1. Introduction
Adipose stem and progenitor cells (ASPCs) are stromal vascular fraction (SVF) cells derived from adipose tissue and do not include immune, endothelial, or red blood cells [
1]. These cells are heterogeneous, and several studies have aimed to characterize and classify them in humans and experimental animals such as mice. Particularly, researchers have targeted cell surface markers to analyze and isolate distinctive cells. Compared to humans and experimental animals, there have been few reports on ASPCs in other species, despite their importance. ASPCs in livestock, in particular, are expected to enhance the fat content and composition in food through in vitro assays of fatty acid metabolism, facilitation of lipogenesis and production of specific fatty acids. SVF cells derived from bovine fat have been reported to exhibit properties resembling mesenchymal stem/stromal cells (MSCs), such as a fibroblast-like morphology and multilineage differentiation [
2]. However, further classification of ASPCs in bovine fat is yet to be conducted. Moreover, MSCs from different species cannot be identified or isolated using the same markers [
3].
Lin−CD29+CD34+Sca-1+CD24+ cells in the white adipose tissue (WAT) of adult mice have been reported to be adipocyte progenitor cells and can reconstruct functional white adipose tissue in vivo [
4]. Moreover, single-cell RNA sequencing (scRNA-seq) has made it possible to classify clusters based on gene expression and identify specific cell surface markers. For example, CD142+ cells in the subcutaneous and visceral adipose tissues of mice were found to inhibit adipogenesis via a paracrine mechanism and were named adipogenesis regulatory cells (Aregs) [
5]. In addition, CD54+ cells in human and mouse WAT have been reported to be committed preadipocytes that differentiate into mature adipocytes with minimal stimulation [
