Differential Regulations of Antioxidant Metabolism and Cold-Responsive Genes in Three Bermudagrass Genotypes under Chilling and Freezing Stress
Abstract
:1. Introduction
2. Results
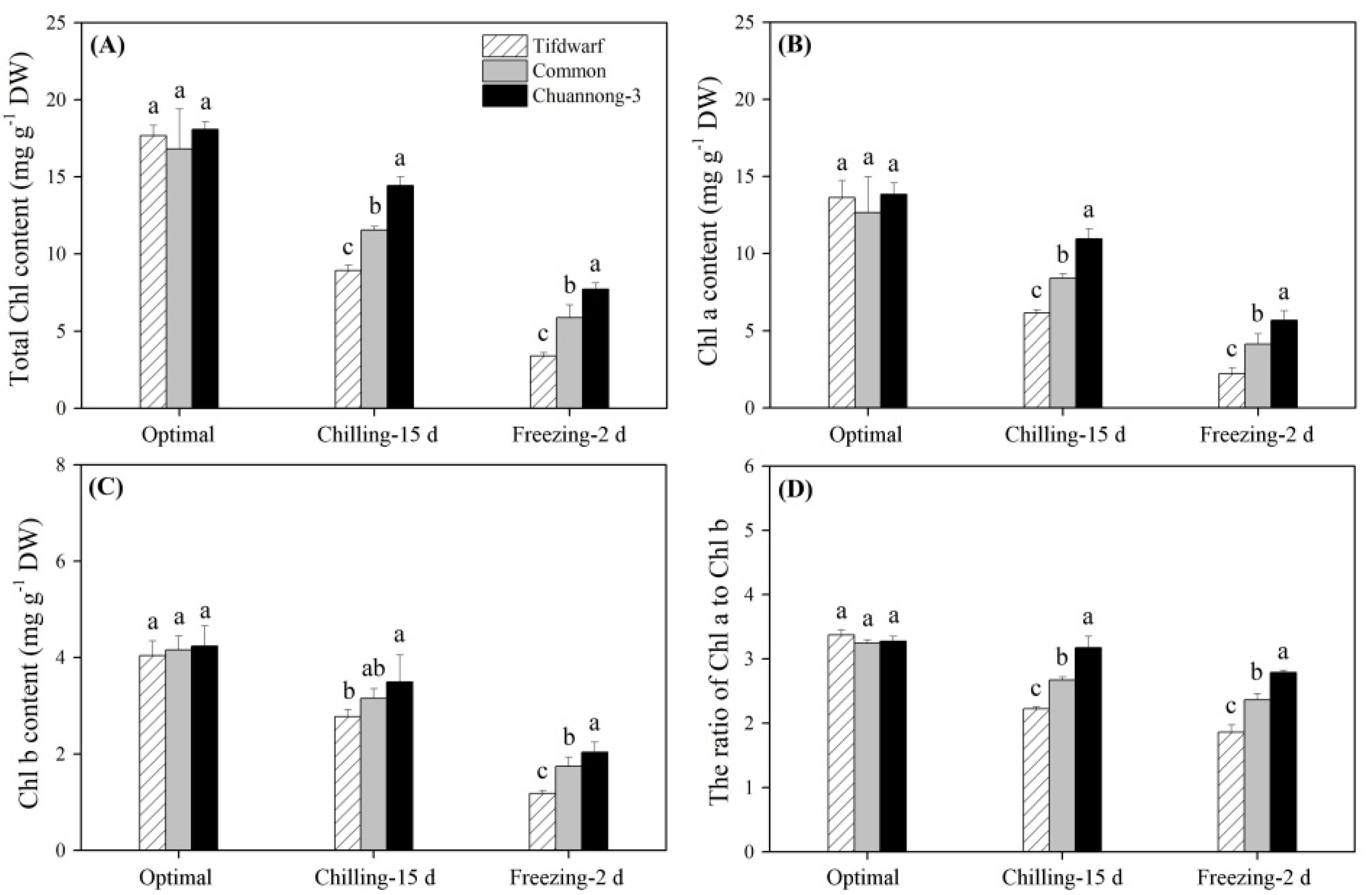
2.1. Chlorophyll Content and Photochemical Efficiency in Response to Chilling and Freezing Stress
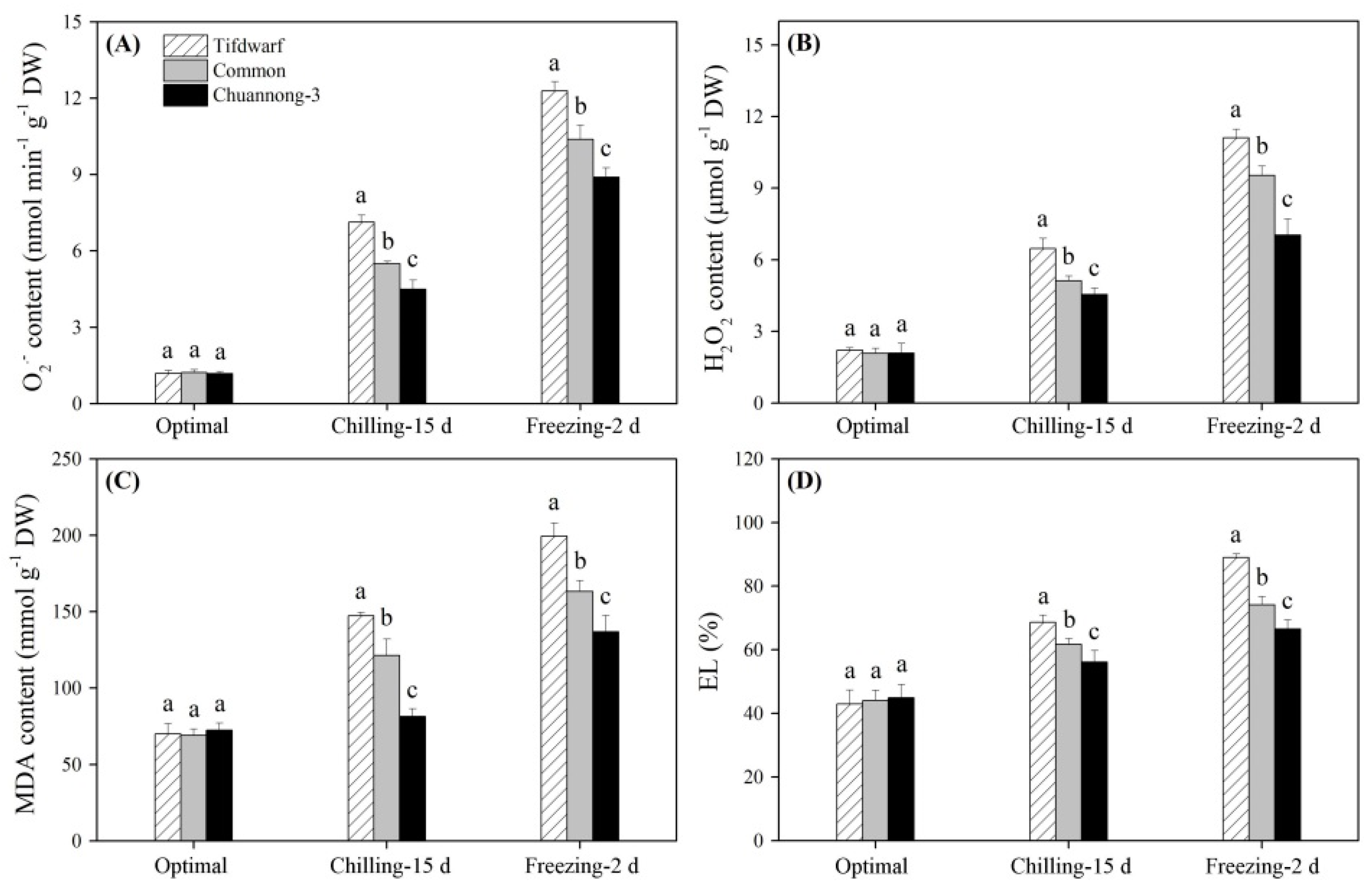
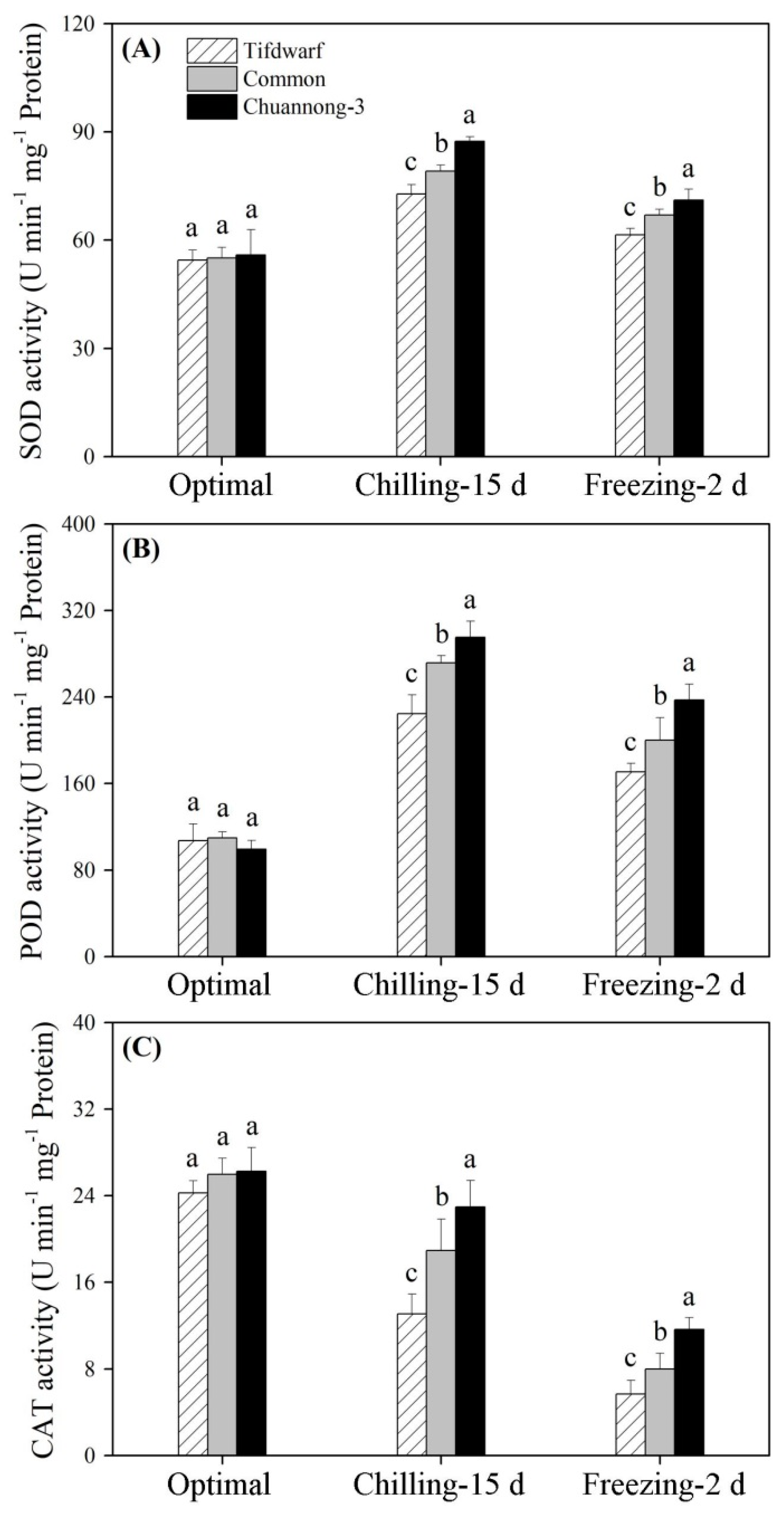
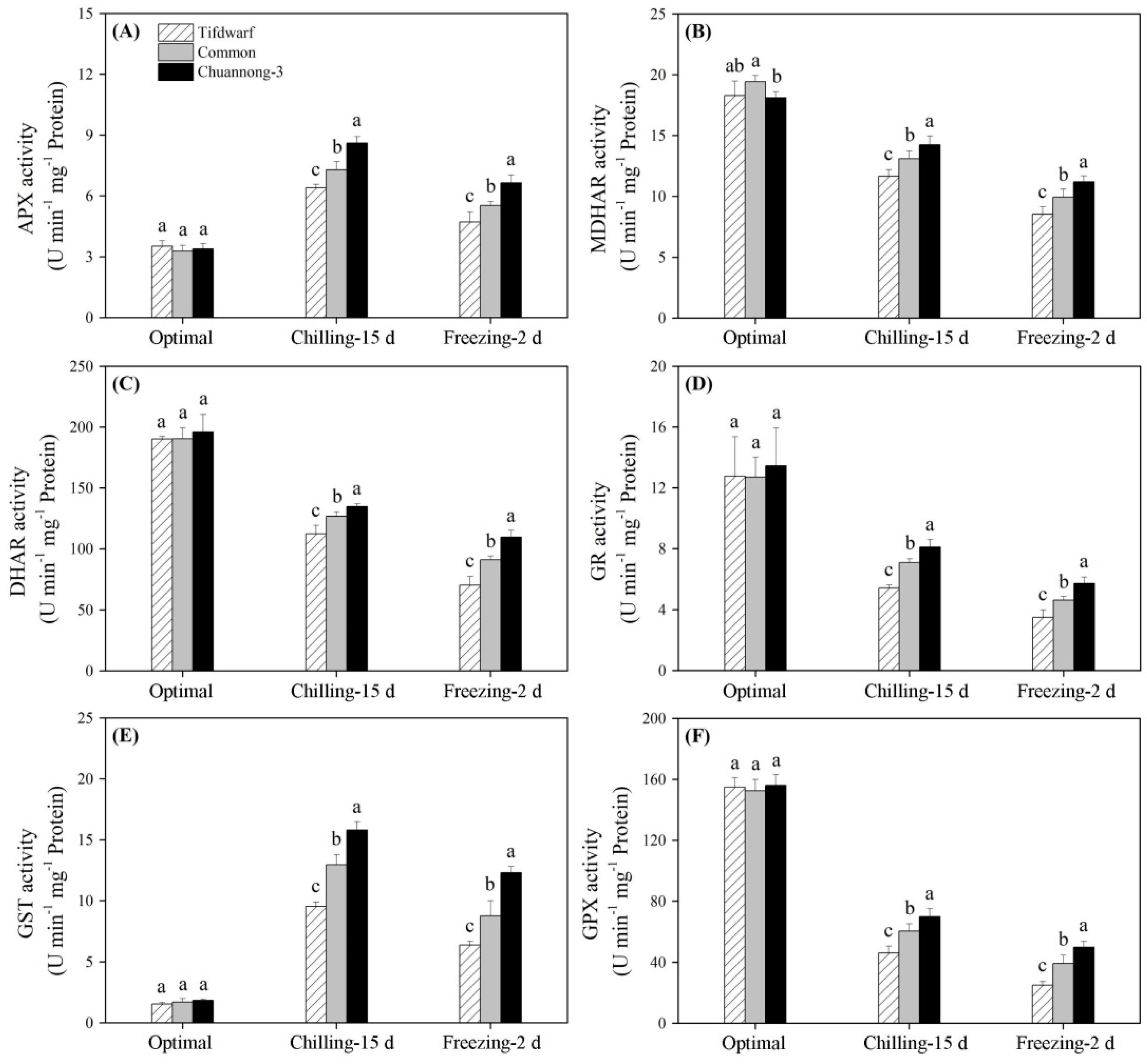
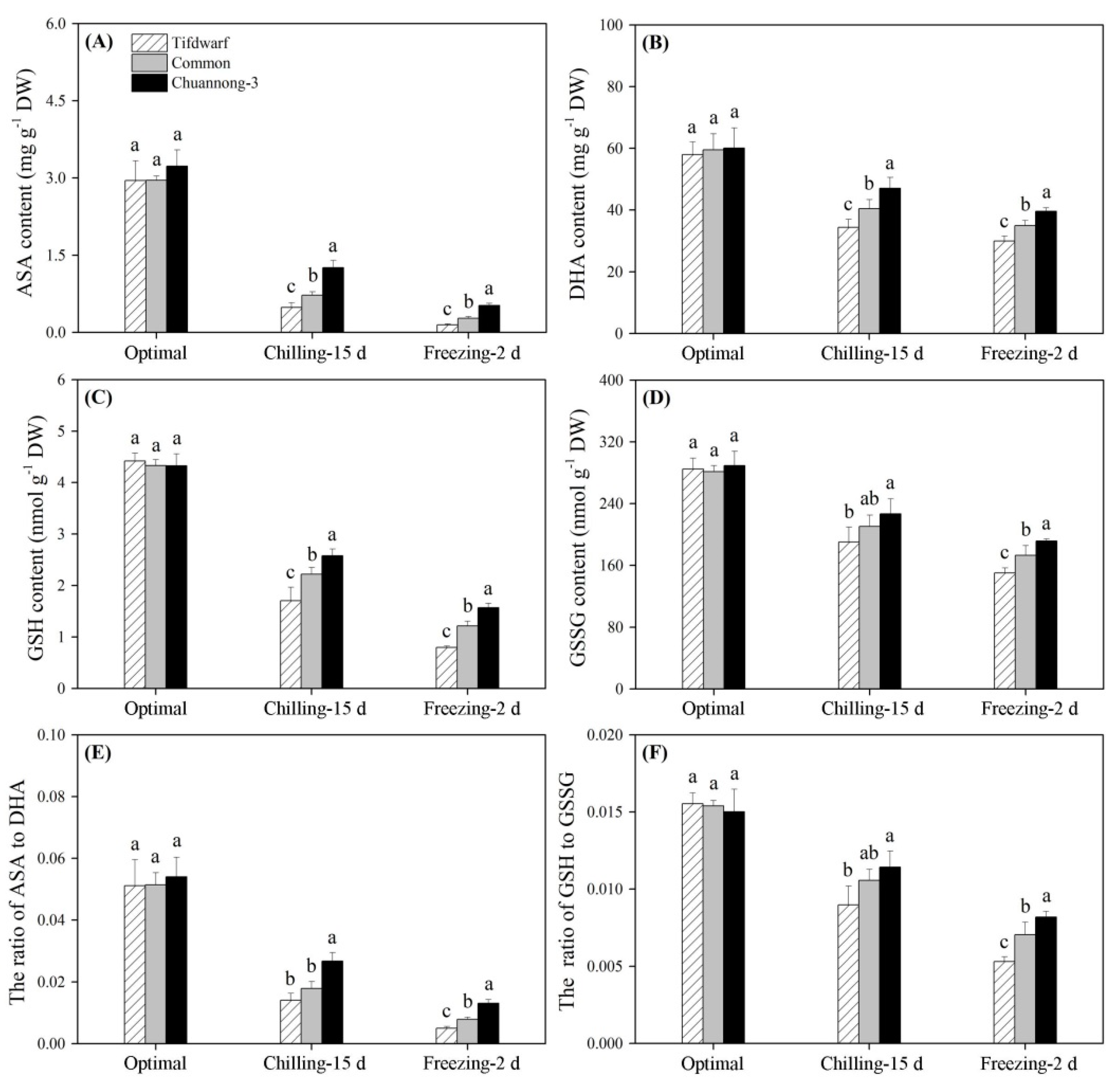
2.2. Antioxidant Metabolism in Response to Chilling and Freezing Stress
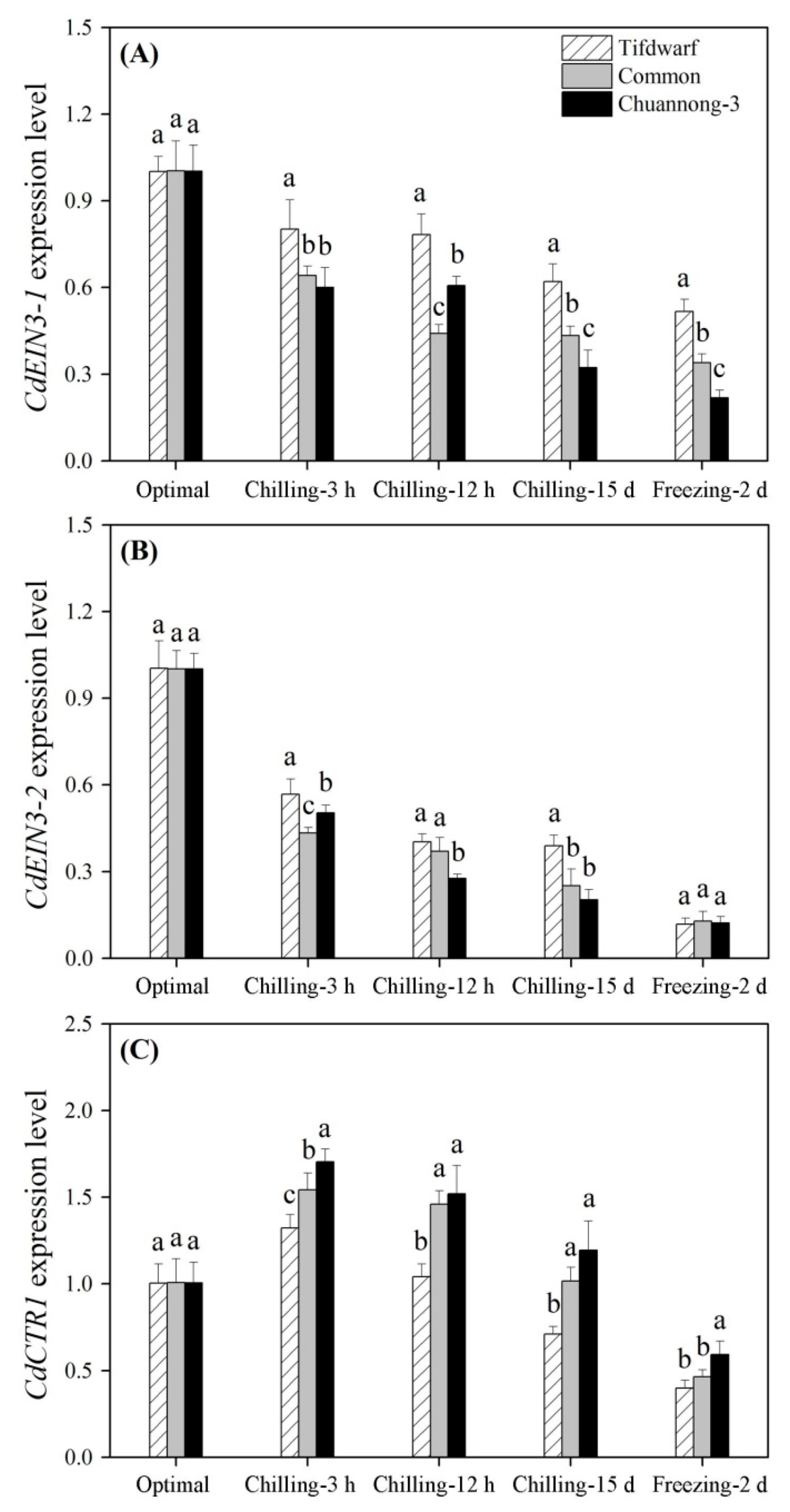
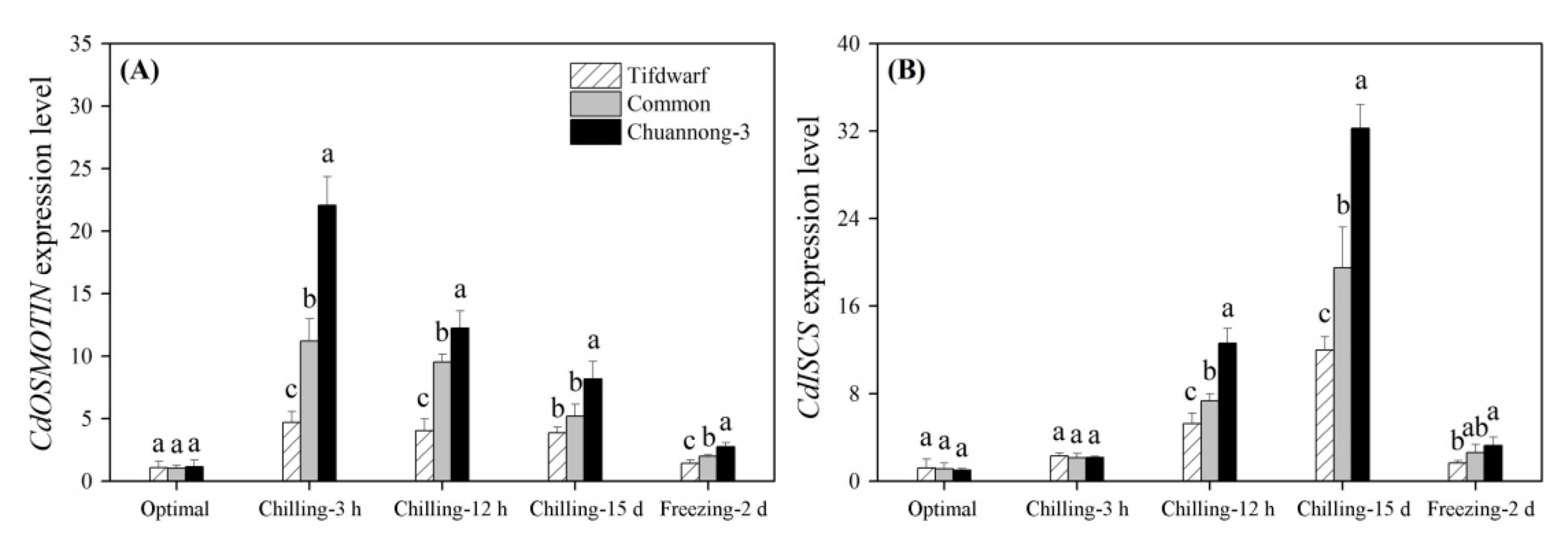
2.3. Differential Expression of Cold-Responsive Genes in Three Bermudagrass Genotypes under Chilling and Freezing Stress
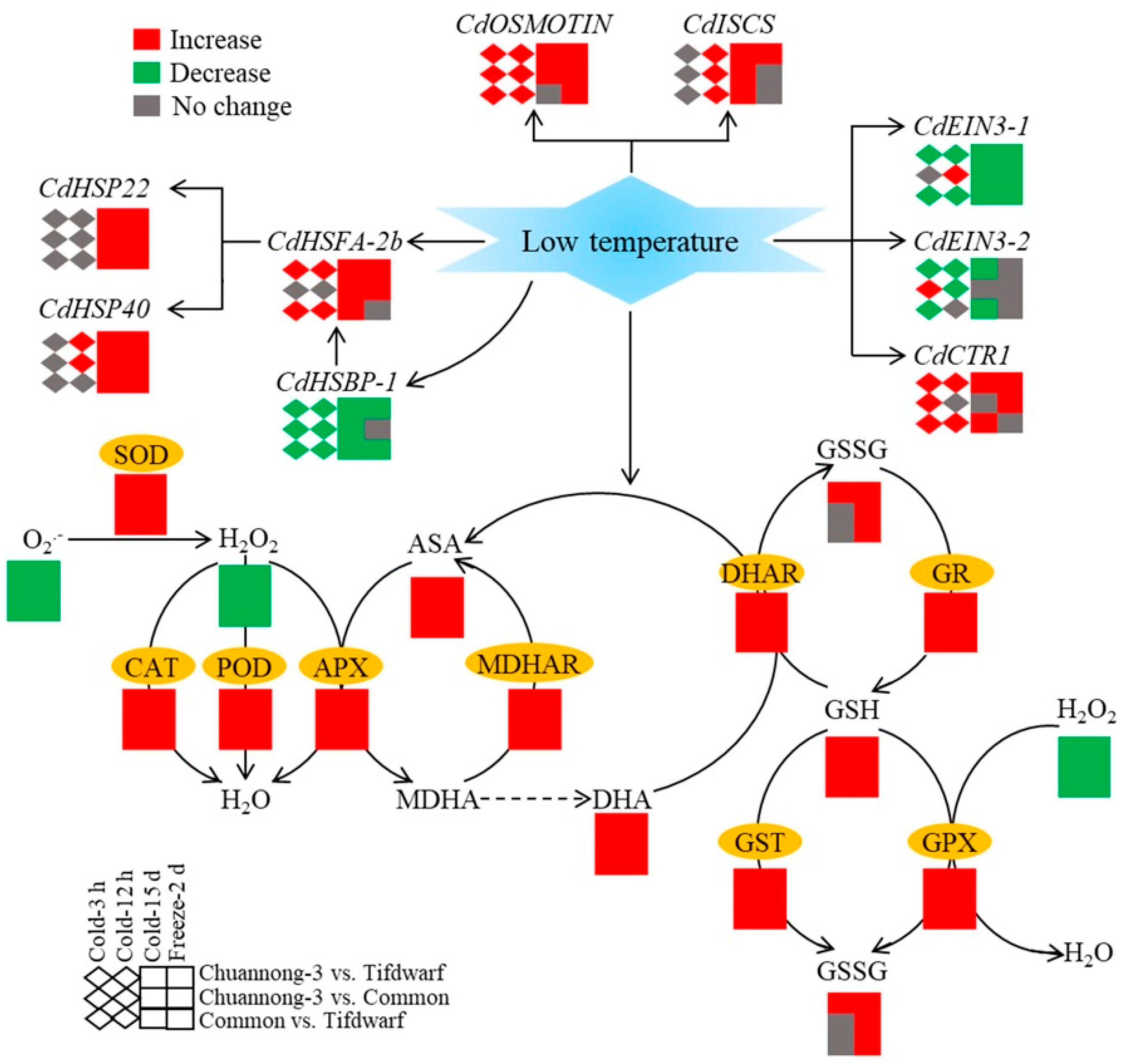
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Determination of Chlorophyll Content, Photochemical Efficiency, and Oxidative Damage
4.3. Determination of Antioxidant Enzyme Activity and Antioxidant Metabolite
4.4. Determination of Gene Expression
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fry, J.; Huang, B. Applied Turfgrass Science and Physiology; John Wiley Sons: Hoboken, NJ, USA, 2004. [Google Scholar]
- Hao, T.; Yang, Z.; Liang, J.; Yu, J.; Liu, J. Foliar application of carnosine and chitosan improving drought tolerance in bermudagrass. Agronomy 2023, 13, 442. [Google Scholar] [CrossRef]
- Bian, Y.; Li, Q.; Zhang, X.; Hao, T.; Liu, N.; Yang, Z.; Yu, J. Lipid composition remodeling plays a critical role during the differential responses of leaves and roots to heat stress in bermudagrass. Environ. Exp. Bot. 2023, 213, 105423. [Google Scholar] [CrossRef]
- Huang, S.; Jiang, S.; Liang, J.; Chen, M.; Shi, Y. Current knowledge of bermudagrass responses to abiotic stresses. Breeding Sci. 2019, 69, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, K.; Ervin, E.H. Bermudagrass freezing tolerance associated with abscisic acid metabolism and dehydrin expression during cold acclimation. J. Amer. Soc. Hort. Sci. 2008, 133, 542–550. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, K.; Ervin, E.H.; Waltz, C.; Murphy, T. Metabolic changes during cold acclimation and deacclimation in five bermudagrass varieties. I. proline, total amino acid, protein, and dehydrin expression. Crop Sci. 2011, 51, 838–846. [Google Scholar] [CrossRef]
