Proline Metabolism Process and Antioxidant Potential of Lycium ruthenicum Murr. in Response to NaCl Treatments
Abstract
:1. Introduction
2. Results
2.1. Growth and Structural Responses of Leaf Surface to Different NaCl Treatments
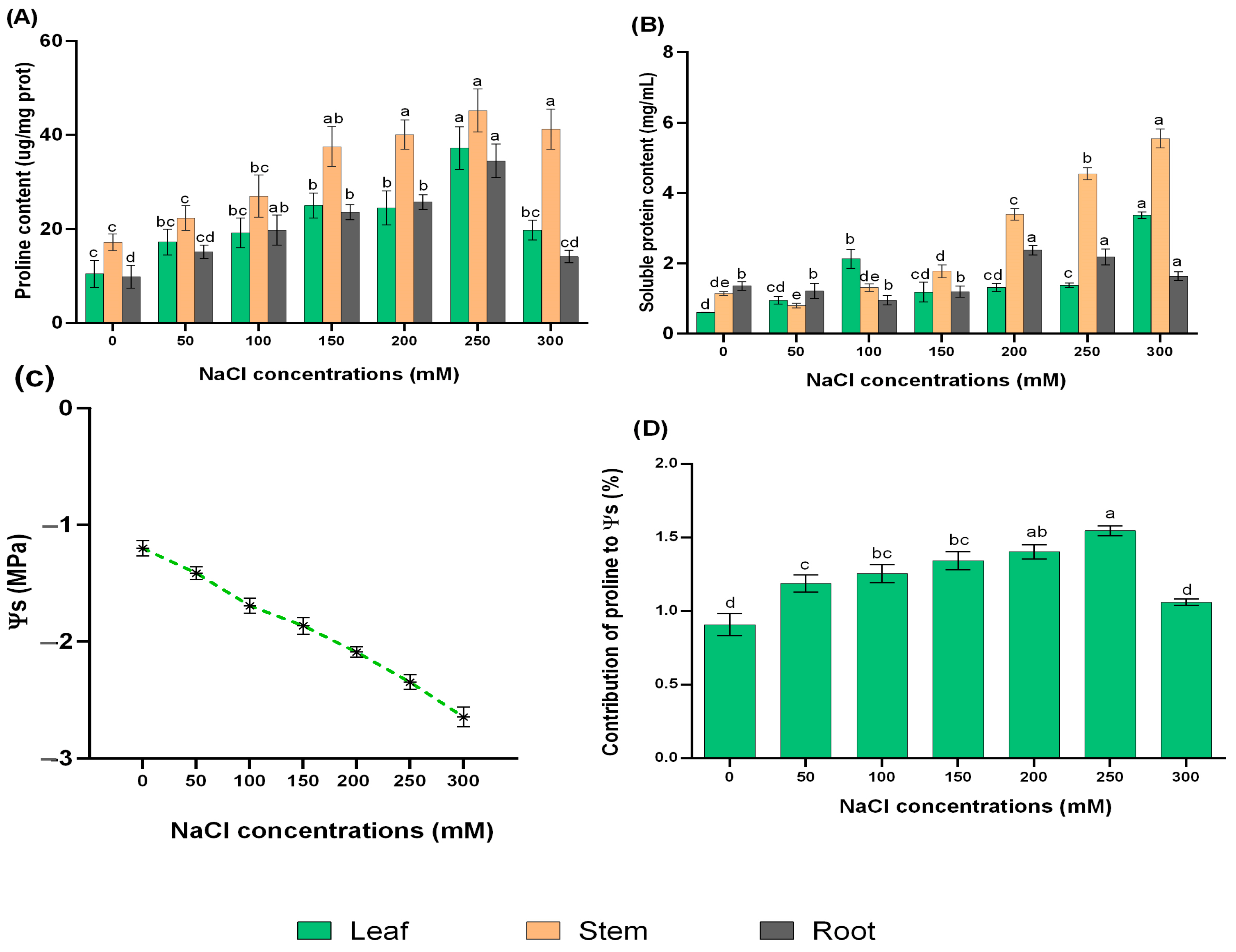
2.2. Responses of Proline, Soluble Protein, and Leaf Osmotic Potential (Ψs) to Different Concentrations of NaCl
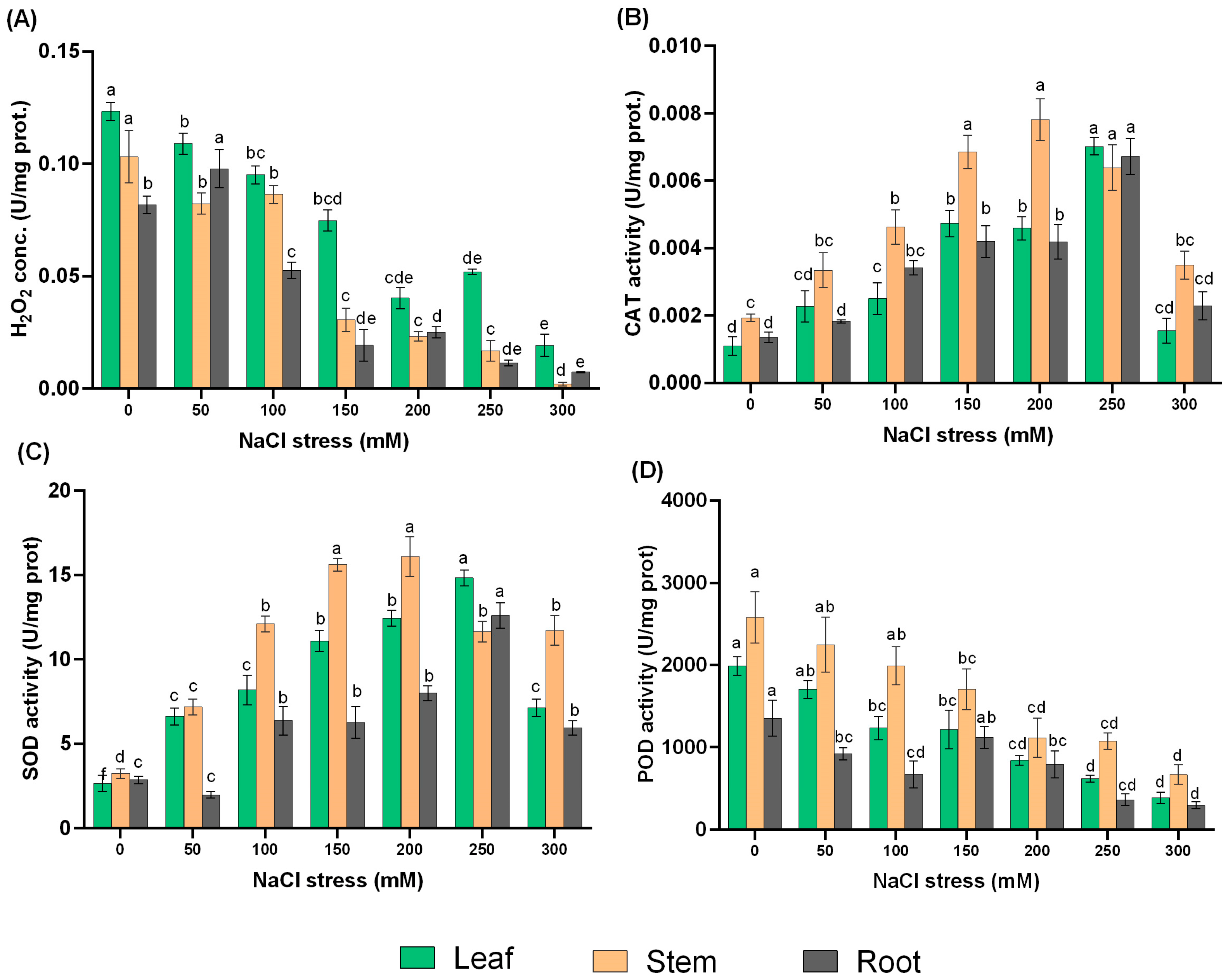
2.3. The Response of Antioxidant Enzymes and H2O2 to NaCl Treatments
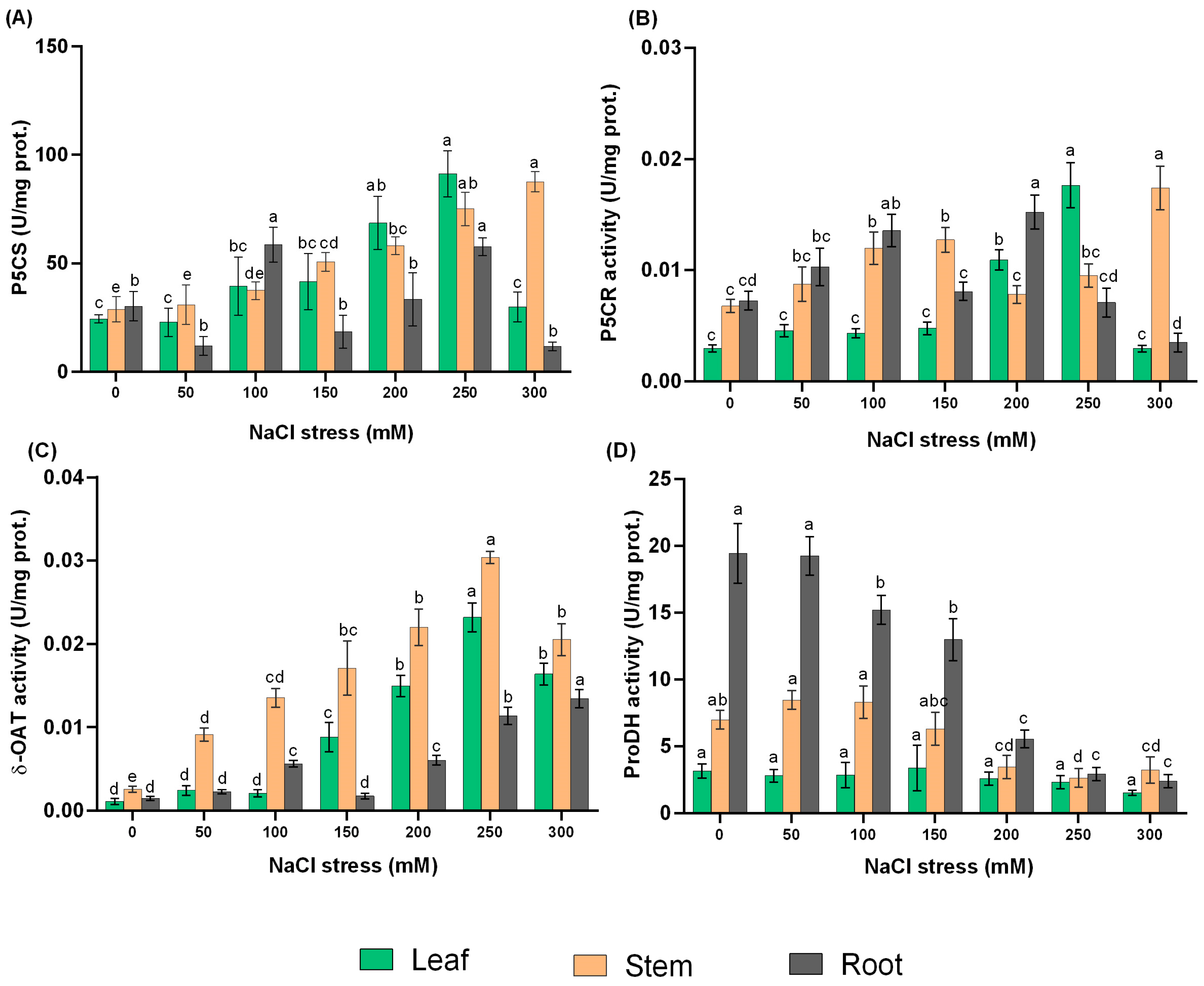
2.4. Effect of NaCl Treatments on the Activities of Proline Metabolic Enzymes
2.5. Bioinformatic Analysis of the Proline Metabolic-Related Genes
2.6. Differential Expression Analysis of the Proline Metabolism-Related Genes
3. Discussion
3.1. The Effects of NaCl Treatments on Growth of L. ruthenicum
3.2. Effects of NaCl Treatments on Antioxidant Enzyme Activities and Proline Content
3.3. Proline Metabolism in L. ruthenicum under NaCl Treatments
4. Materials and Methods
4.1. Plant Material, Growth Condition, and NaCl Treatments
4.2. Leaf Surface Structural and Osmotic Potential (Ψs) Analysis
4.3. Osmolyte Accumulation, Antioxidant Activity, Metabolic Enzyme Content, and H2O2 Content Determination
4.4. Bioinformatic Analysis of Proline Metabolic Enzyme Genes
4.5. RNA Extraction, cDNA Synthesis, and RT-qPCR Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khan, M.; Iqbal, N.; Masood, A.; Khan, N. Variation in salt tolerance of wheat cultivars: Role of glycinebetaine and ethylene. Pedosphere 2012, 22, 746–754. [Google Scholar] [CrossRef]
- Naliwajski, M.; Skłodowska, M. The Relationship between the Antioxidant System and Proline Metabolism in the Leaves of Cucumber Plants Acclimated to Salt Stress. Cells 2021, 10, 609. [Google Scholar] [CrossRef]
- Flowers, T.J.; Munns, R.; Colmer, T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Duhan, S.; Kumari, A.; Lal, M.; Sheokand, S. Oxidative stress and antioxidant defense under combined waterlogging and salinity stresses. In Reactive Oxygen, Nitrogen Sulfur Species in Plants: Production, Metabolism, Signaling Defense Mechanisms; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2019; pp. 113–142. [Google Scholar] [CrossRef]
- Zhang, X.; Yin, H.; Chen, S.; He, J.; Guo, S. Changes in antioxidant enzyme activity and transcript levels of related genes in Limonium sinense Kuntze seedlings under NaCl stress. J. Chem. 2014, 2014, 749047. [Google Scholar] [CrossRef]
- Kumari, M.; Asthir, B.; Verma, D.K.; Singh, V. Biochemical Evaluation of Irrigated Flooded Transplanted and Aerobic Rice (Oryza sativa L.): A Review. In Rice Science: Biotechnological Molecular Advancements; CRC Press: Boca Raton, FL, USA, 2018; p. 109. [Google Scholar]
- Racchi, M.L. Antioxidant Defenses in Plants with Attention to Prunus and Citrus spp. Antioxidants 2013, 2, 340–369. [Google Scholar] [CrossRef] [PubMed]
- Zandi, P.; Schnug, E. Reactive Oxygen Species, Antioxidant Responses and Implications from a Microbial Modulation Perspective. Biology 2022, 11, 155. [Google Scholar] [CrossRef] [PubMed]
