Comparative Transcriptomic Analyses for the Optimization of Thawing Regimes during Conventional Cryopreservation of Mature and Immature Human Testicular Tissue
Abstract
1. Introduction
2. Results
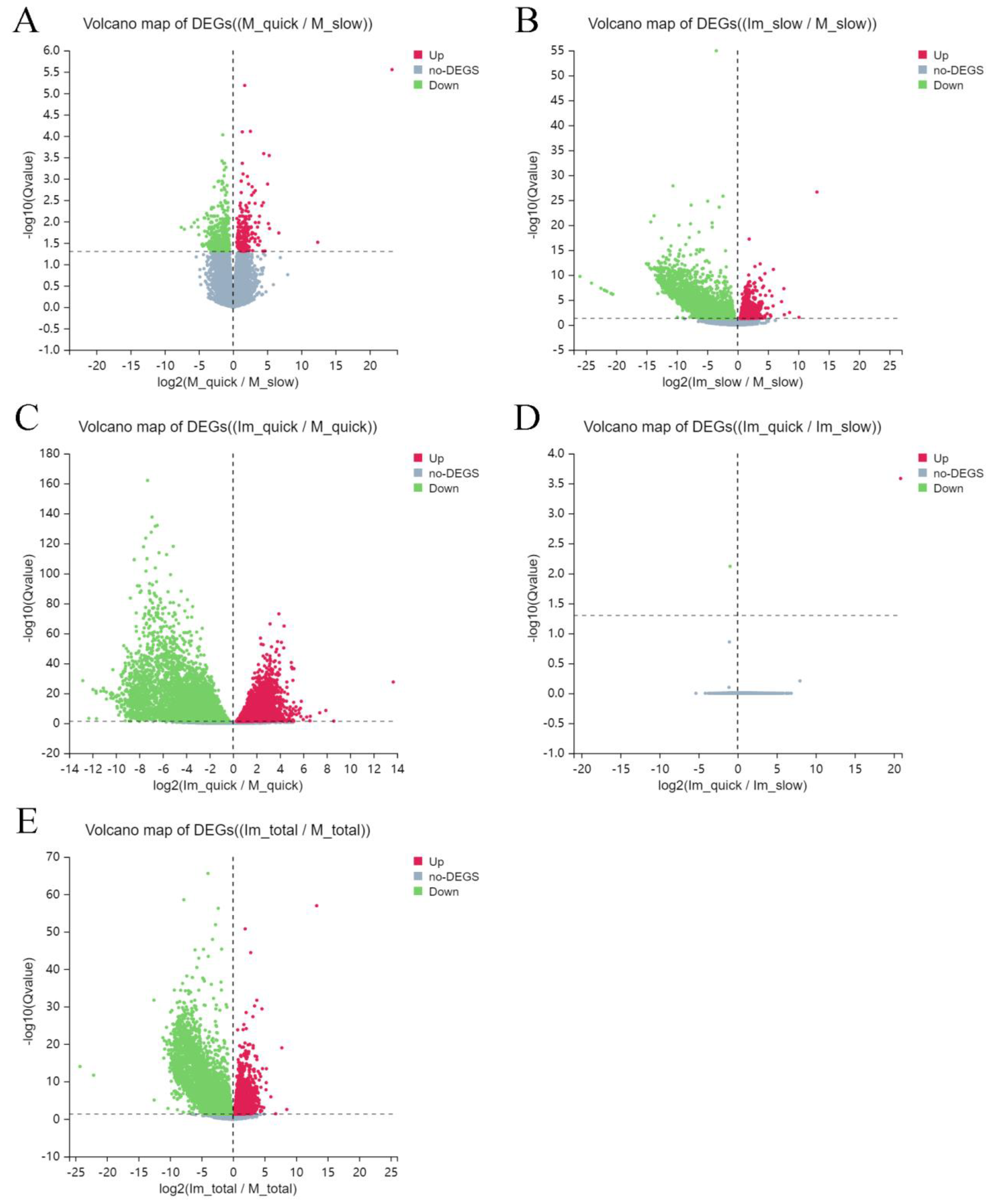
2.1. Differentially Expressed Genes (DEG)
2.2. Differentially Expressed Genes (DEG) through Enrichment Analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways
2.3. Differentially Expressed Genes (DEG) through Gene Ontology (GO) Enrichment Analysis
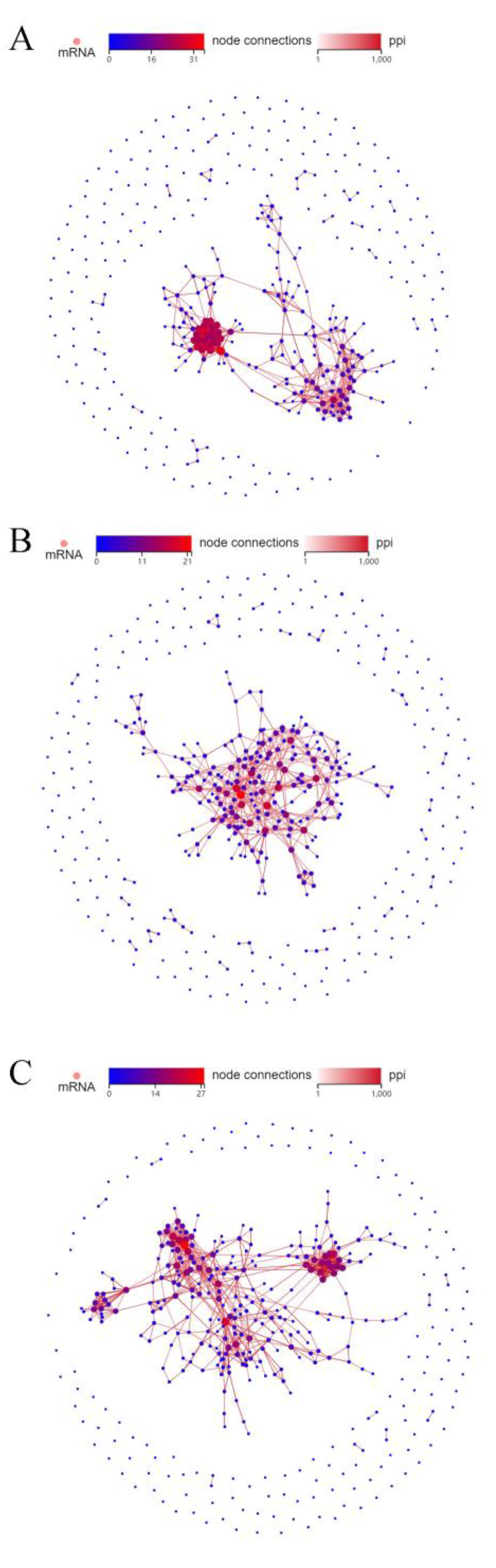
2.4. Protein–Protein Interactions (PPI) Network
3. Discussion
3.1. Differentially Expressed Genes (DEG)
3.2. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways
3.3. Gene Ontology (GO)
3.4. Protein–Protein Interactions (PPI)
3.5. Some Practical Aspects of Described Technology for Cryopreservation
4. Materials and Methods
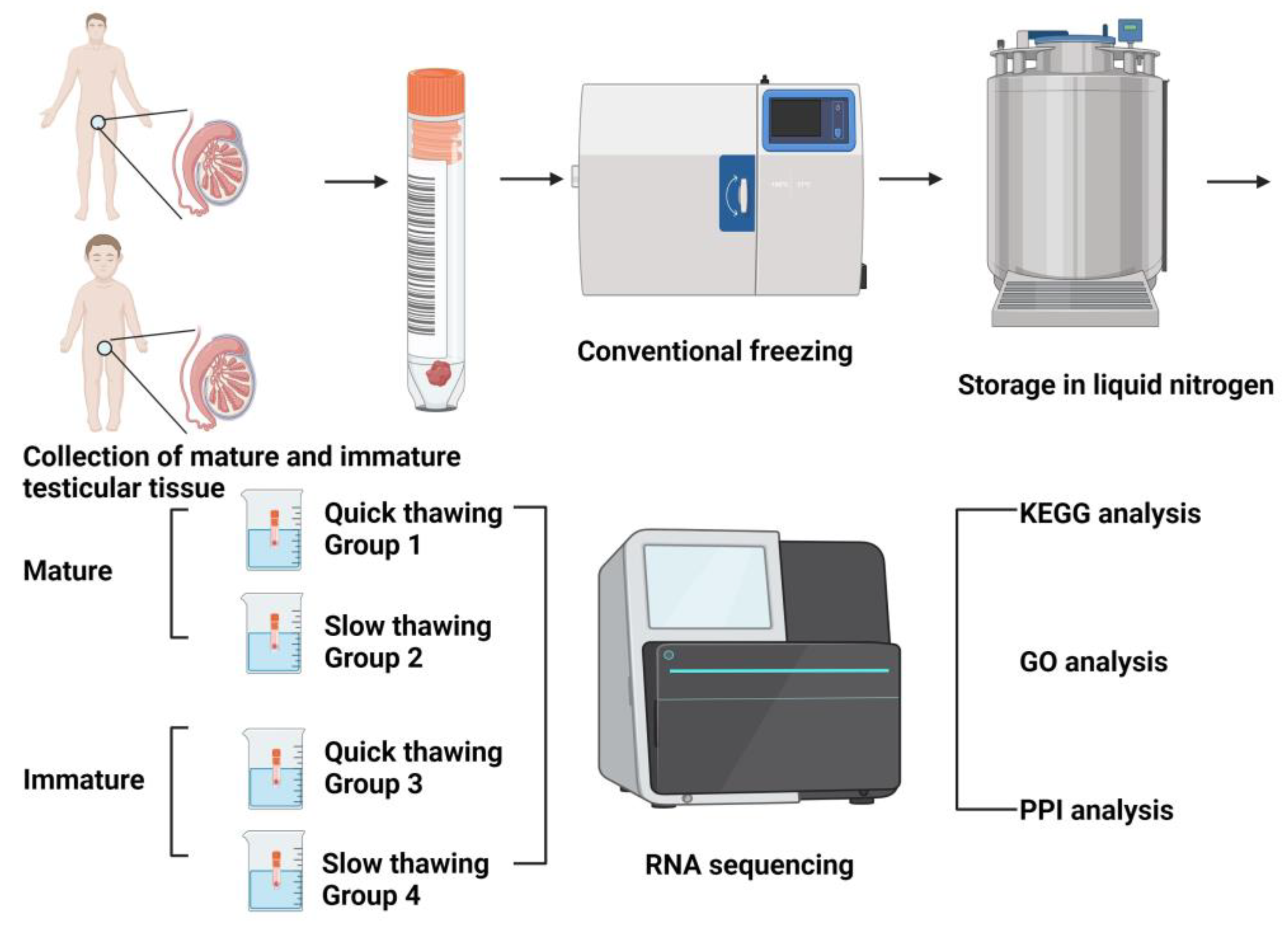
4.1. Design of Experiments
4.2. Extraction and Cryopreservation of Testicular Tissue (Equilibration with Cryoprotectants, Thawing and Removal of Cryoprotectants)
4.3. Sequencing and Data Extraction
4.4. Differentially Expressed Genes (DEG) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Onofre, J.; Baert, Y.; Faes, K.; Goossens, E. Cryopreservation of Testicular Tissue or Testicular Cell Suspensions: A Pivotal Step in Fertility Preservation. Hum. Reprod. Update 2016, 22, 744–761. [Google Scholar] [CrossRef] [PubMed]
- Youlden, D.R.; Baade, P.D.; Moore, A.S.; Pole, J.D.; Valery, P.C.; Aitken, J.F. Childhood Cancer Survival and Avoided Deaths in Australia, 1983–2016. Paediatr. Perinat. Epidemiol. 2023, 37, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Barton, S.E.; Najita, J.S.; Ginsburg, E.S.; Leisenring, W.M.; Stovall, M.; Weathers, R.E.; Sklar, C.A.; Robison, L.L.; Diller, L. Infertility, Infertility Treatment, and Achievement of Pregnancy in Female Survivors of Childhood Cancer: A Report from the Childhood Cancer Survivor Study Cohort. Lancet Oncol. 2013, 14, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Green, D.M.; Liu, W.; Kutteh, W.H.; Ke, R.W.; Shelton, K.C.; Sklar, C.A.; Chemaitilly, W.; Pui, C.H.; Klosky, J.L.; Spunt, S.L.; et al. Cumulative Alkylating Agent Exposure and Semen Parameters in Adult Survivors of Childhood Cancer: A Report from the St Jude Lifetime Cohort Study. Lancet Oncol. 