Cancer Stem Cell Markers—Clinical Relevance and Prognostic Value in High-Grade Serous Ovarian Cancer (HGSOC) Based on The Cancer Genome Atlas Analysis
Abstract
1. Introduction
2. Results
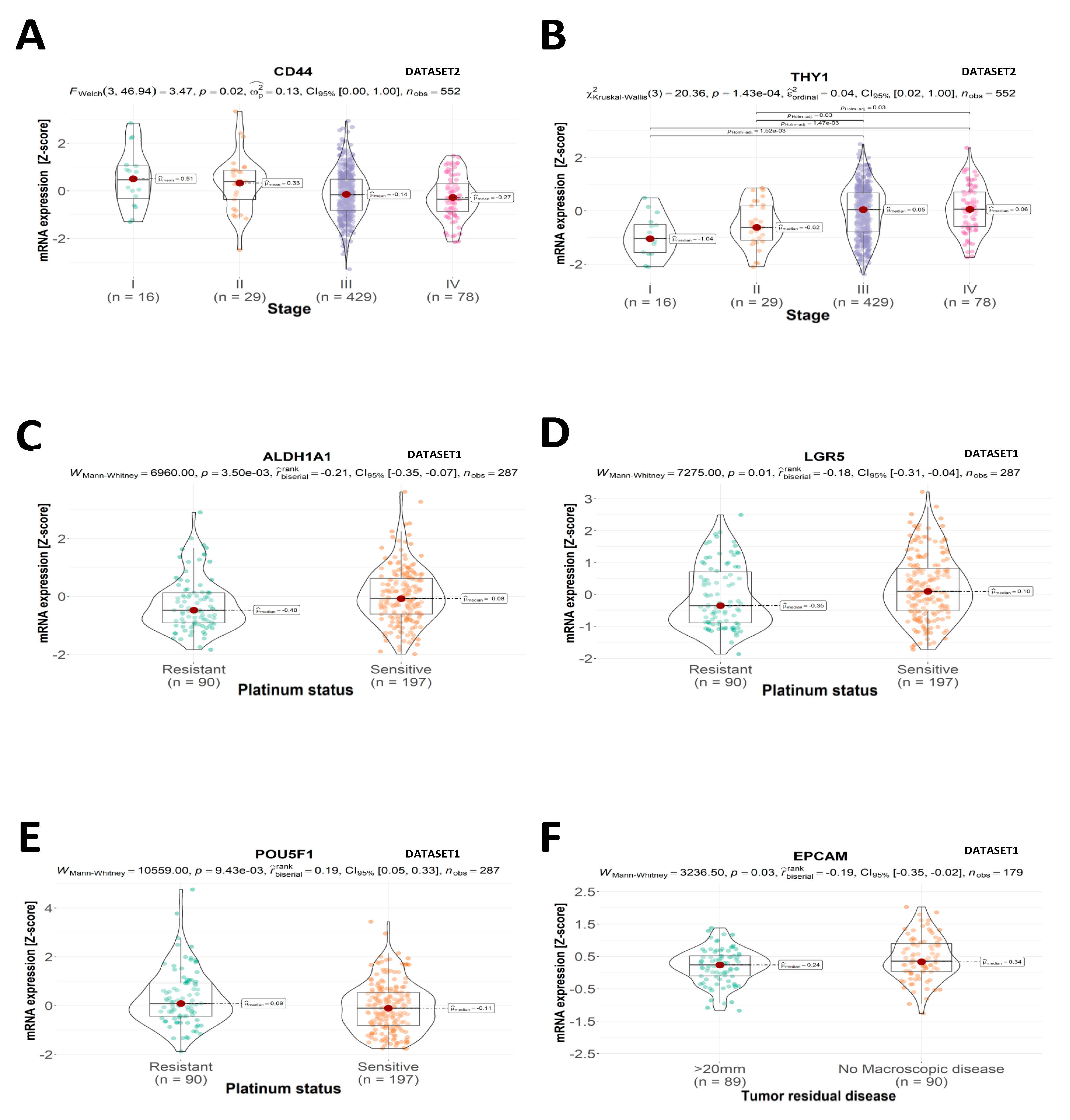
2.1. Clinical and Pathological Features
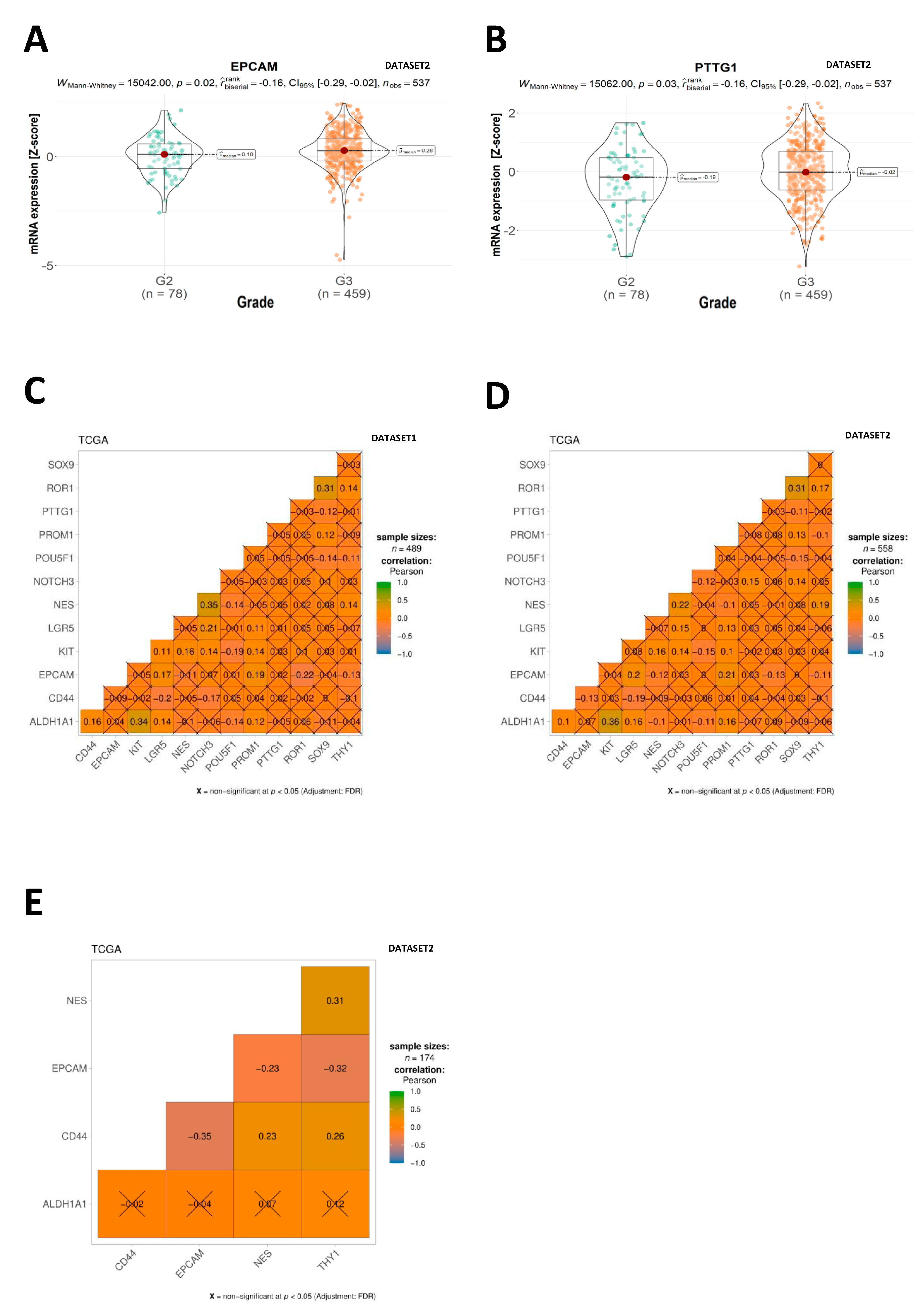
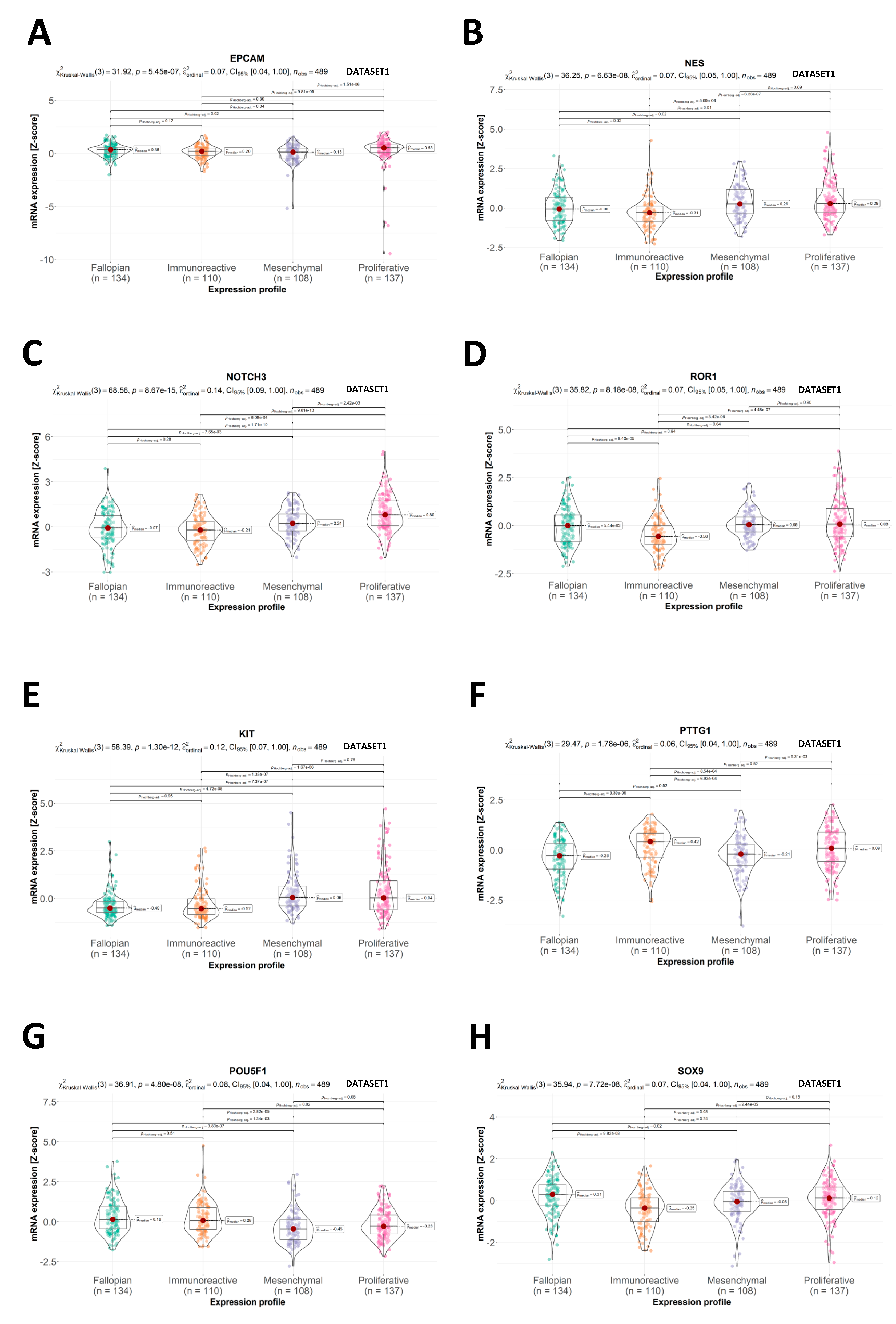
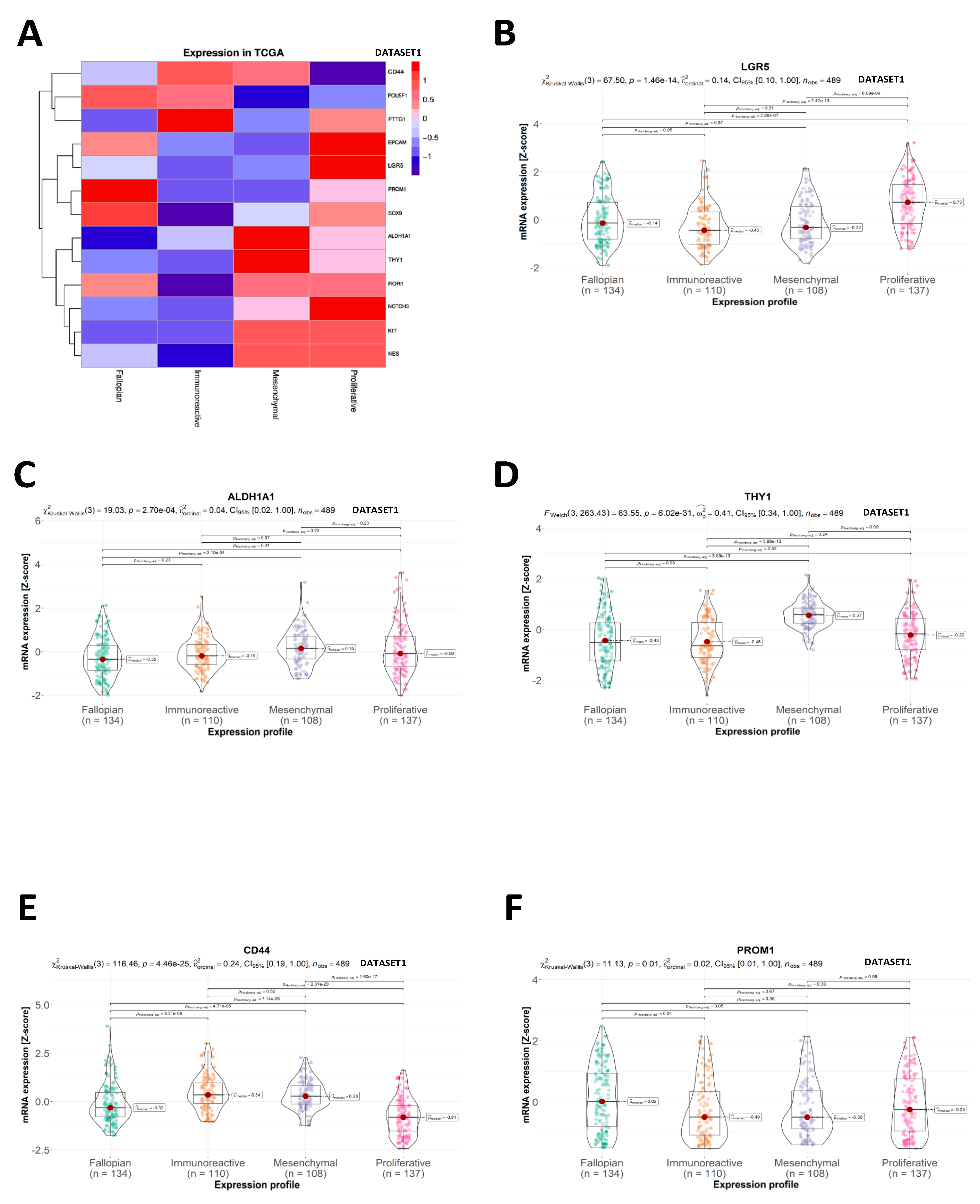
2.2. CSC Expression Profiles
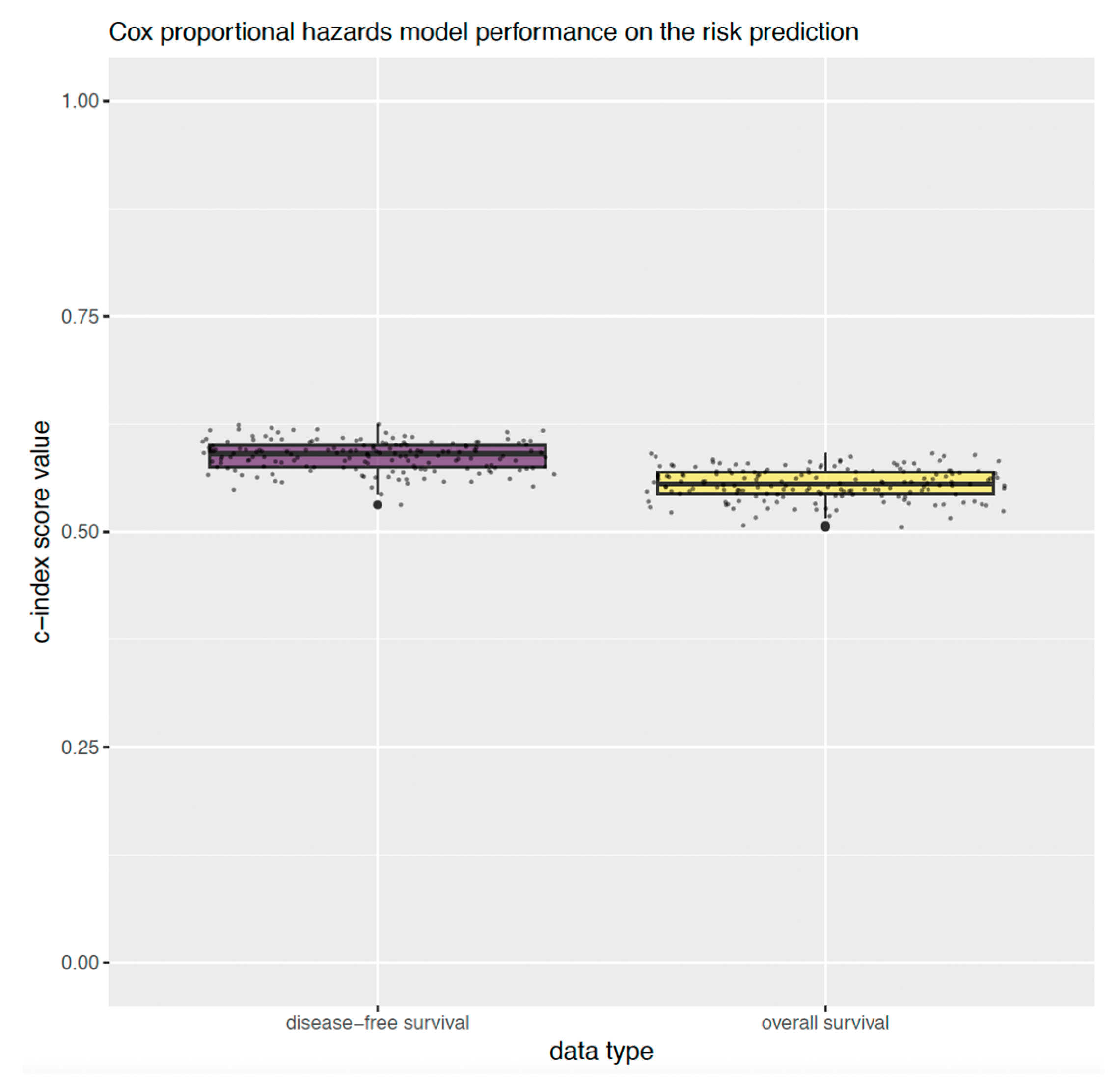
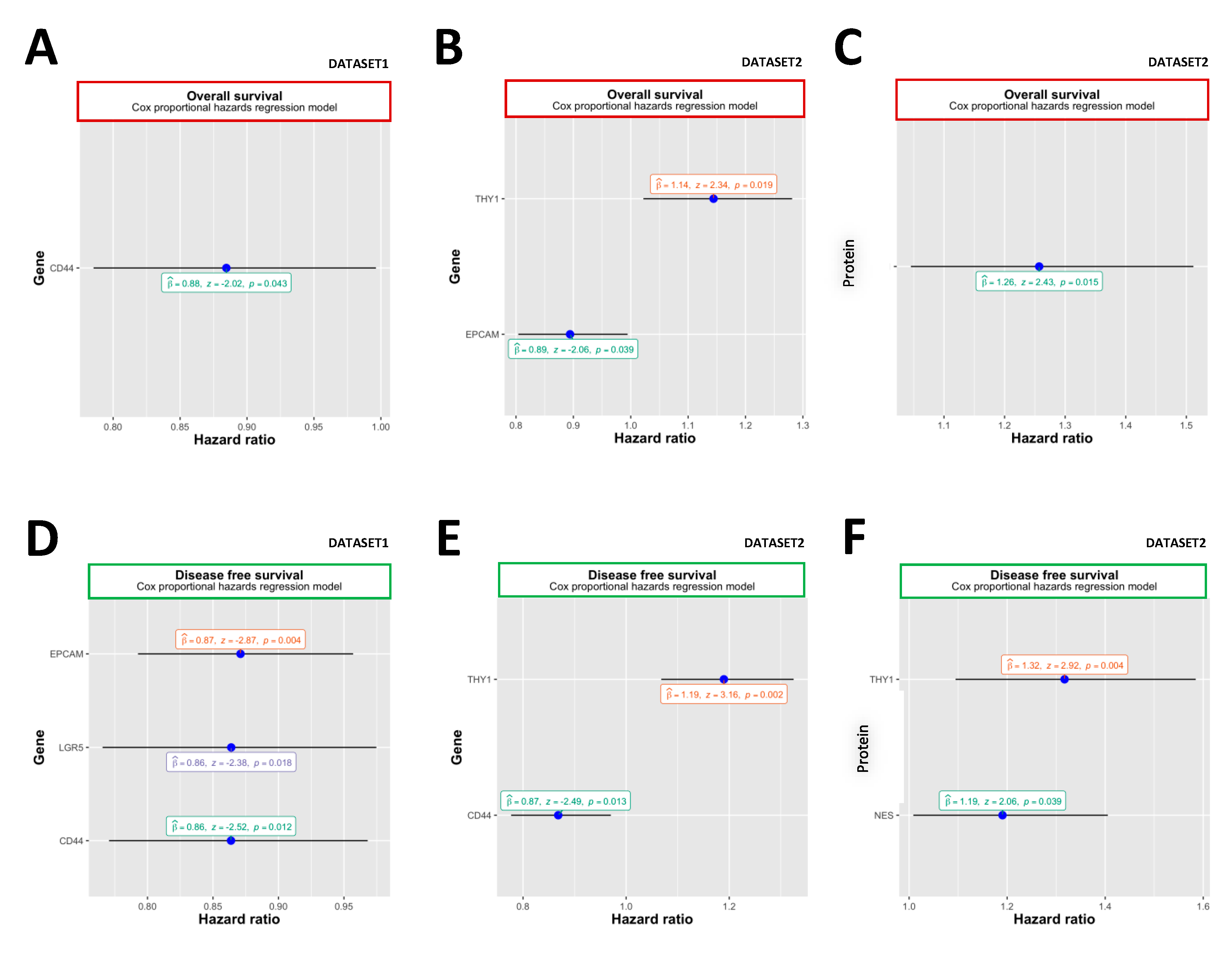
2.3. Overall Survival
2.4. Disease-Free Survival
2.5. Correlation Analysis
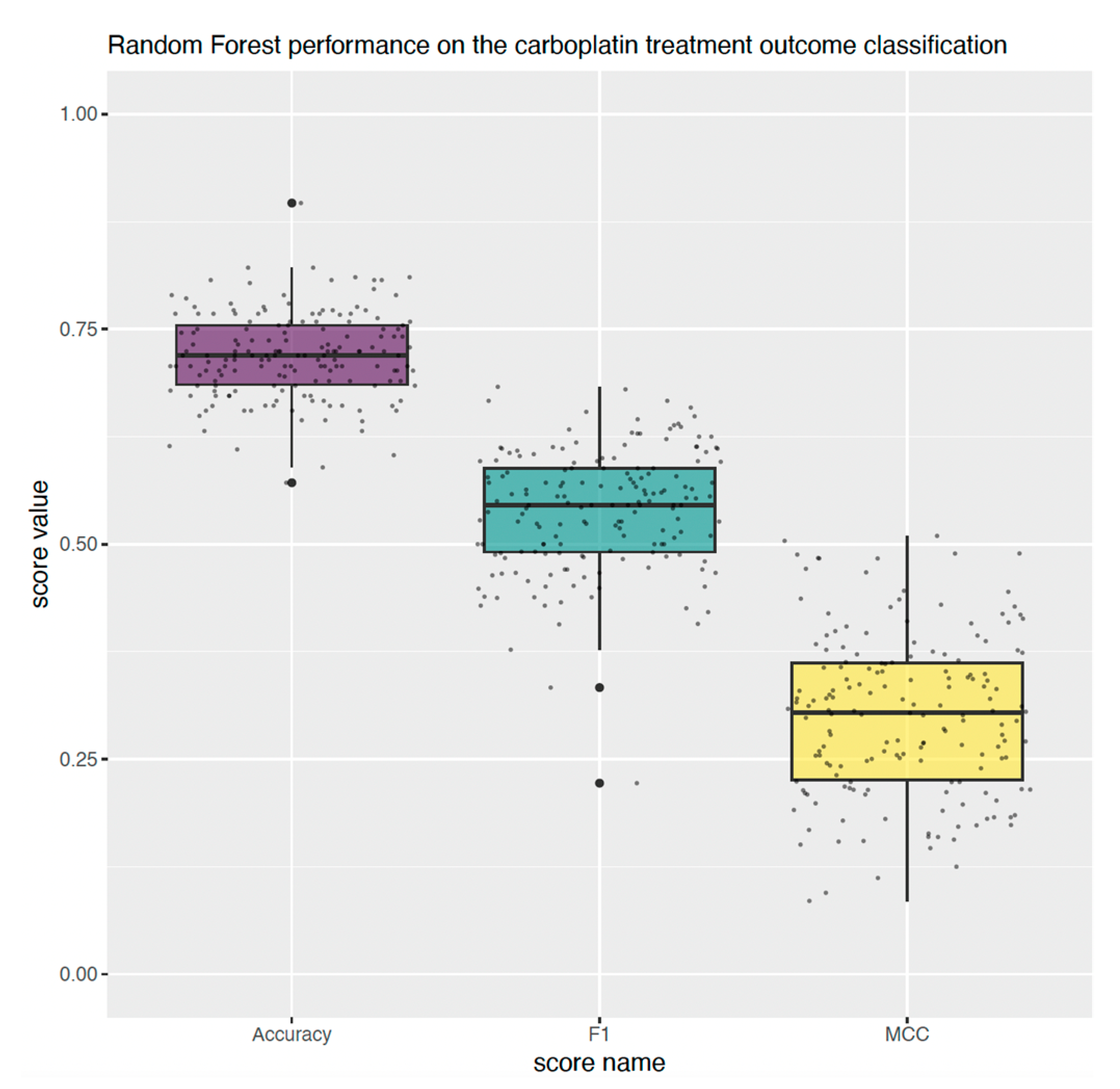
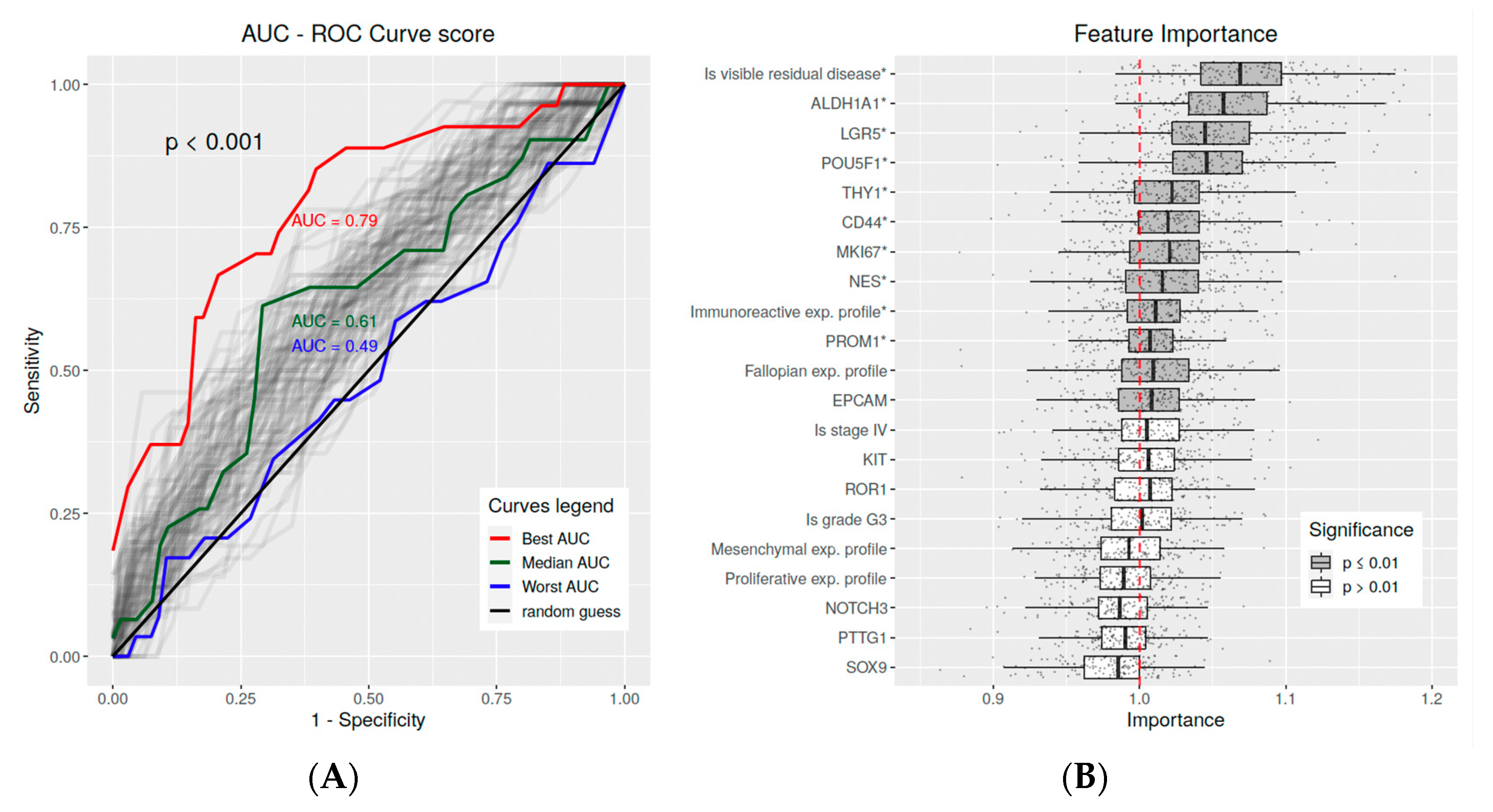
2.6. Multivariate Predictive Analysis
3. Discussion
4. Materials and Methods
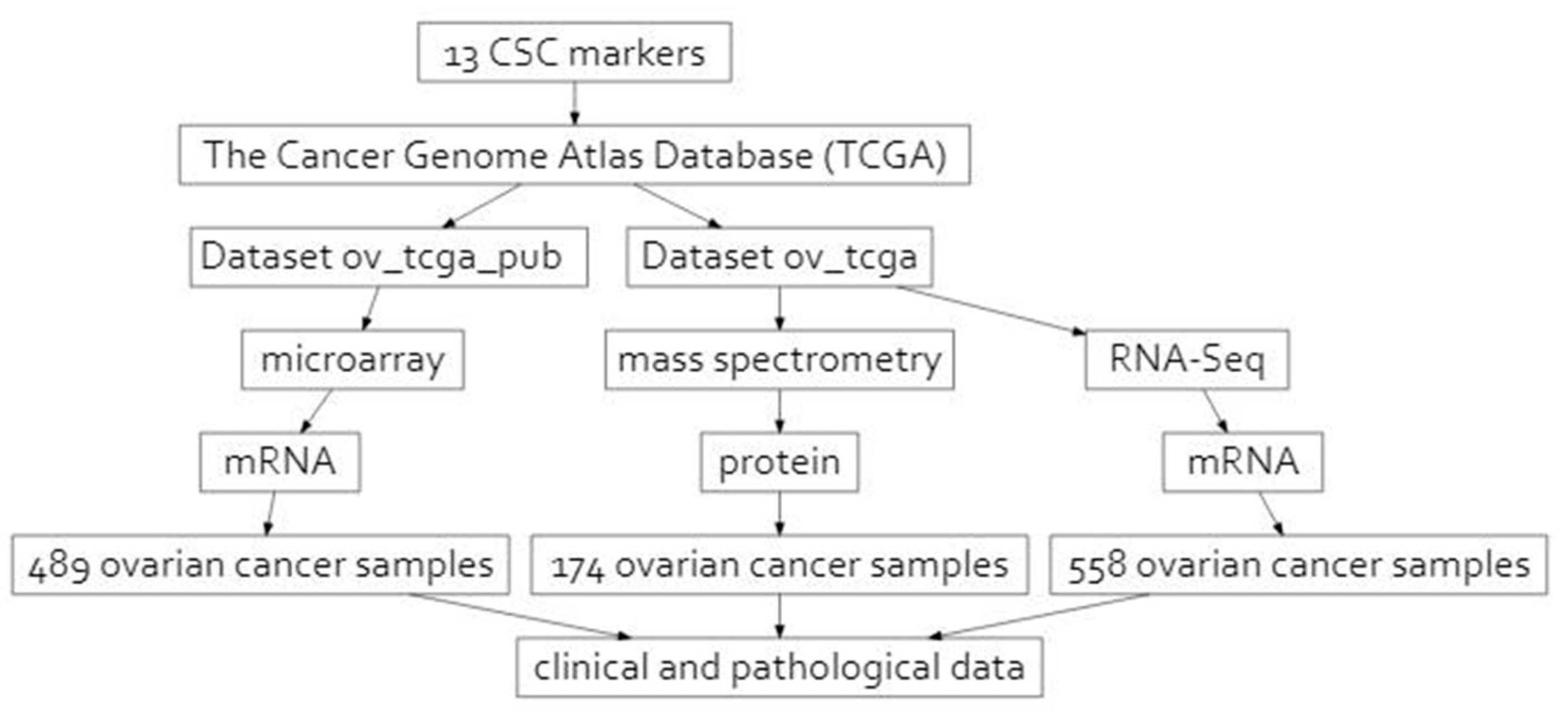
4.1. Data Retrieval
4.2. Clinical and Pathological Features
4.3. Data Preprocessing
4.4. Statistics
4.5. Univariate Analysis of Overall and Disease-Free Survival
4.6. Predictive Analysis of Carboplatin Treatment Outcome
4.7. Multivariate Analysis of Overall and Disease-Free Survival
4.8. Feature Importance Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Zhan, Q.; Wang, C.; Ngai, S. Ovarian Cancer Stem Cells: A New Target for Cancer Therapy. BioMed Res. Int. 2013, 2013, 916819. [Google Scholar]
- Tomao, F.; Papa, A.; Martina, S.; Rossi, L.; Russo, G.L.; Panici, P.B.; Ciabatta, F.R.; Tomao, S. Investigating Molecular Profiles of Ovarian Cancer: An Update on Cancer Stem Cells. J. Cancer 2014, 5, 301–310. [Google Scholar]