- Kusvuran, S.; Kiran, S.; Ellialtioglu, S.S. Antioxidant enzyme activities and abiotic stress tolerance relationship in vegetable crops. In Abiotic Biotic Stress in Plants—Recent Advances and Future Perspectives; Books on Demand: Norderstedt, Germany, 2016; pp. 481–506. [Google Scholar] [CrossRef]
- Lopez-Huertas, E.; Charlton, W.L.; Johnson, B.; Graham, I.A.; Baker, A. Stress induces peroxisome biogenesis genes. EMBO J. 2000, 19, 6770–6777. [Google Scholar] [CrossRef]
- Kavi Kishor, P.B.; Sreenivasulu, N. Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ. 2014, 37, 300–311. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Maggio, A.; Miyazaki, S.; Veronese, P.; Fujita, T.; Ibeas, J.I.; Damsz, B.; Narasimhan, M.L.; Hasegawa, P.M.; Joly, R.J.; Bressan, R.A. Does proline accumulation play an active role in stress-induced growth reduction? Plant J. 2002, 31, 699–712. [Google Scholar] [CrossRef]
- Dar, M.I.; Naikoo, M.I.; Rehman, F.; Naushin, F.; Khan, F.A. Proline accumulation in plants: Roles in stress tolerance and plant development. In Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies; Springer: Berlin/Heidelberg, Germany, 2016; pp. 155–166. [Google Scholar]
- Yu, R.; Zuo, T.; Diao, P.; Fu, J.; Fan, Y.; Wang, Y.; Zhao, Q.; Ma, X.; Lu, W.; Li, A.; et al. Melatonin Enhances Seed Germination and Seedling Growth of Medicago sativa Under Salinity via a Putative Melatonin Receptor MsPMTR1. Front. Plant Sci. 2021, 12, 702875. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Li, S.; Riaz, M.; Jiang, C. Proline metabolism and biosynthesis behave differently in response to boron-deficiency and toxicity in Brassica napus. Plant Physiol. Biochem. 2021, 167, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, Y.; Yadav, V.; Zhao, W.; He, Y.; Zhang, X.; Wei, C. Drought-induced proline is mainly synthesized in leaves and transported to roots in watermelon under water deficit. Hortic. Plant J. 2022, 8, 615–626. [Google Scholar] [CrossRef]
- Alvarez, M.E.; Savouré, A.; Szabados, L. Proline metabolism as regulatory hub. Trends Plant Sci. 2022, 27, 39–55. [Google Scholar] [CrossRef]
- Funck, D.; Stadelhofer, B.; Koch, W. Ornithine-delta-aminotransferase is essential for arginine catabolism but not for proline biosynthesis. BMC Plant Biol. 2008, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Senthil-Kumar, M.; Mysore, K.S. Ornithine-delta-aminotransferase and proline dehydrogenase genes play a role in non-host disease resistance by regulating pyrroline-5-carboxylate metabolism-induced hypersensitive response. Plant Cell Environ. 2012, 35, 1329–1343. [Google Scholar] [CrossRef]
- Anwar, A.; She, M.; Wang, K.; Riaz, B.; Ye, X. Biological Roles of Ornithine Aminotransferase (OAT) in Plant Stress Tolerance: Present Progress and Future Perspectives. Int. J. Mol. Sci. 2018, 19, 3681. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Xu, W.; Sun, N.; Mandal, S.; Wang, H.; Geng, Z. Efficient improvement of soil salinization through phytoremediation induced by chemical remediation in extreme arid land northwest China. Int. J. Phytoremediat. 2020, 22, 334–341. [Google Scholar] [CrossRef]