- Zhang, X.; Ervin, E.H.; Waltz, C.; Murphy, T. Metabolic changes during cold acclimation and deacclimation in five bermudagrass varieties: II. cytokinin and abscisic acid metabolism. Crop Sci. 2011, 51, 847–853. [Google Scholar] [CrossRef]
- Cheng, Z.; Jin, R.; Cao, M.; Liu, X.; Chan, Z. Exogenous application of ABA mimic 1 (AM1) improves cold stress tolerance in bermudagrass (Cynodon dactylon). Plant Cell Tissue Organ Cult. PCTOC 2016, 125, 231–240. [Google Scholar] [CrossRef]
- Baek, K.H. Production of reactive oxygen species by freezing stress and the protective roles of antioxidant enzymes in plants. J. Agr. Chem. Environ. 2012, 1, 34–40. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Zhang, J. Advances in the research on the AsA-GSH cycle in horticultural crops. Front. Agr. China 2010, 4, 84–90. [Google Scholar] [CrossRef]
- Raza, A.; Salehi, H.; Rahman, M.A.; Zahid, Z.; Madadkar Haghjou, M.; Najafi-Kakavand, S.; Charagh, S.; Osman, H.S.; Albaqami, M.; Zhuang, Y.; et al. Plant hormones and neurotransmitter interactions mediate antioxidant defenses under induced oxidative stress in plants. Front. Plant Sci. 2022, 13, 961872. [Google Scholar] [CrossRef]
- Garbero, M.; Pedranzani, H.; Zirulnik, F.; Molina, A.; Pérez-Chaca, M.V.; Vigliocco, A.; Abdala, G. Short-term cold stress in two cultivars of Digitaria eriantha: Effects on stress-related hormones and antioxidant defense system. Acta Physiol. Plant. 2011, 33, 497–507. [Google Scholar] [CrossRef]
- Amini, S.; Maali-Amiri, R.; Kazemi-Shahandashti, S.S.; López-Gómez, M.; Sadeghzadeh, B.; Sobhani-Najafabadi, A.; Kariman, K. Effect of cold stress on polyamine metabolism and antioxidant responses in chickpea. J. Plant Physiol. 2021, 258–259, 153387. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Adhikari, L.; Baral, R.; Paudel, D.; Min, D.; Makaju, S.O.; Poudel, H.P.; Acharya, J.P.; Missaoui, A.M. Cold stress in plants: Strategies to improve cold tolerance in forage species. Plant Stress 2022, 4, 100081. [Google Scholar] [CrossRef]