- Keyvani, V.; Farshchian, M.; Esmaeili, S.A.; Yari, H.; Moghbeli, M.; Nezhad, S.R.K.; Abbaszadegan, M.R. Ovarian Cancer Stem Cells and Targeted Therapy. J. Ovarian Res. 2019, 12, 120. [Google Scholar] [PubMed]
- Gupta, P.B.; Chaffer, C.L.; Weinberg, R.A. Cancer Stem Cells: Mirage or Reality? Nat. Med. 2009, 15, 1010–1012. [Google Scholar]
- Takahashi, A.; Hong, L.; Chefetz, I. Review Open Access Cancer Drug Resistance How to Win the Ovarian Cancer Stem Cell Battle: Destroying the Roots. Cancer Drug Resist. 2020, 3, 1021–1054. [Google Scholar]
- Lupia, M.; Cavallaro, U. Ovarian Cancer Stem Cells: Still an Elusive Entity? Mol. Cancer 2017, 16, 64. [Google Scholar]
- Markowska, A.; Sajdak, S.; Huczyński, A.; Rehlis, S.; Markowska, J. Ovarian Cancer Stem Cells: A Target for Oncological Therapy. Adv. Clin. Exp. Med. 2018, 27, 1017–1020. [Google Scholar] [PubMed]
- Parte, S.C.; Batra, S.K.; Kakar, S.S. Characterization of Stem Cell and Cancer Stem Cell Populations in Ovary and Ovarian Tumors. J. Ovarian Res. 2018, 11, 69. [Google Scholar] [PubMed]
- The Cancer Genome Atlas Research Network. Integrated Genomic Analyses of Ovarian Carcinoma. Nature 2011, 474, 609–615, Erratum in Nature 2012, 490, 292. [Google Scholar]
- Verhaak, R.G.W.; Tamayo, P.; Yang, J.Y.; Hubbard, D.; Zhang, H.; Creighton, C.J.; Fereday, S.; Lawrence, M.; Carter, S.L.; Mermel, C.H.; et al. Prognostically Relevant Gene Signatures of High-Grade Serous Ovarian Carcinoma. J. Clin. Investig. 2013, 123, 517–525. [Google Scholar]