- Li, S.; Gou, W.; Wang, H.; White, J.F.; Wu, G.; Su, P. Trade-Off Relationships of Leaf Functional Traits of Lycium ruthenicum in Response to Soil Properties in the Lower Reaches of Heihe River, Northwest China. Diversity 2021, 13, 453. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Y.; Zeng, S.; Patra, B.; Yuan, L.; Wang, Y. Isolation and characterization of a salt stress-responsive betaine aldehyde dehydrogenase in Lycium ruthenicum Murr. Physiol. Plant 2018, 163, 73–87. [Google Scholar] [CrossRef]
- Dai, F.; Li, A.; Rao, S.; Chen, J. Potassium transporter LrKUP8 is essential for K+ preservation in Lycium ruthenicum, a salt-resistant desert shrub. Genes 2019, 10, 600. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shu, Q.; Wang, L.; Yu, M.; Hu, Y.; Zhang, H.; Tao, Y.; Shao, Y. Genetic diversity of the endangered and medically important Lycium ruthenicum Murr. revealed by sequence-related amplified polymorphism (SRAP) markers. Biochem. Syst. 2012, 45, 86–97. [Google Scholar] [CrossRef]
- Tiika, R.J.; Wei, J.; Ma, R.; Yang, H.; Cui, G.; Duan, H.; Ma, Y. Identification and expression analysis of the WRKY gene family during different developmental stages in Lycium ruthenicum Murr. fruit. PeerJ 2020, 8, e10207. [Google Scholar] [CrossRef]
- Dhar, P.; Tayade, A.; Ballabh, B.; Chaurasia, O.; Bhatt, R.; Srivastava, R. Lycium ruthenicum Murray: A less-explored but high-value medicinal plant from Trans-Himalayan cold deserts of Ladakh, India. Plant Arch. 2011, 11, 583–586. [Google Scholar]
- Mocan, A.; Zengin, G.; Simirgiotis, M.; Schafberg, M.; Mollica, A.; Vodnar, D.C.; Crişan, G.; Rohn, S. Functional constituents of wild and cultivated Goji (L. barbarum L.) leaves: Phytochemical characterization, biological profile, and computational studies. J. Enzym. Inhib. Med. Chem. 2017, 32, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hou, L.-Y.; Li, Q.-M.; Jiang, Z.-P.; Liu, D.; Zhu, Y. The effects of exogenous antioxidant germanium (Ge) on seed germination and growth of Lycium ruthenicum Murr subjected to NaCl stress. Environ. Technol. 2016, 37, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.J.; Duan, H.R.; Zhang, F.; Li, Y.; Yang, H.S.; Tian, F.P.; Zhou, X.H.; Wang, C.M.; Ma, R. Transcriptomic analysis of Lycium ruthenicum Murr. during fruit ripening provides insight into structural and regulatory genes in the anthocyanin biosynthetic pathway. PLoS ONE 2018, 13, e0208627. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Ma, R.; Yang, Y.; Ni, Q.; Ma, Y. Effects of NaCl treatment on the growth and physiological indexes of Lycium ruthenicum. Mol. Plant Breed 2019, 17, 4437–4447. [Google Scholar]
- Mann, A.; Sharma, C.; Kumar, N.; Kumar, A.; Kumar, A.; Sheoran, P. Halophytes as new model plant species for salt tolerance strategies. Front. Plant Sci. 2023, 14, 1483. [Google Scholar] [CrossRef]