- Bhat, K.A.; Mahajan, R.; Pakhtoon, M.M.; Urwat, U.; Bashir, Z.; Shah, A.A.; Agrawal, A.; Bhat, B.; Sofi, P.A.; Masi, A.; et al. Low Temperature stress tolerance: An insight into the omics approaches for legume crops. Front. Plant Sci. 2022, 13, 888710. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, C.M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015, 82, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, X.; Bürger, M.; Chory, J.; Wang, X. The role of ethylene in plant temperature stress response. Trends Plant Sci. 2023, 28, 808–824. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef]
- Hu, Z.; Fan, J.; Chen, K.; Amombo, E.; Chen, L.; Fu, J. Effects of ethylene on photosystem II and antioxidant enzyme activity in Bermuda grass under low temperature. Photosynth. Res. 2016, 128, 59–72. [Google Scholar] [CrossRef]
- Mishra, S.; Chowdhary, A.A.; Bhau, B.S.; Srivastava, V. Hydrogen sulphide-mediated alleviation and its interplay with other signalling molecules during temperature stress. Plant Biol. 2022, 24, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Wang, W.; Hou, L.; Liu, X. Hydrogen sulfide is involved in the chilling stress response in Vitis vinifera L. Acta Soc. Bot. Pol. 2013, 82, 295–302. [Google Scholar] [CrossRef]
- Shi, H.; Ye, T.; Han, N.; Bian, H.; Liu, X.; Chan, Z. Hydrogen sulfide regulates abiotic stress tolerance and biotic stress resistance in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Bashir, M.A.; Silvestri, C.; Ahmad, T.; Hafiz, I.A.; Abbasi, N.A.; Manzoor, A.; Cristofori, V.; Rugini, E. Osmotin: A cationic protein leads to improve biotic and abiotic stress tolerance in plants. Plants 2020, 9, 992. [Google Scholar] [CrossRef]
- Hu, W.; Hu, G.; Han, B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009, 176, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Shashikumar, K.; Nus, J.L. Cultivar and winter cover effects on bermudagrass cold acclimation and crown moisture content. Crop Sci. 1993, 33, 813–817. [Google Scholar] [CrossRef]