- Riester, M.; Wei, W.; Waldron, L.; Culhane, A.C.; Trippa, L.; Oliva, E.; Kim, S.H.; Michor, F.; Huttenhower, C.; Parmigiani, G.; et al. Risk Prediction for Late-Stage Ovarian Cancer by Meta-Analysis of 1525 Patient Samples. J. Natl. Cancer Inst. 2014, 106, dju048. [Google Scholar] [PubMed]
- Waldron, L.; Riester, M.; Birrer, M. Molecular Subtypes of High-Grade Serous Ovarian Cancer: The Holy Grail? J. Natl. Cancer Inst. 2014, 106, dju297. [Google Scholar] [PubMed]
- Tan, T.Z.; Miow, Q.H.; Huang, R.Y.J.; Wong, M.K.; Ye, J.; Lau, J.A.; Wu, M.C.; Bin Abdul Hadi, L.H.; Soong, R.; Choolani, M.; et al. Functional Genomics Identifies Five Distinct Molecular Subtypes with Clinical Relevance and Pathways for Growth Control in Epithelial Ovarian Cancer. EMBO Mol. Med. 2013, 5, 1051–1066. [Google Scholar] [PubMed]
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel Molecular Subtypes of Serous and Endometrioid Ovarian Cancer Linked to Clinical Outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar]
- Cioffi, M.; Dalterio, C.; Camerlingo, R.; Tirino, V.; Consales, C.; Riccio, A.; Ieranò, C.; Cecere, S.C.; Losito, N.S.; Greggi, S.; et al. Identification of a Distinct Population of CD133+ CXCR4+ Cancer Stem Cells in Ovarian Cancer. Sci. Rep. 2015, 5, 10357. [Google Scholar]
- Napoli, J.L.; Boerman, M.H.E.M.; Chai, X.; Zhai, Y.; Fiorella, P.D. Enzymes and Binding Proteins Affecting Retinoic Acid Concentrations. J. Steroid Biochem. Mol. Biol. 1995, 53, 497–502. [Google Scholar]
- Martincuks, A.; Li, P.C.; Zhao, Q.; Zhang, C.; Li, Y.J.; Yu, H.; Rodriguez-Rodriguez, L. CD44 in Ovarian Cancer Progression and Therapy Resistance—A Critical Role for STAT3. Front. Oncol. 2020, 10, 2551. [Google Scholar]