- Hu, J.; Hu, X.; Zhang, H.; Yu, Q. Moderate NaCl alleviates osmotic stress in Lycium ruthenicum. Plant Growth Regul. 2022, 96, 25–35. [Google Scholar] [CrossRef]
- Wang, L.-Q.; Min, Y.-W.; Lin, H.-M. Effect of salt stress on ion absorption and distribution of two Lycium seedlings. Acta Pratacult. Sin. 2011, 20, 129–136. [Google Scholar]
- Xianzhao, L.; Chunzhi, W.; Qing, S. Screening for Salt Tolerance in Eight Halophyte Species from Yellow River Delta at the Two Initial Growth Stages. ISRN Agron. 2013, 2013, 592820. [Google Scholar] [CrossRef]
- Dimitrova, V.; Georgieva, T.; Markovska, Y. Influence of salt stress on some physiological characteristics of two Lycium varieties grown ex vitro in hydroponics. In Proceedings of the Youth Scientific Conference Kliment’s Days, Sofia, Bulgaria, 3–5 November 2016; pp. 141–148. [Google Scholar]
- Mann, A.; Kumar, N.; Lata, C.; Kumar, A.; Kumar, A.; Meena, B.L. Functional annotation of differentially expressed genes under salt stress in Dichanthium annulatum. Plant Physiol. Rep. 2019, 24, 104–111. [Google Scholar] [CrossRef]
- Sozharajan, R.; Natarajan, S. Influence of NaCl salinity on plant growth and nutrient assimilation of Zea mays L. J. Appl. Adv. Res. 2016, 1, 54. [Google Scholar] [CrossRef]
- Li, W.; Rao, S.; Du, C.; Liu, L.; Dai, G.; Chen, J. Strategies used by two goji species, Lycium ruthenicum and Lycium barbarum, to defend against salt stress. Sci. Hortic. 2022, 306, 111430. [Google Scholar] [CrossRef]
- Guo, H.; Cui, Y.-N.; Pan, Y.-Q.; Wang, S.-M.; Bao, A.-K. Sodium chloride facilitates the secretohalophyte Atriplex canescens adaptation to drought stress. Plant Physiol. Biochem. 2020, 150, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.Q.; Guo, H.; Wang, S.M.; Zhao, B.; Zhang, J.L.; Ma, Q.; Yin, H.J.; Bao, A.K. The Photosynthesis, Na(+)/K(+) Homeostasis and Osmotic Adjustment of Atriplex canescens in Response to Salinity. Front. Plant Sci. 2016, 7, 848. [Google Scholar] [CrossRef] [PubMed]
- Tahjib-Ui-Arif, M.; Sohag, A.A.; Afrin, S.; Bashar, K.K.; Afrin, T.; Mahamud, A.G.M.S.; Polash, M.A.; Hossain, M.T.; Sohel, M.A.; Brestic, M.; et al. Differential Response of Sugar Beet to Long-Term Mild to Severe Salinity in a Soil–Pot Culture. Agriculture 2019, 9, 223. [Google Scholar] [CrossRef]
- He, X.; Xu, L.; Pan, C.; Gong, C.; Wang, Y.; Liu, X.; Yu, Y. Drought resistance of Camellia oleifera under drought stress: Changes in physiology and growth characteristics. PLoS ONE 2020, 15, e0235795. [Google Scholar] [CrossRef]
- Hao, S.; Wang, Y.; Yan, Y.; Liu, Y.; Wang, J.; Chen, S. A Review on Plant Responses to Salt Stress and Their Mechanisms of Salt Resistance. Horticulturae 2021, 7, 132. [Google Scholar] [CrossRef]
- Blum, A. Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Environ. 2017, 40, 4–10. [Google Scholar] [CrossRef] [PubMed]
- El Moukhtari, A.; Cabassa-Hourton, C.; Farissi, M.; Savouré, A. How Does Proline Treatment Promote Salt Stress Tolerance During Crop Plant Development? Front. Plant Sci. 2020, 11, 1127. [Google Scholar] [CrossRef]