- Munshaw, G.C.; Ervin, E.H.; Shang, C.; Askew, S.D.; Zhang, X.; Lemus, R.W. Influence of late-season iron, nitrogen, and seaweed extract on fall color retention and cold tolerance of four bermudagrass cultivars. Crop Sci. 2006, 46, 273–283. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Zhao, J.; Yu, G.; Huang, T.; Peng, Y.; Hassan, M.J. A bermudagrass variant exhibits strong tolerance to low temperature associated with enhanced sugar metabolism and cold-responsive pathways. Crop Sci. 2023, 63, 2553–2568. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Chen, S. Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, X.T.; Wang, F.; Shao, Y.L.; Zhang, A.M.; Chang, W. The effects of chilling stress on antioxidant enzymes activities and proline, malondialdehyde, soluble sugar contents in three Paphiopedilum Species. Russ. J. Plant Physiol. 2023, 70, 61. [Google Scholar] [CrossRef]
- Ihtisham, M.; Hasanuzzaman, M.; El-Sappah, A.H.; Zaman, F.; Khan, N.; Raza, A.; Sarraf, M.; Khan, S.; Abbas, M.; Hassan, M.J.; et al. Primary plant nutrients modulate the reactive oxygen species metabolism and mitigate the impact of cold stress in overseeded perennial ryegrass. Front. Plant Sci. 2023, 14, 1149823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ervin, E.H.; LaBranche, A.J. Metabolic defense responses of seeded bermudagrass during acclimation to freezing stress. Crop Sci. 2006, 46, 2598–2605. [Google Scholar] [CrossRef]
- Fan, J.; Ren, J.; Zhu, W.; Amombo, E.; Fu, J.; Chen, L. Antioxidant responses and gene expression in bermudagrass under cold stress. J. Amer. Soc. Hort. Sci. 2014, 139, 699–705. [Google Scholar] [CrossRef]
- Hu, Z.; Fan, J.; Xie, Y.; Amombo, E.; Liu, A.; Gitau, M.M.; Khaldun, A.B.M.; Chen, L.; Fu, J. Comparative photosynthetic and metabolic analyses reveal mechanism of improved cold stress tolerance in bermudagrass by exogenous melatonin. Plant Physiol. Biochem. 2016, 100, 94–104. [Google Scholar] [CrossRef]