- Mizrak, D.; Brittan, M.; Alison, M.R. CD 133: Molecule of the Moment. J. Pathol. 2008, 214, 3–9. [Google Scholar]
- Roy, L.; Bobbs, A.; Sattler, R.; Kurkewich, J.L.; Dausinas, P.B.; Nallathamby, P.; Dahl, K.D.C. CD133 Promotes Adhesion to the Ovarian Cancer Metastatic Niche. Cancer Growth Metastasis 2018, 11, 117906441876788. [Google Scholar] [CrossRef]
- Jiang, J.; Chen, Y.; Zhang, M.; Zhou, H.; Wu, H. Relationship between CD177 and the Vasculogenic Mimicry, Clinicopathological Parameters, and Prognosis of Epithelial Ovarian Cancer. Ann. Palliat. Med. 2020, 9, 3985–3992. [Google Scholar]
- Robinson, M.; Gilbert, S.F.; Waters, J.A.; Lujano-olazaba, O.; Lara, J.; Alexander, L.J.; Green, S.E.; Burkeen, G.A.; Patrus, O.; Sarwar, Z.; et al. Characterization of SOX2, OCT4 and NANOG in Ovarian Cancer Tumor-Initiating Cells. Cancers 2021, 13, 262. [Google Scholar] [PubMed]
- Price, J.C.; Azizi, E.; Naiche, L.A.; Parvani, J.G.; Shukla, P.; Kim, S.; Slack-Davis, J.K.; Pe’er, D.; Kitajewski, J.K. Notch3 Signaling Promotes Tumor Cell Adhesion and Progression in a Murine Epithelial Ovarian Cancer Model. PLoS ONE 2020, 15, e0233962. [Google Scholar]
- Aguilar-Medina, M.; Avendaño-Félix, M.; Lizárraga-Verdugo, E.; Bermúdez, M.; Romero-Quintana, J.G.; Ramos-Payan, R.; Ruíz-García, E.; López-Camarillo, C. SOX9 Stem-Cell Factor: Clinical and Functional Relevance in Cancer. J. Oncol. 2019, 2019, 6754040. [Google Scholar] [PubMed]