- Wang, H.; Tang, X.; Wang, H.; Shao, H.B. Proline accumulation and metabolism-related genes expression profiles in Kosteletzkya virginica seedlings under salt stress. Front. Plant Sci. 2015, 6, 792. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, R.; Corpas, F.J.; Carreras, A.; Gómez-Rodríguez, M.V.; Chaki, M.; Pedrajas, J.R.; Fernández-Ocaña, A.; Del Río, L.A.; Barroso, J.B. The dehydrogenase-mediated recycling of NADPH is a key antioxidant system against salt-induced oxidative stress in olive plants. Plant Cell Environ. 2006, 29, 1449–1459. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yang, H.; Chang, Y.; Cheng, J.; Bai, S.; Yin, J. Effects of nitric oxide on reactive oxygen species and antioxidant capacity in Chinese Bayberry during storage. Sci. Hortic. 2012, 135, 106–111. [Google Scholar] [CrossRef]
- Mo, S.; Biao, A.; Wang, Z.; Lin, S.; Yang, T.; Pan, L.; Wang, Y.; Zeng, S. Spatio transcriptome uncover novel insight into the Lycium ruthenicum seedling tolerant to salt stress. Ind. Crops Prod. 2022, 177, 114502. [Google Scholar] [CrossRef]
- Li, C.; Mur, L.A.J.; Wang, Q.; Hou, X.; Zhao, C.; Chen, Z.; Wu, J.; Guo, Q. ROS scavenging and ion homeostasis is required for the adaptation of halophyte Karelinia caspia to high salinity. Front. Plant Sci. 2022, 13, 979956. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Gao, Y.; Sun, S.; Lu, X.; Li, Q.; Li, L.; Wang, K.; Liu, J. Effects of Salt Stress on the Antioxidant Activity and Malondialdehyde, Solution Protein, Proline, and Chlorophyll Contents of Three Malus Species. Life 2022, 12, 1929. [Google Scholar] [CrossRef]
- Guo, S.; Ma, X.; Cai, W.; Wang, Y.; Gao, X.; Fu, B.; Li, S. Exogenous Proline Improves Salt Tolerance of Alfalfa through Modulation of Antioxidant Capacity, Ion Homeostasis, and Proline Metabolism. Plants 2022, 11, 2994. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-T.; Qiu, Z.-B.; Zhang, X.-W.; Wang, L.-S. Exogenous hydrogen peroxide can enhance tolerance of wheat seedlings to salt stress. Acta Physiol. Plant. 2011, 33, 835–842. [Google Scholar] [CrossRef]
- Li, T.; Hu, Y.; Du, X.; Tang, H.; Shen, C.; Wu, J. Salicylic Acid Alleviates the Adverse Effects of Salt Stress in Torreya grandis cv. Merrillii Seedlings by Activating Photosynthesis and Enhancing Antioxidant Systems. PLoS ONE 2014, 9, e109492. [Google Scholar] [CrossRef]
- Ben Ahmed, C.; Ben Rouina, B.; Sensoy, S.; Boukhriss, M.; Ben Abdullah, F. Exogenous proline effects on photosynthetic performance and antioxidant defense system of young olive tree. J. Agric. Food Chem. 2010, 58, 4216–4222. [Google Scholar] [CrossRef] [PubMed]
- Trovato, M.; Mattioli, R.; Costantino, P. Multiple roles of proline in plant stress tolerance and development. Rend. Lincei 2008, 19, 325–346. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Pandian, S.K.; Ramesh, M. Transgenic indica rice cv. ADT 43 expressing a Δ1-pyrroline-5-carboxylate synthetase (P5CS) gene from Vigna aconitifolia demonstrates salt tolerance. Plant Cell Tissue Organ Cult. 2011, 107, 383–395. [Google Scholar] [CrossRef]