- Fan, J.; Hu, Z.; Xie, Y.; Chan, Z.; Chen, K.; Amombo, E.; Chen, L.; Fu, J. Alleviation of cold damage to photosystem II and metabolisms by melatonin in Bermudagrass. Front. Plant Sci. 2015, 6, 925. [Google Scholar] [CrossRef]
- Jibiao; Fan; Ke; Chen; Erick; Amombo; Zhengrong; Hu; Liang; Jinmin, Physiological and molecular mechanism of nitric oxide (NO) involved in bermudagrass response to cold stress. PLoS ONE 2015, 10, e0132991.
- Shin, S.Y.; Kim, I.S.; Kim, Y.S.; Lee, H.; Yoon, H.S. Ectopic expression of Brassica rapa L. MDHAR increased tolerance to freezing stress by enhancing antioxidant systems of host plants. S. Afr. J. Bot. 2013, 88, 388–400. [Google Scholar] [CrossRef]
- Robison, J.D.; Yamasaki, Y.; Randall, S.K. The ethylene signaling pathway negatively impacts CBF/DREB-regulated cold response in soybean (Glycine max). Front. Plant Sci. 2019, 10, 121. [Google Scholar] [CrossRef]
- Pandey, A.K.; Gautam, A. Stress responsive gene regulation in relation to hydrogen sulfide in plants under abiotic stress. Physiol. Plant. 2020, 168, 511–525. [Google Scholar] [CrossRef]
- Chao, Q.; Rothenberg, M.; Solano, R.; Roman, G.; Terzaghi, W.; Ecker, J.R. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 1997, 89, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Develop. 1998, 12, 3703–3714. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, H.; Hutchison, C.E.; Laskey, J.; Kieber, J.J. Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 2003, 33, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Jin, Z.; Liu, D.; Yang, G.; Pei, Y. Hydrogen sulfide alleviates the cold stress through MPK4 in Arabidopsis thaliana. Plant Physiol. Biochem. 2017, 120, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Feng, L.; Zhang, S.; Li, L.; Guan, R.; Long, W.; Xian, Z.; Zhang, J.; Shen, W. Ammonia borane positively regulates cold tolerance in Brassica napus via hydrogen sulfide signaling. BMC Plant Biol. 2022, 22, 585. [Google Scholar] [CrossRef]
- Aghdam, M.S.; Mahmoudi, R.; Razavi, F.; Rabiei, V.; Soleimani, A. Hydrogen sulfide treatment confers chilling tolerance in hawthorn fruit during cold storage by triggering endogenous H2S accumulation, enhancing antioxidant enzymes activity and promoting phenols accumulation. Sci. Hort. 2018, 238, 264–271. [Google Scholar] [CrossRef]