2014, 15, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Patra, T.; Pathak, D.; Gupta, M.K. Strategies for Cryopreservation of Testicular Cells and Tissues in Cancer and Genetic Diseases. Cell Tissue Res. 2021, 385, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Wyns, C.; Curaba, M.; Petit, S.; Vanabelle, B.; Laurent, P.; Wese, J.F.; Donnez, J. Management of Fertility Preservation in Prepubertal Patients: 5 Years’ Experience at the Catholic University of Louvain. Hum. Reprod. 2011, 26, 737–747. [Google Scholar] [CrossRef]
- Lecluze, E.; Jégou, B.; Rolland, A.D.; Chalmel, F. New Transcriptomic Tools to Understand Testis Development and Functions. Mol. Cell Endocrinol. 2018, 468, 47–59. [Google Scholar] [CrossRef]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grützner, F.; Kaessmann, H. The Evolution of Lncrna Repertoires and Expression Patterns in Tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef]
- Dumont, L.; Maestre, H.L.; Chalmel, F.; Huber, L.; Rives-Feraille, A.; Moutard, L.; Bateux, F.; Rondanino, C.; Rives, N. Throughout in Vitro First Spermatogenic Wave: Next-Generation Sequencing Gene Expression Patterns of Fresh and Cryopreserved Prepubertal Mice Testicular Tissue Explants. Front. Endocrinol. 2023, 14, 1112834. [Google Scholar] [CrossRef]
- Amelkina, O.; Silva, A.M.D.; Silva, A.R.; Comizzoli, P. Transcriptome Dynamics in Developing Testes of Domestic Cats and Impact of Age on Tissue Resilience to Cryopreservation. BMC Genom. 2021, 22, 847. [Google Scholar] [CrossRef]
- Wang, M.; Todorov, P.; Wang, W.; Isachenko, E.; Rahimi, G.; Mallmann, P.; Isachenko, V. Cryoprotectants-Free Vitrification and Conventional Freezing of Human Spermatozoa: A Comparative Transcript Profiling. Int. J. Mol. Sci. 2022, 23, 3047. [Google Scholar] [CrossRef] [PubMed]
- Bonenfant, D.; Coulot, M.; Towbin, H.; Schindler, P.; van Oostrum, J. Characterization of Histone H2a and H2b Variants and Their Post-Translational Modifications by Mass Spectrometry. Mol. Cell Proteom. 2006, 5, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhai, Y.; Zhang, D.; Che, C.; Zhang, Y.; Li, Q.; Zhang, X.; Zhao, L. Rnaseq Analysis of the Drug Jian-Yan-Ling (Jyl) Using Both in Vivo and in Vitro Models. Heliyon 2023, 9, e16143. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wei, Y.; Li, H.; Li, W.; Gu, C.; Sun, J.; Xia, H.; Zhang, J.; Chen, F.; Liu, Q. The Ubiquitination and Acetylation of Histones Are Associated with Male Reproductive Disorders Induced by Chronic Exposure to Arsenite. Toxicol. Appl. Pharmacol. 2020, 408, 115253. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Liu, Z.; Xiong, K.; Duan, H.; Yang, J.; Liao, P. Gadd45b Predicts Lung Squamous Cell Carcinoma Survival and Impacts Immune Infiltration, and T Cell Exhaustion. Autoimmunity 2023, 56, 2209706. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, M.E. Gadd45b: At the Intersection of Transcriptional and Epigenetic Changes Associated with Cocaine-Related Behaviors. Neuropsychopharmacology 2021, 46, 685–686. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Cai, L.; Li, G.; Cai, J.; Yi, X. Gadd45b Facilitates Metastasis of Ovarian Cancer through Epithelial-Mesenchymal Transition. OncoTargets Ther. 2021, 14, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Yazdani, M.; Shahdadfar, A.; Reppe, S.; Sapkota, D.; Vallenari, E.M.; Lako, M.; Connon, C.J.; Figueiredo, F.C.; Utheim, T.P. Response of Human Oral Mucosal Epithelial Cells to Different Storage Temperatures: A Structural and Transcriptional Study. PLoS ONE 2020, 15, e0243914. [Google Scholar] [CrossRef]
- Hoesel, B.; Schmid, J.A. The Complexity of NF-κB Signaling in Inflammation and Cancer. Mol. Cancer 2013, 12, 86. [Google Scholar] [CrossRef]
- Pentikäinen, V.; Suomalainen, L.; Erkkilä, K.; Martelin, E.; Parvinen, M.; Pentikäinen, M.O.; Dunkel, L. Nuclear Factor-Kappa B Activation in Human Testicular Apoptosis. Am. J. Pathol. 2002, 160, 205–218. [Google Scholar] [CrossRef]
- Du, S.; Zheng, H. Role of Foxo Transcription Factors in Aging and Age-Related Metabolic and Neurodegenerative Diseases. Cell Biosci. 2021, 11, 188. [Google Scholar] [CrossRef] [PubMed]
- Farhan, M.; Wang, H.; Gaur, U.; Little, P.J.; Xu, J.; Zheng, W. Foxo Signaling Pathways as Therapeutic Targets in Cancer. Int. J. Biol. Sci. 2017, 13, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Zheng, Y.; Zeng, W.; Chen, L.; Yang, S.; Du, P.; Wang, Y.; Yu, X.; Zhang, X. Comparison of Proteomic Profiles from the Testicular Tissue of Males with Impaired and Normal Spermatogenesis. Syst. Biol. Reprod. Med. 2021, 67, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Huang, Y.; Wei, G.; Pang, Y.; Yuan, H.; Zou, X.; Xie, Y.; Chen, W. Testicular Toxicity in Rats Exposed to Alcl(3): A Proteomics Study. Biol. Trace Elem. Res. 2023. ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.L.; Zaher, H.S. Ribosome Quality Control Antagonizes the Activation of the Integrated Stress Response on Colliding Ribosomes. Mol. Cell 2021, 81, 614–628.e4. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.; Cluet, D.; Ricci, E.P. Ribosome Dynamics and Mrna Turnover, a Complex Relationship under Constant Cellular Scrutiny. Wiley Interdiscip. Rev. RNA 2021, 12, e1658. [Google Scholar] [CrossRef]
- Wu, C.C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 2020, 182, 404–416.e14. [Google Scholar] [CrossRef]
- Aksak, T.; Satar, D.A.; Bağci, R.; Gülteki, N.E.; Coşkun, A.; Rdelen, U.D. Investigation of the Effect of COVID-19 on Sperm Count, Motility, and Morphology. J. Med. Virol. 2022, 94, 5201–5205. [Google Scholar] [CrossRef]
- Nie, X.; Qian, L.; Sun, R.; Huang, B.; Dong, X.; Xiao, Q.; Zhang, Q.; Lu, T.; Yue, L.; Chen, S.; et al. Multi-Organ Proteomic Landscape of COVID-19 Autopsies. Cell 2021, 184, 775–791.e14. [Google Scholar] [CrossRef]
- Li, H.; Xiao, X.; Zhang, J.; Zafar, M.I.; Wu, C.; Long, Y.; Lu, W.; Pan, F.; Meng, T.; Zhao, K.; et al. Impaired Spermatogenesis in COVID-19 Patients. EClinicalMedicine 2020, 28, 100604. [Google Scholar] [CrossRef]
- Joazeiro, C.A.P. Mechanisms and Functions of Ribosome-Associated Protein Quality Control. Nat. Rev. Mol. Cell Biol. 2019, 20, 368–383. [Google Scholar] [CrossRef] [PubMed]
- Akopian, D.; Shen, K.; Zhang, X.; Shan, S.O. Signal Recognition Particle: An Essential Protein-Targeting Machine. Annu. Rev. Biochem. 2013, 82, 693–721. [Google Scholar] [CrossRef] [PubMed]
- Keenan, R.J.; Freymann, D.M.; Stroud, R.M.; Walter, P. The Signal Recognition Particle. Annu. Rev. Biochem. 2001, 70, 755–775. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.K.; Gupta, N.; Sankhwar, S.N.; Rajender, S. Differential Genes Expression between Fertile and Infertile Spermatozoa Revealed by Transcriptome Analysis. PLoS ONE 2015, 10, e0127007. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhao, H.; Chen, L.; Liu, M. Multifaceted Functions of Rps27a: An Unconventional Ribosomal Protein. J. Cell Physiol. 2023, 238, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, T.R.S.; Simões, R.; Mendes, C.M.; Goissis, M.D.; Nakajima, E.; Martins, E.A.L.; Visintin, J.A.; Assumpção, M. Detection of Protamine 2 in Bovine Spermatozoa and Testicles. Andrology 2019, 7, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Amjad, S.; Mushtaq, S.; Rehman, R.; Zahid, N.; Munir, A.; Siddiqui, P.Q.R. Spermatozoa Retrieval in Azoospermia and Expression Profile of Jmjd1a, Tnp2, and Prm2 in a Subset of the Karachi Population. Andrology 2021, 9, 1934–1942. [Google Scholar] [CrossRef] [PubMed]
- Sawaied, A.; Arazi, E.; AbuElhija, A.; Lunenfeld, E.; Huleihel, M. The Presence of Colony-Stimulating Factor-1 and Its Receptor in Different Cells of the Testis; It Involved in the Development of Spermatogenesis in Vitro. Int. J. Mol. Sci. 2021, 22, 2325. [Google Scholar] [CrossRef]
- Mazur, P. Principles of Cryobiology. In Life in the Frozen State; CRC Press: Boca Raton, FL, USA, 2004; pp. 29–92. [Google Scholar]
- Chaytor, J.L.; Tokarew, J.M.; Wu, L.K.; Leclère, M.; Tam, R.Y.; Capicciotti, C.J.; Guolla, L.; von Moos, E.; Findlay, C.S.; Allan, D.S.; et al. Inhibiting Ice Recrystallization and Optimization of Cell Viability after Cryopreservation. Glycobiology 2012, 22, 123–133. [Google Scholar] [CrossRef]
- Prickett, R.C.; Marquez-Curtis, L.A.; Elliott, J.A.; McGann, L.E. Effect of Supercooling and Cell Volume on Intracellular Ice Formation. Cryobiology 2015, 70, 156–163. [Google Scholar] [CrossRef]
- Donnez, J.; Kim, S.S. Principles and Practice of Fertility Preservation; Cambridge University Press: Cambridge, UK, 2011. [Google Scholar]
- Tournaye, H.; Camus, M.; Goossens, A.; Liu, J.; Nagy, P.; Silber, S.; Van Steirteghem, A.C.; Devroey, P. Recent Concepts in the Management of Infertility Because of Non-Obstructive Azoospermia. Hum. Reprod. 1995, 10 (Suppl. S1), 115–119. [Google Scholar] [CrossRef] [PubMed]
- Tournaye, H.; Liu, J.; Nagy, P.Z.; Camus, M.; Goossens, A.; Silber, S.; Van Steirteghem, A.C.; Devroey, P. Correlation between Testicular Histology and Outcome after Intracytoplasmic Sperm Injection Using Testicular Spermatozoa. Hum. Reprod. 1996, 11, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, P.N.; Su, L.M. Physiological Consequences of Testicular Sperm Extraction. Hum. Reprod. 1997, 12, 1688–1692. [Google Scholar] [CrossRef] [PubMed]