- Guerzoni, J.T.S.; Belintani, N.G.; Moreira, R.M.P.; Hoshino, A.A.; Domingues, D.S.; Vieira, L.G.E. Stress-induced Δ1-pyrroline-5-carboxylate synthetase (P5CS) gene confers tolerance to salt stress in transgenic sugarcane. Acta Physiol. Plant. 2014, 36, 2309–2319. [Google Scholar] [CrossRef]
- Wei, C.; Cui, Q.; Zhang, X.-Q.; Zhao, Y.-Q.; Jia, G.-X. Three P5CS genes including a novel one from Lilium regale play distinct roles in osmotic, drought and salt stress tolerance. J. Plant Biol. 2016, 59, 456–466. [Google Scholar] [CrossRef]
- Armengaud, P.; Thiery, L.; Buhot, N.; Grenier-De March, G.; Savouré, A. Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiol. Plant 2004, 120, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Verbruggen, N.; Hermans, C. Proline accumulation in plants: A review. Amino Acids 2008, 35, 753–759. [Google Scholar] [CrossRef]
- Misra, N.; Gupta, A.K. Effect of salt stress on proline metabolism in two high yielding genotypes of green gram. Plant Sci. 2005, 169, 331–339. [Google Scholar] [CrossRef]
- Kumar, S.G.; Reddy, A.M.; Sudhakar, C. NaCl effects on proline metabolism in two high yielding genotypes of mulberry (Morus alba L.) with contrasting salt tolerance. Plant Sci. 2003, 165, 1245–1251. [Google Scholar] [CrossRef]
- de Freitas, P.A.F.; de Carvalho, H.H.; Costa, J.H.; Miranda, R.S.; Saraiva, K.; de Oliveira, F.D.B.; Coelho, D.G.; Prisco, J.T.; Gomes-Filho, E. Salt acclimation in sorghum plants by exogenous proline: Physiological and biochemical changes and regulation of proline metabolism. Plant Cell Rep. 2019, 38, 403–416. [Google Scholar] [CrossRef]
- Funck, D.; Eckard, S.; Müller, G. Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol. 2010, 10, 70. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Tiika, R.J.; Ma, Y.; Yang, H.; Cui, G.; Tian, F.; Zhang, Y.; Duan, H.; Ma, R. Transcriptome-wide identification and analysis of the KT/HAK/KUP family in black goji under NaCl stress. Agron. J. 2022, 114, 2069–2080. [Google Scholar] [CrossRef]
- Schmidt, R.; Mieulet, D.; Hubberten, H.M.; Obata, T.; Hoefgen, R.; Fernie, A.R.; Fisahn, J.; San Segundo, B.; Guiderdoni, E.; Schippers, J.H.; et al. Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 2013, 25, 2115–2131. [Google Scholar] [CrossRef]
- Li, Q.; Li, B.; Wang, J.; Chang, X.; Mao, X.; Jing, R. TaPUB15, a U-Box E3 ubiquitin ligase gene from wheat, enhances salt tolerance in rice. Food Energy Secur. 2021, 10, e250. [Google Scholar] [CrossRef]
- Guerrier, G. Fluxes of Na+, K+ and Cl−, and osmotic adjustment in Lycopersicon pimpinellifolium and L. esculentum during short-and long-term exposures to NaCl. Physiol. Plant. 1996, 97, 583–591. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.a.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q. Guidance of Plant Physiology and Biochemistry Experiment; China Agriculture Press: Beijing, China, 1995; pp. 59–99. [Google Scholar]
- Hammerschmidt, R.; Nuckles, E.M.; Kuć, J. Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiol. Plant Pathol. 1982, 20, 73–82. [Google Scholar] [CrossRef]