- Abeysinghe, H.; Cao, Q.; Xu, J.; Pollock, S.; Veyberman, Y.; Guckert, N.; Keng, P.; Wang, N. THY1 Expression Is Associated with Tumor Suppression of Human Ovarian Cancer. Cancer Genet. Cytogenet. 2003, 143, 125–132. [Google Scholar]
- Abeysinghe, H.; Pollock, S.; Guckert, N.; Veyberman, Y.; Keng, P.; Halterman, M.; Federoff, H.; Rosenblatt, J.; Wang, N. The Role of the THY1 Gene in Human Ovarian Cancer Suppression Based on Transfection Studies. Cancer Genet. Cytogenet. 2004, 149, 1–10. [Google Scholar]
- Connor, E.V.; Saygin, C.; Braley, C.; Wiechert, A.C.; Karunanithi, S.; Crean-Tate, K.; Abdul-Karim, F.W.; Michener, C.M.; Rose, P.G.; Lathia, J.D.; et al. Thy-1 Predicts Poor Prognosis and Is Associated with Self-Renewal in Ovarian Cancer. J. Ovarian Res. 2019, 12, 112. [Google Scholar]
- Tayama, S.; Motohara, T.; Narantuya, D.; Li, C.; Fujimoto, K.; Sakaguchi, I.; Tashiro, H.; Saya, H.; Nagano, O.; Katabuchi, H.; et al. The Impact of EpCAM Expression on Response to Chemotherapy and Clinical Outcomes in Patients with Epithelial Ovarian Cancer. Oncotarget 2017, 8, 44312–44325. [Google Scholar]
- Liu, W.; Zhang, J.; Gan, X.; Shen, F.; Yang, X.; Du, N.; Xia, D.; Liu, L.; Qiao, L.; Pan, J.; et al. LGR5 Promotes Epithelial Ovarian Cancer Proliferation, Metastasis, and Epithelial–Mesenchymal Transition through the Notch1 Signaling Pathway. Cancer Med. 2018, 7, 3132. [Google Scholar]
- Parte, S.; Virant-Klun, I.; Patankar, M.; Batra, S.K.; Straughn, A.; Kakar, S.S. PTTG1: A Unique Regulator of Stem/Cancer Stem Cells in the Ovary and Ovarian Cancer. Stem Cell Rev. Rep. 2019, 15, 866–879. [Google Scholar]
