential Organ Left-Right Patterning in Zebrafish. Front. Cell Dev. Biol. 2022, 10, 952844. [Google Scholar] [CrossRef] [PubMed]
- Butti, Z.; Pan, Y.E.; Giacomotto, J.; Patten, S.A. Reduced C9orf72 Function Leads to Defective Synaptic Vesicle Release and Neuromuscular Dysfunction in Zebrafish. Commun. Biol. 2021, 4, 792. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lin, J.; Liu, X.; Li, W.; Ding, Y.; Zhang, Y.; Zhou, S.; Guo, N.; Li, Q. Characterization of the Locomotor Activities of Zebrafish Larvae under the Influence of Various Neuroactive Drugs. Ann. Transl. Med. 2018, 6, 173. [Google Scholar] [CrossRef]
- Oliveira, N.A.S.; Pinho, B.R.; Oliveira, J.M.A. Swimming against ALS: How to Model Disease in Zebrafish for Pathophysiological and Behavioral Studies. Neurosci. Biobehav. Rev. 2023, 148, 105138. [Google Scholar] [CrossRef]
- Shimomura, T.; Kawakami, M.; Tatsumi, K.; Tanaka, T.; Morita-Takemura, S.; Kirita, T.; Wanaka, A. The Role of the Wnt Signaling Pathway in Upper Jaw Development of Chick Embryo. Acta Histochem. Cytochem. 2019, 52, 19–26. [Google Scholar] [CrossRef]
- Reeck, J.C.; Oxford, J.T. The Shape of the Jaw-Zebrafish Col11a1a Regulates Meckel’s Cartilage Morphogenesis and Mineralization. J. Dev. Biol. 2022, 10, 40. [Google Scholar] [CrossRef]
- Sasano, Y.; Zhu, J.X.; Kamakura, S.; Kusunoki, S.; Mizoguchi, I.; Kagayama, M. Expression of Major Bone Extracellular Matrix Proteins during Embryonic Osteogenesis in Rat Mandibles. Anat. Embryol. 2000, 202, 31–37. [Google Scholar] [CrossRef]
- Raterman, S.T.; Metz, J.R.; Wagener, F.A.D.T.G.; Von den Hoff, J.W. Zebrafish Models of Craniofacial Malformations: Interactions of Environmental Factors. Front. Cell Dev. Biol. 2020, 8, 1346. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 3rd ed.; University of Oregon Press: Eugene, OR, USA, 1995. [Google Scholar]
- Leal, M.C.; Cardoso, E.R.; Nóbrega, R.H.; Batlouni, S.R.; Bogerd, J.; França, L.R.; Schulz, R.W. Histological and Stereological Evaluation of Zebrafish (Danio Rerio) Spermatogenesis with an Emphasis on Spermatogonial Generations. Biol. Reprod. 2009, 81, 177–187. [Google Scholar] [CrossRef]
- Marcon, M.; Mocelin, R.; Benvenutti, R.; Costa, T.; Herrmann, A.P.; de Oliveira, D.L.; Koakoski, G.; Barcellos, L.J.G.; Piato, A. Environmental Enrichment Modulates the Response to Chronic Stress in Zebrafish. J. Exp. Biol. 2018, 221, jeb176735. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Valcarce, D.G.; Vuelta, E.; Robles, V.; Herráez, M.P. Paternal Exposure to Environmental 17-Alpha-Ethinylestradiol Concentrations Modifies Testicular Transcription, Affecting the Sperm Transcript Content and the Offspring Performance in Zebrafish. Aquat. Toxicol. 2017, 193, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
| Gene | Accession Number | Oligo | Sequence (5′ to 3′) | Amplicon (bp) |
|---|---|---|---|---|
| atad5 | XM_003198109.4 | F | GGTGTTTGGCGCCATTTG | 98 |
| R | TCTGAAGTGCGCTTGAACCT | |||
| col8a1a | NM_001142374.1 | F | AAGGTGTGTGTTTTCGGGGT | 185 |
| R | TTGCTGAGGATGCGACTTGT | |||
| si:ch211-203k16.3 | XM_021468697.1 | F | GCAGGCGTTTTGGAAGGTTT | 206 |
| R | ACCCCATCTTAAGAAACATGTGC | |||
| col9a1b | NM_213264.2 | F | GATCTGGGACTCCTGGTCCT | 165 |
| R | TGGGGCCTATGAGACCATCA | |||
| e2f7 | NM_001045147.2 | F | ACAGGGACTTTTACAGGCCG | 282 |
| R | TACAAACGCCGCACCTTAGT | |||
| emilin3a | XM_021474148.1 | F | ACGAGTGGAAAGTGCAGAGG | 132 |
| R | GCCCTCAATCTTTCAGCCCT | |||
| map1b | NM_001128201.2 | F | AACCACACCACCAAAGCTCA | 188 |
| R | CCATAGGCGTTGGGACACTT | |||
| nefmb | NM_001123280.1 | F | CGGTGCGCAATAACGAGAAG | 83 |
| R | CTCGAGGAAGCGAACCTTGT | |||
| tpx2 | NM_001327745.1 | F | AGATCAGTCATGCAGGTGCT | 72 |
| R | AGCCAACTCTCCTCTAGGCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riesco, M.F.; Valcarce, D.G.; Sellés-Egea, A.; Esteve-Codina, A.; Herráez, M.P.; Robles, V. miR-29a Is Downregulated in Progenies Derived from Chronically Stressed Males. Int. J. Mol. Sci. 2023, 24, 14107. https://doi.org/10.3390/ijms241814107
Riesco MF, Valcarce DG, Sellés-Egea A, Esteve-Codina A, Herráez MP, Robles V. miR-29a Is Downregulated in Progenies Derived from Chronically Stressed Males. International Journal of Molecular Sciences. 2023; 24(18):14107. https://doi.org/10.3390/ijms241814107
Chicago/Turabian StyleRiesco, Marta F., David G. Valcarce, Alba Sellés-Egea, Anna Esteve-Codina, María Paz Herráez, and Vanesa Robles. 2023. "miR-29a Is Downregulated in Progenies Derived from Chronically Stressed Males" International Journal of Molecular Sciences 24, no. 18: 14107. https://doi.org/10.3390/ijms241814107
APA StyleRiesco, M. F., Valcarce, D. G., Sellés-Egea, A., Esteve-Codina, A., Herráez, M. P., & Robles, V. (2023). miR-29a Is Downregulated in Progenies Derived from Chronically Stressed Males. International Journal of Molecular Sciences, 24(18), 14107. https://doi.org/10.3390/ijms241814107