- Rajeswari, V.; Paliwal, K. Peroxidase and catalase changes during in vitro adventitious shoot organogenesis from hypocotyls of Albizia odoratissima L.f. (Benth). Acta Physiol. Plant. 2008, 30, 825–832. [Google Scholar] [CrossRef]
- Satterfield, C.N.; Bonnell, A.H. Interferences in Titanium Sulfate Method for Hydrogen Peroxide. Anal. Chem. 1955, 27, 1174–1175. [Google Scholar] [CrossRef]
- Han, P.L.; Dong, Y.H.; Gu, K.D.; Yu, J.Q.; Hu, D.G.; Hao, Y.J. The apple U-box E3 ubiquitin ligase MdPUB29 contributes to activate plant immune response to the fungal pathogen Botryosphaeria dothidea. Planta 2019, 249, 1177–1188. [Google Scholar] [CrossRef]
- Campanile, C.; Forlani, G.; Basso, A.L.; Marasco, R.; Ricca, E.; Sacco, M.; Ferrara, L.; De Felice, M. Identification and characterization of the proBA operon of Streptococcus bovis. Appl. Environ. Microbiol. 1993, 59, 519–522. [Google Scholar] [CrossRef]
- Rena, A.B.; Splittstoesser, W.E. Proline dehydrogenase and pyrroline-5-carboxylate reductase from pumpkin cotyledons. Phytochemistry 1975, 14, 657–661. [Google Scholar] [CrossRef]
- Charest, C.; Ton Phan, C. Cold acclimation of wheat (Triticum aestivum): Properties of enzymes involved in proline metabolism. Physiol. Plant. 1990, 80, 159–168. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Chang, E.; Shi, S.; Liu, J.; Cheng, T.; Xue, L.; Yang, X.; Yang, W.; Lan, Q.; Jiang, Z. Selection of Reference Genes for Quantitative Gene Expression Studies in Platycladus orientalis (Cupressaceae) Using Real-Time PCR. PLoS ONE 2012, 7, e33278. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
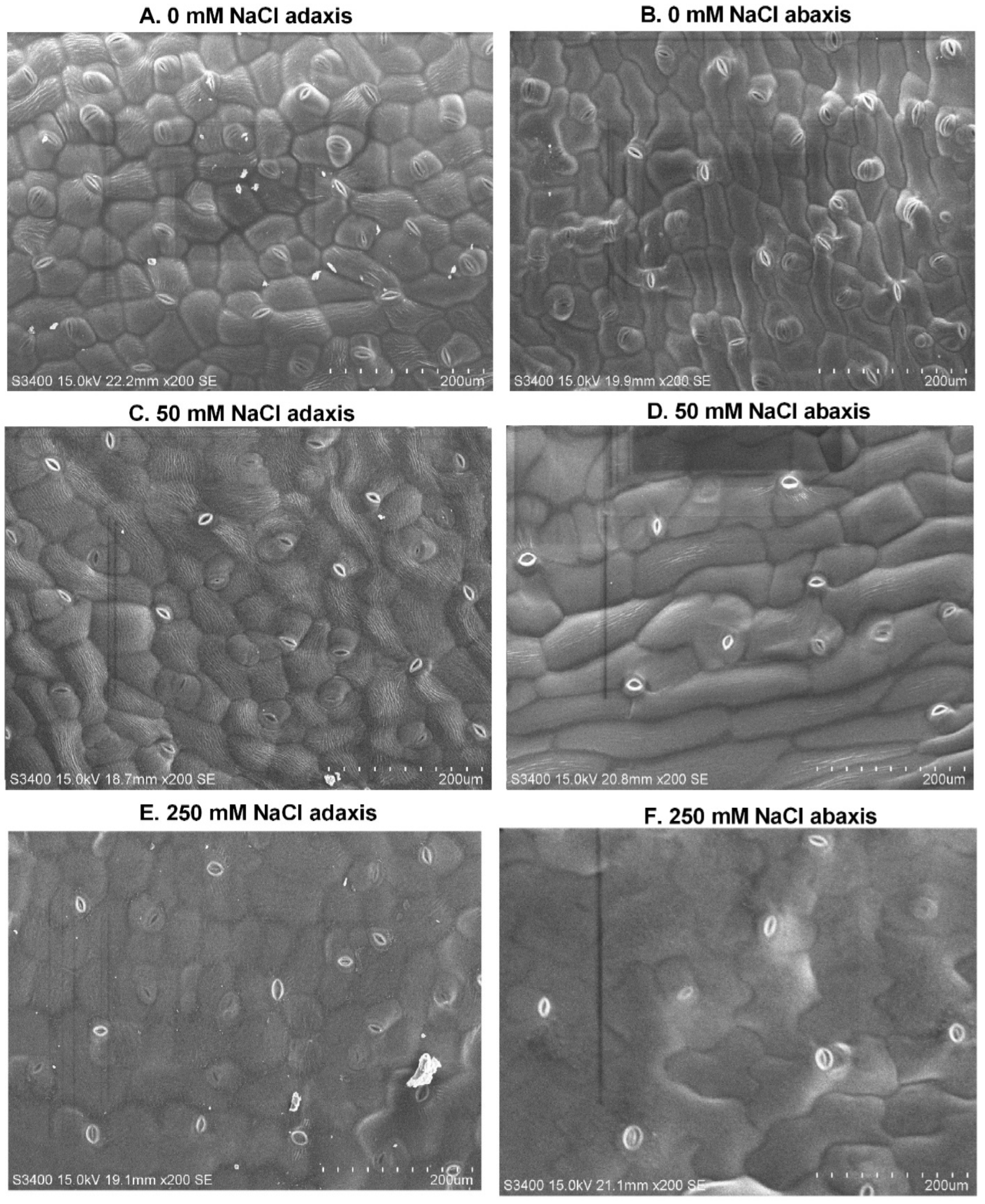

| NaCl (mM) | Shoot Length (cm) | Root Length (cm) | Leaf Fresh Weight (g) | Stem Fresh Weight (g) | Root Fresh Weight (g) |
|---|---|---|---|---|---|
| 0 | 19.14 ± 0.18 b | 18.72 ± 0.37 b | 2.03 ± 0.23 c | 0.69 ± 0.01 c | 1.11 ± 0.07 b |
| 50 | 19.22 ± 0.14 ab | 19.84 ± 0.24 a | 2.59 ± 0.18 b | 0.74 ± 0.03 bc | 1.18 ± 0.12 b |
| 100 | 19.77 ± 0.19 a | 19.56 ± 0.30 a | 2.69 ± 0.09 b | 0.78 ± 0.01 ab | 1.45 ± 0.05 a |
| 150 | 19.15 ± 0.32 b | 17.02 ± 0.26 c | 3.34 ± 0.15 a | 0.82 ± 0.04 a | 1.04 ± 0.14 c |
| 200 | 18.42 ± 0.15 c | 16.40 ± 0.10 cd | 1.66 ± 0.11 cd | 0.36 ± 0.02 d | 0.43 ± 0.10 cd |
| 250 | 16.16 ± 0.19 d | 15.74 ± 0.20 d | 1.35 ± 0.13 de | 0.15 ± 0.02 e | 0.20 ± 0.06 d |
| 300 | 14.24 ± 0.25 e | 14.38 ± 0.11 e | 1.05 ± 0.11 e | 0.13 ± 0.01 e | 0.13 ± 0.04 d |
| NaCl (mM) | Stomata Density | Stomata Size (μm2) | Guard Cell Size (μm2) | Epidermal Cell Size (μm2) | ||||
|---|---|---|---|---|---|---|---|---|
| Adax | Abax | Adax | Abax | Adax | Abax | Adax | Abax | |
| 0 | 39 | 33 | 26.93 ± 1.9 b | 28.48 ± 3.8 b | 36.64 ± 1.0 bc | 40.92 ± 0.8 b | 19,600 ± 469 c | 21,620 ± 738 c |
| 50 | 27 | 21 | 42.68 ± 3.5 a | 38.73 ± 1.9 a | 39.48 ± 0.7 b | 35.26 ± 1.6 bc | 25,716 ± 3169 c | 33,550 ± 735 b |
| 250 | 20 | 14 | 3.93 ± 0.6 c | 2.63 ± 0.3 c | 60.98 ± 3.9 a | 32.13 ± 2.4 c | 36,794 ± 3565 b | 52,322 ± 3105 a |