- Henry, C.; Llamosas, E.; Knipprath-Meszaros, A.; Schoetzau, A.; Obermann, E.; Fuenfschilling, M.; Caduff, R.; Fink, D.; Hacker, N.; Ward, R.; et al. Targeting the ROR1 and ROR2 Receptors in Epithelial Ovarian Cancer Inhibits Cell Migration and Invasion. Oncotarget 2015, 6, 40310–40326. [Google Scholar]
- Osman, W.M.; Shash, L.S.; Ahmed, N.S. Emerging Role of Nestin as an Angiogenesis and Cancer Stem Cell Marker in Epithelial Ovarian Cancer: Immunohistochemical Study. Appl. Immunohistochem. Mol. Morphol. 2017, 25, 571–580. [Google Scholar] [PubMed]
- Yoon, H.J.; Oh, Y.L.; Ko, E.J.; Kang, A.; Eo, W.K.; Kim, K.H.; Lee, J.Y.; Kim, A.; Chun, S.; Kim, H.; et al. Effects of Thymosin Β4-Derived Peptides on Migration and Invasion of Ovarian Cancer Cells. Genes Genom. 2021, 43, 987–993. [Google Scholar]
- Abd El-Fattah, G.A.; Ibrahim, E.; Nasif, S.N. Significance of Tumoral and Stromal ALDH 1A1 Expression in Breast Invasive Duct Carcinoma in Egyptian Female Patients. Egypt. J. Pathol. 2019, 39, 151–158. [Google Scholar]
- Woopen, H.; Pietzner, K.; Richter, R.; Fotopoulou, C.; Joens, T.; Braicu, E.I.; Mellstedt, H.; Mahner, S.; Lindhofer, H.; Darb-Esfahani, S.; et al. Overexpression of the Epithelial Cell Adhesion Molecule Is Associated with a More Favorable Prognosis and Response to Platinum-Based Chemotherapy in Ovarian Cancer. J. Gynecol. Oncol. 2014, 25, 221–228. [Google Scholar] [PubMed]
- EPCAM Protein Expression Summary—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000119888-EPCAM (accessed on 8 June 2023).