- Caubrière, D.; Moseler, A.; Rouhier, N.; Couturier, J. Diversity and roles of cysteine desulfurases in photosynthetic organisms. J. Exp. Bot. 2023, 74, 3345–3360. [Google Scholar] [CrossRef]
- Hu, Z.; Liu, A.; Bi, A.; Amombo, E.; Gitau, M.M.; Huang, X.; Chen, L.; Fu, J. Identification of differentially expressed proteins in bermudagrass response to cold stress in the presence of ethylene. Environ. Exp. Bot. 2017, 139, 67–78. [Google Scholar] [CrossRef]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. BBA-Gene Regul. Mech. 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Rana, R.M.; Dong, S.; Tang, H.; Ahmad, F.; Zhang, H. Functional analysis of OsHSBP1 and OsHSBP2 revealed their involvement in the heat shock response in rice (Oryza sativa L.). J. Exp. Bot. 2012, 63, 6003–6016. [Google Scholar] [CrossRef]
- Hsu, S.F.; Lai, H.C.; Jinn, T.L. Cytosol-localized heat shock factor-binding protein, AtHSBP, functions as a negative regulator of heat shock response by translocation to the nucleus and is required for seed development in arabidopsis. Plant Physiol. 2010, 153, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.; Dong, D.; Li, Y.; Wang, X.; Guo, L.; Liu, L.; Dong, X.; Li, X.; Yuan, X.; Ren, S.; et al. Heat shock-induced cold acclimation in cucumber through CsHSFA1d-activated JA biosynthesis and signaling. Plant J. 2022, 111, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Gan, L.; Zhang, J.; Shen, Y.; Qian, J.; Han, M.; Zhang, C.; Fan, J.; Sun, S.; Yan, X. A regulatory network of heat shock modules-photosynthesis-redox systems in response to cold stress across a latitudinal gradient in bermudagrass. Front. Plant Sci. 2021, 12, 751901. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, Y.; Tao, X.; Wang, J.Z.; Cheng, H.Y.; Yang, H.; Ma, X.R. Heat shock factor genes of tall fescue and perennial ryegrass in response to temperature stress by RNA-Seq analysis. Front. Plant Sci. 2016, 6, 1226. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Jiang, T.; Zhang, C.; Li, X.; Wang, C.; Zhang, Y.; Li, T.; Dirk, L.M.A.; Downie, A.B.; Zhao, T. Maize HSFA2 and HSBP2 antagonistically modulate raffinose biosynthesis and heat tolerance in Arabidopsis. Plant J. 2019, 100, 128–142. [Google Scholar] [CrossRef]