| Protein Name | Protein ID | Aa Residues | MW (kDa) | pI | Subcellular Localization Prediction |
|---|---|---|---|---|---|
| LrP5CS1 | TRINITY_DN21284_c1_g3_i9_3 | 387 | 41.94 | 5.72 | cyto |
| LrP5CS2 | TRINITY_DN21540_c0_g2_i1_2 | 717 | 77.59 | 6.11 | cyto |
| LrP5CS3 | TRINITY_DN25963_c0_g2_i9_10 | 588 | 63.52 | 5.55 | cyto |
| LrP5CR | TRINITY_DN15477_c0_g1_i2_5 | 274 | 28.44 | 8.92 | cyto |
| LrOAT1 | TRINITY_DN11125_c0_g1_i4_8 | 320 | 35.11 | 8.53 | mito |
| LrOAT2 | TRINITY_DN13448_c0_g1_i1_6 | 319 | 35.02 | 8.53 | mito |
| LrProDH1 | TRINITY_DN36679_c0_g1_i1_7 | 501 | 56.20 | 8.24 | mito |
| LrProDH2 | TRINITY_DN50_c0_g1_i2_8 | 489 | 54.92 | 7.97 | mito |
| LrP5CDH | TRINITY_DN23202_c0_g1_i3_9 | 448 | 50.15 | 6.38 | mito |
| Gene | Primer Name | Sequence (5′–3′) | E(%) | R2 |
|---|---|---|---|---|
| LrP5CS1 | LrP5CS1-F | CATCATCTCCATTGGGTGTTCT | 99 | 0.99 |
| LrP5CS1-R | CCTCCTTCCCTCCTTTCAGC | |||
| LrP5CS2 | LrP5CS2-F | GCGGAGGTGGGCATTAGTA | 93 | 0.99 |
| LrP5CS2-R | CTTGCGAGCCATTTAGTTGTTA | |||
| LrP5CS3 | LrP5CS3-F | CATTGGCCGCATTTTAAAGAG | 105 | 0.99 |
| LrP5CS3-R | TGTACGAGTGCATCAGGTCGTG | |||
| LrP5CR | LrP5CR-F | CACCAGGGGGAACAACCATT | 99 | 0.99 |
| LrP5CR-R | TGCGTTTAGCTGCACCAACAA | |||
| LrOAT1 | LrOAT1-F | TCTTCCCCTTCTCAAAATCTCA | 100 | 0.99 |
| LrOAT1-R | CCCAGACGGATGAACCCTTAG | |||
| LrOAT2 | LrOAT2-F | GTCAGCATTGATGTCCAGTTTCT | 107 | 0.99 |
| LrOAT2-R | TGTCCACATTGCTAGCTGGT | |||
| LrProDH1 | LrProDH1-F | ATTCAGGGAGGTAATAATGGGG | 103 | 0.99 |
| LrProDH1-R | GTAACCGTCCTTCGTACTTCTTCT | |||
| LrProDH2 | LrProDH2-F | CTCTAACGGCAGAGGAAGAGC | 101 | 0.99 |
| LrProDH2-R | CCGCATCAATGAGTAAGGGA | |||
| LrP5CDH | LrP5CDH-F | TGCCGTCAATCATGTATCG | 100 | 0.99 |
| LrP5CDH-R | ATTGTGGTTTGACCTTGC | |||
| LrEF1-α | LrEF1a-F | CCATACCAGCATCACCATTCTTC | 100 | 0.99 |
| LrEF1a-R | GTCACACTTCCCACATTGCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiika, R.J.; Duan, H.; Yang, H.; Cui, G.; Tian, F.; He, Y.; Ma, Y.; Li, Y. Proline Metabolism Process and Antioxidant Potential of Lycium ruthenicum Murr. in Response to NaCl Treatments. Int. J. Mol. Sci. 2023, 24, 13794. https://doi.org/10.3390/ijms241813794
Tiika RJ, Duan H, Yang H, Cui G, Tian F, He Y, Ma Y, Li Y. Proline Metabolism Process and Antioxidant Potential of Lycium ruthenicum Murr. in Response to NaCl Treatments. International Journal of Molecular Sciences. 2023; 24(18):13794. https://doi.org/10.3390/ijms241813794
Chicago/Turabian StyleTiika, Richard John, Huirong Duan, Hongshan Yang, Guangxin Cui, Fuping Tian, Yongtao He, Yanjun Ma, and Yi Li. 2023. "Proline Metabolism Process and Antioxidant Potential of Lycium ruthenicum Murr. in Response to NaCl Treatments" International Journal of Molecular Sciences 24, no. 18: 13794. https://doi.org/10.3390/ijms241813794
APA StyleTiika, R. J., Duan, H., Yang, H., Cui, G., Tian, F., He, Y., Ma, Y., & Li, Y. (2023). Proline Metabolism Process and Antioxidant Potential of Lycium ruthenicum Murr. in Response to NaCl Treatments. International Journal of Molecular Sciences, 24(18), 13794. https://doi.org/10.3390/ijms241813794