- Sauzay, C.; Voutetakis, K.; Chatziioannou, A.A.; Chevet, E.; Avril, T. CD90/Thy-1, a Cancer-Associated Cell Surface Signaling Molecule. Front. Cell Dev. Biol. 2019, 7, 66. [Google Scholar]
- Zeng, L.; Peng, Z.; Duan, Z. Expression of THY1 Gene in Epithelial Ovarian Cancer. Chin. J. Oncol. 2009, 31, 118–120. [Google Scholar]
- Abeysinghe, H.R.; Li, Q.L.; Guckert, N.L.; Reeder, J.; Wang, N. THY-1 Induction Is Associated with up-Regulation of Fibronectin and Thrombospondin-1 in Human Ovarian Cancer. Cancer Genet. Cytogenet. 2005, 161, 151–158. [Google Scholar]
- Haeryfar, S.M.M.; Hoskin, D.W. Thy-1: More than a Mouse Pan-T Cell Marker. J. Immunol. 2004, 173, 3581–3588. [Google Scholar]
- Xie, W.; Yu, J.; Yin, Y.; Zhang, X.; Zheng, X.; Wang, X. OCT4 Induces EMT and Promotes Ovarian Cancer Progression by Regulating the PI3K/AKT/MTOR Pathway. Front. Oncol. 2022, 12, 876257. [Google Scholar]
- Ruan, Z.; Yang, X.; Cheng, W. OCT4 Accelerates Tumorigenesis through Activating JAK/STAT Signaling in Ovarian Cancer Side Population Cells. Cancer Manag. Res. 2019, 11, 389. [Google Scholar]
- Kim, H.; Lee, D.H.; Park, E.; Myung, J.K.; Park, J.H.; Kim, D.I.; Kim, S.I.; Lee, M.; Kim, Y.; Park, C.M.; et al. Differential Epithelial and Stromal LGR5 Expression in Ovarian Carcinogenesis. Sci. Rep. 2022, 12, 11200. [Google Scholar] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [PubMed]
- Van Den Goorbergh, R.; Van Smeden, M.; Timmerman, D.; Van Calster, B. The Harm of Class Imbalance Corrections for Risk Prediction Models: Illustration and Simulation Using Logistic Regression. J. Am. Med. Inf. Assoc. 2022, 29, 1525–1534. [Google Scholar]
- Patil, I. Visualizations with Statistical Details: The “ggstatsplot” Approach. J. Open Source Softw. 2021, 6, 3167. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by Random Forest. R News 2002, 2, 18–22. [Google Scholar]
- Package “pracma” Title Practical Numerical Math Functions Depends R (≥3.1.0). 2022. Available online: https://cran.r-project.org/web/packages/pracma/index.html (accessed on 8 June 2023).
- Marquardt, D.W.; Snee, R.D. Ridge Regression in Practice. Am. Stat. 1975, 29, 3–20. [Google Scholar]
- Simon, N.; Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. J. Stat. Softw. 2011, 39, 1–13. [Google Scholar]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar]
- Drawing Survival Curves Using “ggplot2” [R Package Survminer Version 0.4.9]. 2021. Available online: https://rpkgs.datanovia.com/survminer/ (accessed on 8 June 2023).
- Fisher, A.; Rudin, C.; Dominici, F. All Models Are Wrong, but Many Are Useful: Learning a Variable’s Importance by Studying an Entire Class of Prediction Models Simultaneously. J. Mach. Learn. Res. 2019, 20, 1–81. [Google Scholar]
- Molnar, C.; Casalicchio, G.; Bischl, B. Iml: An R Package for Interpretable Machine Learning. J. Open Source Softw. 2018, 3, 786. [Google Scholar]
- Stauder, R.; Günthert, U. CD44 Isoforms. Impact on Lymphocyte Activation and Differentiation. Immunologist 1995, 3, 78–83. [Google Scholar]
- CD44 Expression Indicates Favorable Prognosis in Epithelial Ovarian Cancer|Clinical Cancer Research|American Association for Cancer Research. Available online: https://aacrjournals.org/clincancerres/article/9/14/5318/202149/CD44-Expression-Indicates-Favorable-Prognosis-in (accessed on 8 June 2023).
- Sosulski, A.; Horn, H.; Zhang, L.; Coletti, C.; Vathipadiekal, V.; Castro, C.M.; Birrer, M.J.; Nagano, O.; Saya, H.; Lage, K.; et al. CD44 Splice Variant V8-10 as a Marker of Serous Ovarian Cancer Prognosis. PLoS ONE 2016, 11, e0156595. [Google Scholar]
- Mao, M.; Zheng, X.; Jin, B.; Zhang, F.; Zhu, L.; Cui, L. Effects of CD44 and E-Cadherin Overexpression on the Proliferation, Adhesion and Invasion of Ovarian Cancer Cells. Exp. Ther. Med. 2017, 14, 5557. [Google Scholar] [PubMed]
- Zhou, J.; Du, Y.; Lu, Y.; Luan, B.; Xu, C.; Yu, Y.; Zhao, H. CD44 Expression Predicts Prognosis of Ovarian Cancer Patients through Promoting Epithelial-Mesenchymal Transition (EMT) by Regulating Snail, ZEB1, and Caveolin-1. Front. Oncol. 2019, 9, 802. [Google Scholar]
- Hu, Y.; Taylor-Harding, B.; Raz, Y.; Haro, M.; Recouvreux, M.S.; Taylan, E.; Lester, J.; Millstein, J.; Walts, A.E.; Karlan, B.Y.; et al. Are Epithelial Ovarian Cancers of the Mesenchymal Subtype Actually Intraperitoneal Metastases to the Ovary? Front. Cell Dev. Biol. 2020, 8, 647. [Google Scholar]
- Izycka, N.; Rucinski, M.; Andrzejewska, M.; Szubert, S.; Nowak-Markwitz, E.; Sterzynska, K. The Prognostic Value of Cancer Stem Cell Markers (CSCs) Expression-ALDH1A1, CD133, CD44-For Survival and Long-Term Follow-Up of Ovarian Cancer Patients. Int. J. Mol. Sci. 2023, 24, 2400. [Google Scholar] [PubMed]
- Kaipio, K.; Chen, P.; Roering, P.; Huhtinen, K.; Mikkonen, P.; Östling, P.; Lehtinen, L.; Mansuri, N.; Korpela, T.; Potdar, S.; et al. ALDH1A1-Related Stemness in High-Grade Serous Ovarian Cancer Is a Negative Prognostic Indicator but Potentially Targetable by EGFR/MTOR-PI3K/Aurora Kinase Inhibitors. J. Pathol. 2020, 250, 159–169. [Google Scholar] [PubMed]
- Ruscito, I.; Darb-Esfahani, S.; Kulbe, H.; Bellati, F.; Zizzari, I.G.; Rahimi Koshkaki, H.; Napoletano, C.; Caserta, D.; Rughetti, A.; Kessler, M.; et al. The Prognostic Impact of Cancer Stem-like Cell Biomarker Aldehyde Dehydrogenase-1 (ALDH1) in Ovarian Cancer: A Meta-Analysis. Gynecol. Oncol. 2018, 150, 151–157. [Google Scholar]
- Nowacka, M.; Ginter-Matuszewska, B.; Świerczewska, M.; Sterzyńska, K.; Nowicki, M.; Januchowski, R. Effect of ALDH1A1 Gene Knockout on Drug Resistance in Paclitaxel and Topotecan Resistant Human Ovarian Cancer Cell Lines in 2D and 3D Model. Int. J. Mol. Sci. 2022, 23, 3036. [Google Scholar]