- Ezeh, U.I.; Moore, H.D.; Cooke, I.D. A Prospective Study of Multiple Needle Biopsies Versus a Single Open Biopsy for Testicular Sperm Extraction in Men with Non-Obstructive Azoospermia. Hum. Reprod. 1998, 13, 3075–3080. [Google Scholar] [CrossRef] [PubMed]
- Hauser, R.; Botchan, A.; Amit, A.; Yosef, D.B.; Gamzu, R.; Paz, G.; Lessing, J.B.; Yogev, L.; Yavetz, H. Multiple Testicular Sampling in Non-Obstructive Azoospermia—Is It Necessary? Hum. Reprod. 1998, 13, 3081–3085. [Google Scholar] [CrossRef] [PubMed]
- Ostad, M.; Liotta, D.; Ye, Z.; Schlegel, P.N. Testicular Sperm Extraction for Nonobstructive Azoospermia: Results of a Multibiopsy Approach with Optimized Tissue Dispersion. Urology 1998, 52, 692–696. [Google Scholar] [CrossRef]
- Ramasamy, R.; Yagan, N.; Schlegel, P.N. Structural and Functional Changes to the Testis after Conventional Versus Microdissection Testicular Sperm Extraction. Urology 2005, 65, 1190–1194. [Google Scholar] [CrossRef]
- Ishikawa, T. Surgical Recovery of Sperm in Non-Obstructive Azoospermia. Asian J. Androl. 2012, 14, 109–115. [Google Scholar] [CrossRef]
- Schiewe, M.C.; Rothman, C.; Spitz, A.; Werthman, P.E.; Zeitlin, S.I.; Anderson, R.E. Validation-Verification of a Highly Effective, Practical Human Testicular Tissue in Vitro Culture-Cryopreservation Procedure Aimed to Optimize Pre-Freeze and Post-Thaw Motility. J. Assist. Reprod. Genet. 2016, 33, 519–528. [Google Scholar] [CrossRef]
- Isachenko, V.; Morgenstern, B.; Todorov, P.; Isachenko, E.; Mallmann, P.; Hanstein, B.; Rahimi, G. Patient with Ovarian Insufficiency: Baby Born after Anticancer Therapy and Re-Transplantation of Cryopreserved Ovarian Tissue. J. Ovarian Res. 2020, 13, 118. [Google Scholar] [CrossRef]
- Isachenko, V.; Mallmann, P.; Petrunkina, A.M.; Rahimi, G.; Nawroth, F.; Hancke, K.; Felberbaum, R.; Genze, F.; Damjanoski, I.; Isachenko, E. Comparison of in Vitro- and Chorioallantoic Membrane (Cam)-Culture Systems for Cryopreserved Medulla-Contained Human Ovarian Tissue. PLoS ONE 2012, 7, e32549. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Isachenko, V.; Todorov, P.; Isachenko, E.; Rahimi, G.; Hanstein, B.; Salama, M.; Mallmann, P.; Tchorbanov, A.; Hardiman, P.; Getreu, N.; et al. Cryopreservation and Xenografting of Human Ovarian Fragments: Medulla Decreases the Phosphatidylserine Translocation Rate. Reprod. Biol. Endocrinol. 2016, 14, 79. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, W.; Todorov, P.; Pei, C.; Isachenko, E.; Rahimi, G.; Mallmann, P.; Nawroth, F.; Isachenko, V. RNA Transcripts in Human Ovarian Cells: Two-Time Cryopreservation Does Not Affect Developmental Potential. Int. J. Mol. Sci. 2023, 24, 6880. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for Rna-Seq Data with Deseq2. Genome Biol. 2014, 15, 550. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, C.; Todorov, P.; Cao, M.; Kong, Q.; Isachenko, E.; Rahimi, G.; Mallmann-Gottschalk, N.; Uribe, P.; Sanchez, R.; Isachenko, V. Comparative Transcriptomic Analyses for the Optimization of Thawing Regimes during Conventional Cryopreservation of Mature and Immature Human Testicular Tissue. Int. J. Mol. Sci. 2024, 25, 214. https://doi.org/10.3390/ijms25010214
Pei C, Todorov P, Cao M, Kong Q, Isachenko E, Rahimi G, Mallmann-Gottschalk N, Uribe P, Sanchez R, Isachenko V. Comparative Transcriptomic Analyses for the Optimization of Thawing Regimes during Conventional Cryopreservation of Mature and Immature Human Testicular Tissue. International Journal of Molecular Sciences. 2024; 25(1):214. https://doi.org/10.3390/ijms25010214
Chicago/Turabian StylePei, Cheng, Plamen Todorov, Mengyang Cao, Qingduo Kong, Evgenia Isachenko, Gohar Rahimi, Nina Mallmann-Gottschalk, Pamela Uribe, Raul Sanchez, and Volodimir Isachenko. 2024. "Comparative Transcriptomic Analyses for the Optimization of Thawing Regimes during Conventional Cryopreservation of Mature and Immature Human Testicular Tissue" International Journal of Molecular Sciences 25, no. 1: 214. https://doi.org/10.3390/ijms25010214
APA StylePei, C., Todorov, P., Cao, M., Kong, Q., Isachenko, E., Rahimi, G., Mallmann-Gottschalk, N., Uribe, P., Sanchez, R., & Isachenko, V. (2024). Comparative Transcriptomic Analyses for the Optimization of Thawing Regimes during Conventional Cryopreservation of Mature and Immature Human Testicular Tissue. International Journal of Molecular Sciences, 25(1), 214. https://doi.org/10.3390/ijms25010214