- Hakim; Ullah, A.; Hussain, A.; Shaban, M.; Khan, A.H.; Alariqi, M.; Gul, S.; Jun, Z.; Lin, S.; Li, J.; et al. Osmotin: A plant defense tool against biotic and abiotic stresses. Plant Physiol. Biochem. 2018, 123, 149–159. [Google Scholar] [CrossRef]
- D’Angeli, S.; Altamura, M.M. Osmotin induces cold protection in olive trees by affecting programmed cell death and cytoskeleton organization. Planta 2007, 225, 1147–1163. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, T.H.H.; Li, P.H. Expression of an ABA-responsive osmotin-like gene during the induction of freezing tolerance in Solanum commersonii. Plant Mol. Biol. 1993, 21, 729–735. [Google Scholar] [CrossRef]
- Patade, V.Y.; Khatri, D.; Kumari, M.; Grover, A.; Mohan Gupta, S.; Ahmed, Z. Cold tolerance in Osmotin transgenic tomato (Solanum lycopersicum L.) is associated with modulation in transcript abundance of stress responsive genes. SpringerPlus 2013, 2, 117. [Google Scholar] [CrossRef]
- Patade, V.Y.; Meena, H.; Grover, A.; Gupta, S.M.; Nasim, M. Containment evaluation, cold tolerance and toxicity analysis in Osmotin transgenic tomato (Solanum lycopersicum L. cv. Pusa Ruby). 3 Biotech 2018, 8, 410. [Google Scholar] [CrossRef]
- Le, T.T.T.; Williams, B.; Mundree, S.G. An osmotin from the resurrection plant Tripogon loliiformis (TlOsm) confers tolerance to multiple abiotic stresses in transgenic rice. Physiol. Plant. 2018, 162, 13–34. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Calif. Agr. Exp. Sta. Circ. 1950, 347, 32. [Google Scholar]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Zeng, W.; Hassan, M.J.; Kang, D.; Peng, Y.; Li, Z. Photosynthetic maintenance and heat shock protein accumulation relating to γ-aminobutyric acid (GABA)-regulated heat tolerance in creeping bentgrass (Agrostis stolonifera). S. Afr. J. Bot. 2021, 141, 405–413. [Google Scholar] [CrossRef]