- Sterzyńska, K.; Klejewski, A.; Wojtowicz, K.; Świerczewska, M.; Nowacka, M.; Kaźmierczak, D.; Andrzejewska, M.; Rusek, D.; Brązert, M.; Brązert, J.; et al. Mutual Expression of ALDH1A1, LOX, and Collagens in Ovarian Cancer Cell Lines as Combined CSCs- and ECM-Related Models of Drug Resistance Development. Int. J. Mol. Sci. 2018, 20, 54. [Google Scholar]
- Mohyeldin, A.; Garzón-Muvdi, T.; Quiñones-Hinojosa, A. Oxygen in Stem Cell Biology: A Critical Component of the Stem Cell Niche. Cell Stem Cell 2010, 7, 150–161. [Google Scholar] [PubMed]

| CSC Marker | Full Name | Function | References |
|---|---|---|---|
| ALDH1A1 | Aldehyde Dehydrogenase 1 Family Member A1 | Enzyme belonging to the aldehyde dehydrogenase family of proteins participating in the biosynthesis of retinoic acid and allowing for the regulation of proper proliferation and differentiation of cancer stem cells. | [16] |
| CD44 | CD44 molecule | A cell-surface glycoprotein that promotes metastasis, stem cell-like phenotypes, and chemoresistance. | [17] |
| PROM1 (CD133) | Prominin-1 | Transmembrane protein that promotes stemness and strengthens adhesion and clearance of mesothelial cells; it causes an increase in peritoneal adhesion. It entails improved adherence to the metastatic niche and infiltration of the peritoneal tissue during metastases. | [18,19] |
| c-Kit (CD117) | KIT proto-oncogene receptor tyrosine kinase | This cytokine receptor is closely related to the neovascularization of epithelial OC tissue and the formation of vasculogenic mimicry. CSCs are associated with the formation of vasculogenic mimicry, which therefore acts as a molecular CSC marker. | [20] |
| POU5F1 (OCT4) | POU class 5 homeobox 1 (octamer-binding transcription factor 4) | It encodes embryonic transcription factors that are vital for quiescence, pluripotency, and long-term self-renewal—properties that are characteristic of CSCs. | [21] |
| NOTCH3 | Notch Receptor 3 | A protein whose activation increases the adhesion between ovarian tumor cells and collagen-rich peritoneal surfaces. | [22] |
| SOX9 | SRY-box transcription factor 9 | A protein that regulates apoptotic and proliferative properties. It allows OC to survive in hypoxic conditions. | [23] |
| SNORD89 | Small nucleolar RNA, C/D box 89 | A small nucleolar RNA that promotes stem-cell-like characteristics via the Notch1/c-Myc pathway. | [23,24,25] |
| SNORA72 | Small nucleolar RNA, H/ACA box 72 | ||
| Thy-1 (CD90) | Thy-1 cell surface antigen | GPI-anchored protein located on the cell surface correlated with increased self-renewal and proliferative ability of OC cells. It acts as a tumor suppressor in OC. Thy-1 is overexpressed in CSCs. | [23,24,25] |
| EpCAM | Epithelial cell adhesion molecule | Transmembrane glycoprotein mediating Ca2+-independent homotypic cell–cell adhesion in epithelia. EpCAM regulates chemoresistance by activating the PI3K/Akt/mTOR signaling pathway. | [26] |
| LGR5 | Leucine-rich repeat containing G protein-coupled receptor 5 | Protein that may promote epithelial OC development through regulation of the Notch1 signaling pathway, which is associated with CSC self-renewal and drug resistance. | [27] |
| PTTG1 (Securin) | PTTG1 regulator of sister chromatid separation, securin | Protein with the ability to regulate CSC-associated self-renewal and epithelial–mesenchymal transition pathways. | [28] |
| ROR1 | Receptor tyrosine kinase like orphan receptor 1 | A surface antigen playing an important role in the Wnt signaling pathway. ROR1 increases tumor cell proliferation, migration, invasion, and oncogenicity and triggers the formation of spheroids, the invasion of the extracellular matrix, or the development of tumor xenografts, which are functional features associated with CSC. | [29,30] |
| NES (Nestin) | Neuroepithelial stem cell protein | Intermediate filament protein involved in ne ovascularization and CSCs and closely related to vasculogenic mimicry formation. | [31] |
| TMSB4X (Tβ4) | Thymosin β4 | A G-actin-sequestering peptide associated with the metastatic potential of tumor cells by stimulating cell migration. Tβ4 expression is strongly associated with CD133 expression and is characteristic of CSCs. | [32] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iżycka, N.; Zaborowski, M.P.; Ciecierski, Ł.; Jaz, K.; Szubert, S.; Miedziarek, C.; Rezler, M.; Piątek-Bajan, K.; Synakiewicz, A.; Jankowska, A.; et al. Cancer Stem Cell Markers—Clinical Relevance and Prognostic Value in High-Grade Serous Ovarian Cancer (HGSOC) Based on The Cancer Genome Atlas Analysis. Int. J. Mol. Sci. 2023, 24, 12746. https://doi.org/10.3390/ijms241612746
Iżycka N, Zaborowski MP, Ciecierski Ł, Jaz K, Szubert S, Miedziarek C, Rezler M, Piątek-Bajan K, Synakiewicz A, Jankowska A, et al. Cancer Stem Cell Markers—Clinical Relevance and Prognostic Value in High-Grade Serous Ovarian Cancer (HGSOC) Based on The Cancer Genome Atlas Analysis. International Journal of Molecular Sciences. 2023; 24(16):12746. https://doi.org/10.3390/ijms241612746
Chicago/Turabian StyleIżycka, Natalia, Mikołaj Piotr Zaborowski, Łukasz Ciecierski, Kamila Jaz, Sebastian Szubert, Cezary Miedziarek, Marta Rezler, Kinga Piątek-Bajan, Aneta Synakiewicz, Anna Jankowska, and et al. 2023. "Cancer Stem Cell Markers—Clinical Relevance and Prognostic Value in High-Grade Serous Ovarian Cancer (HGSOC) Based on The Cancer Genome Atlas Analysis" International Journal of Molecular Sciences 24, no. 16: 12746. https://doi.org/10.3390/ijms241612746
APA StyleIżycka, N., Zaborowski, M. P., Ciecierski, Ł., Jaz, K., Szubert, S., Miedziarek, C., Rezler, M., Piątek-Bajan, K., Synakiewicz, A., Jankowska, A., Figlerowicz, M., Sterzyńska, K., & Nowak-Markwitz, E. (2023). Cancer Stem Cell Markers—Clinical Relevance and Prognostic Value in High-Grade Serous Ovarian Cancer (HGSOC) Based on The Cancer Genome Atlas Analysis. International Journal of Molecular Sciences, 24(16), 12746. https://doi.org/10.3390/ijms241612746