- Elstner, E.F.; Heupel, A. Inhibition of nitrite formation from hydroxylammoniumchloride: A simple assay for superoxide dismutase. Anal. Biochem 1976, 70, 616–620. [Google Scholar] [CrossRef]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: Protective role of exogenous polyamines. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Blum, A.; Ebercon, A. Cell membrane stability as a measure of drought and heat tolerance in wheat. Crop Sci. 1981, 21, 43–47. [Google Scholar] [CrossRef]
- Dhindsa, R.; Plumb-Dhindsa, P.; Thorpe, T. Leaf senescence: Correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Chance, B.; Maehly, A.C. Assay of catalases and peroxidases. Meth. Enzymol. 1955, 1, 764–775. [Google Scholar]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Cakmak, I.; Strbac, D.; Marschner, H. Activities of hydrogen peroxide-scavenging enzymes in germinating wheat seeds. J. Exp. Bot. 1993, 44, 127–132. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Huang, C.; Han, L. Differential Regulations of Antioxidant Metabolism and Cold-Responsive Genes in Three Bermudagrass Genotypes under Chilling and Freezing Stress. Int. J. Mol. Sci. 2023, 24, 14070. https://doi.org/10.3390/ijms241814070
Li Z, Huang C, Han L. Differential Regulations of Antioxidant Metabolism and Cold-Responsive Genes in Three Bermudagrass Genotypes under Chilling and Freezing Stress. International Journal of Molecular Sciences. 2023; 24(18):14070. https://doi.org/10.3390/ijms241814070
Chicago/Turabian StyleLi, Zhou, Cheng Huang, and Liebao Han. 2023. "Differential Regulations of Antioxidant Metabolism and Cold-Responsive Genes in Three Bermudagrass Genotypes under Chilling and Freezing Stress" International Journal of Molecular Sciences 24, no. 18: 14070. https://doi.org/10.3390/ijms241814070
APA StyleLi, Z., Huang, C., & Han, L. (2023). Differential Regulations of Antioxidant Metabolism and Cold-Responsive Genes in Three Bermudagrass Genotypes under Chilling and Freezing Stress. International Journal of Molecular Sciences, 24(18), 14070. https://doi.org/10.3390/ijms241814070