6]. Furthermore, CD26+ cells are highly proliferative and multipotent progenitors capable of differentiating into CD54+ and CD142+ cells [
6]. Ferrero et al. reviewed the heterogeneity of ASPCs throughout previous works and classified them into “adipogenic stem cells (ASCs): CD26+”, “preadipocytes (PreAs): CD54+”, or “adipogenesis regulators (Aregs): CD142+” [
1].
The mechanisms underlying adipogenesis in ASPCs, particularly those associated with cell surface markers, have been investigated. CD146 (a melanoma cell adhesion molecule, MCAM) has been identified as a ligand that interacts with more than 10 molecules, including components of the extracellular matrix, proangiogenic factor receptors, and growth factors [
7]. Among these, angiopoietin-like protein 2 (ANGPTL2) binds to CD146 and facilitates adipogenesis, lipid accumulation, and adipose inflammation through the activation of cAMP response element-binding protein [
8]. Consistent with this report, human CD146+ cells are highly adipogenic stem and progenitor cells [
9,
10]. However, it is unclear whether bovine CD146+ cells share this capacity.
In this study, we isolated novel ASPC populations based on the expression patterns of cell surface markers and confirmed whether the markers and stemness of human and mouse ASPCs, such as ASCs, PreAs, and Aregs, are conserved in cattle. Specifically, we performed prospective isolation using flow cytometry before primary culture. We then evaluated the self-renewal and adipogenic capacities of the isolated ASPCs.
3. Discussion
This study aimed to isolate novel ASPC populations from bovine fat and confirm the conservation of markers and stemness in ASPCs, such as ASCs, PreAs, and Aregs, between cattle and other previously reported species. Our findings indicate similarities and differences, as described below.
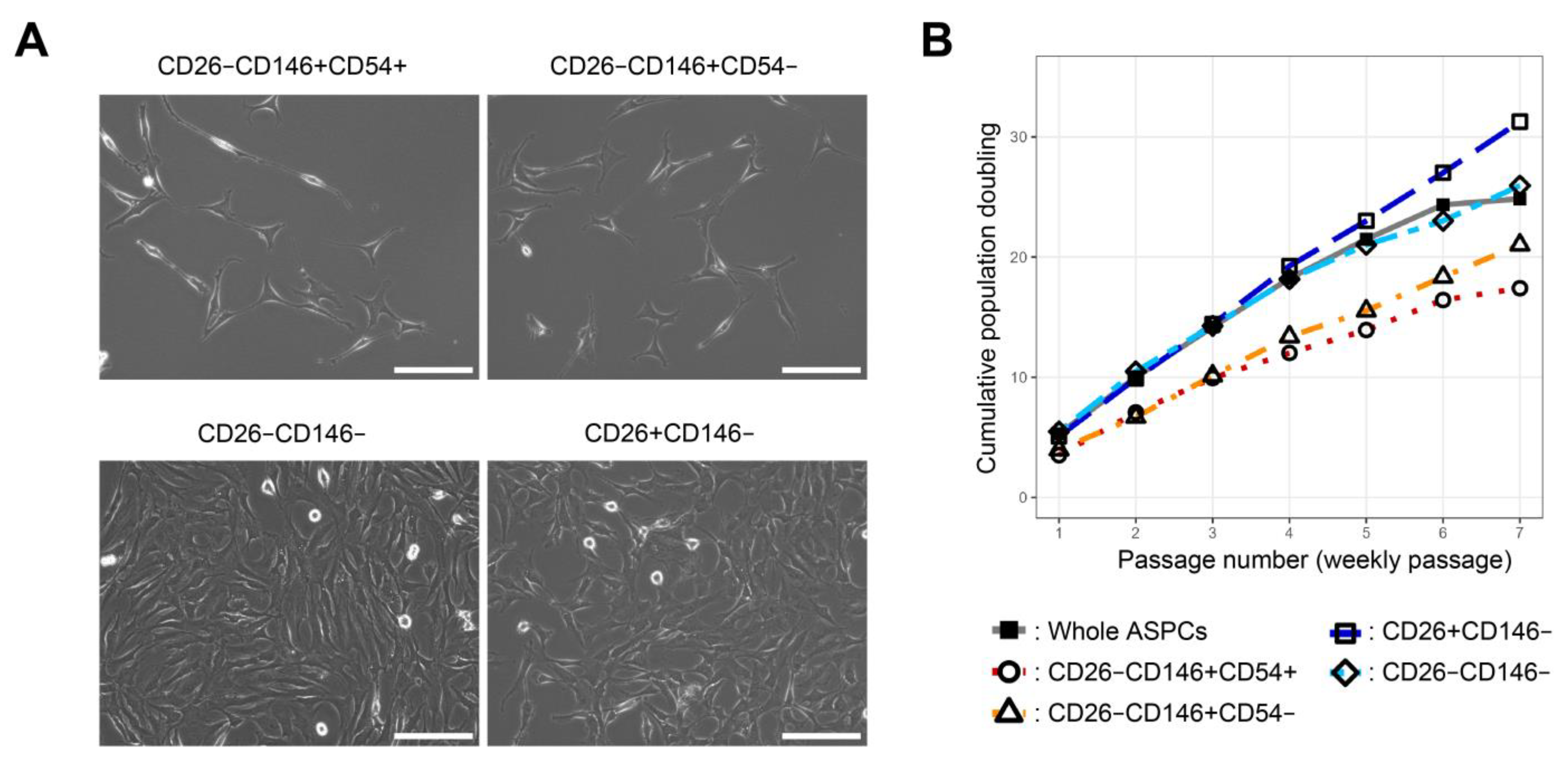
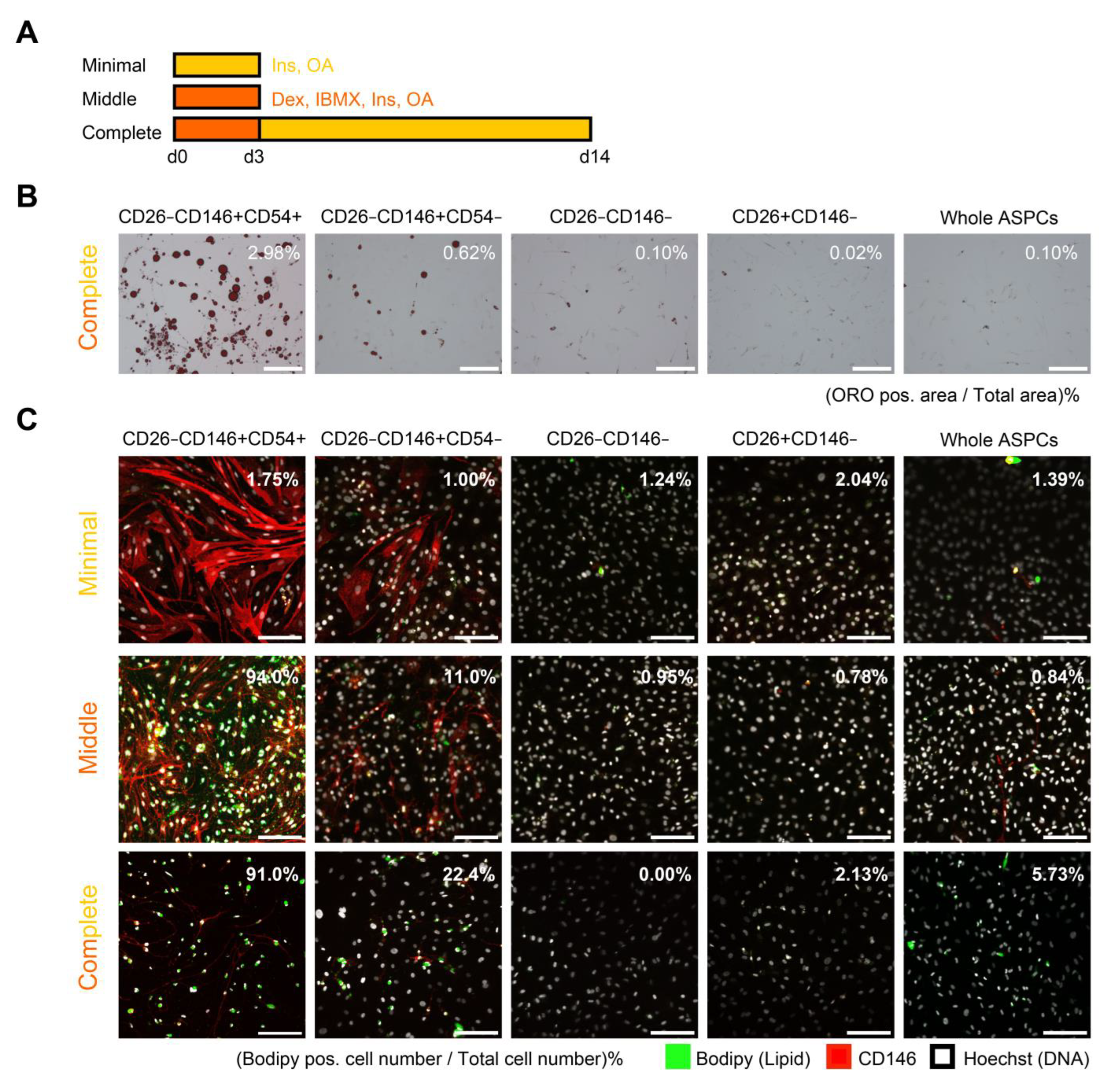
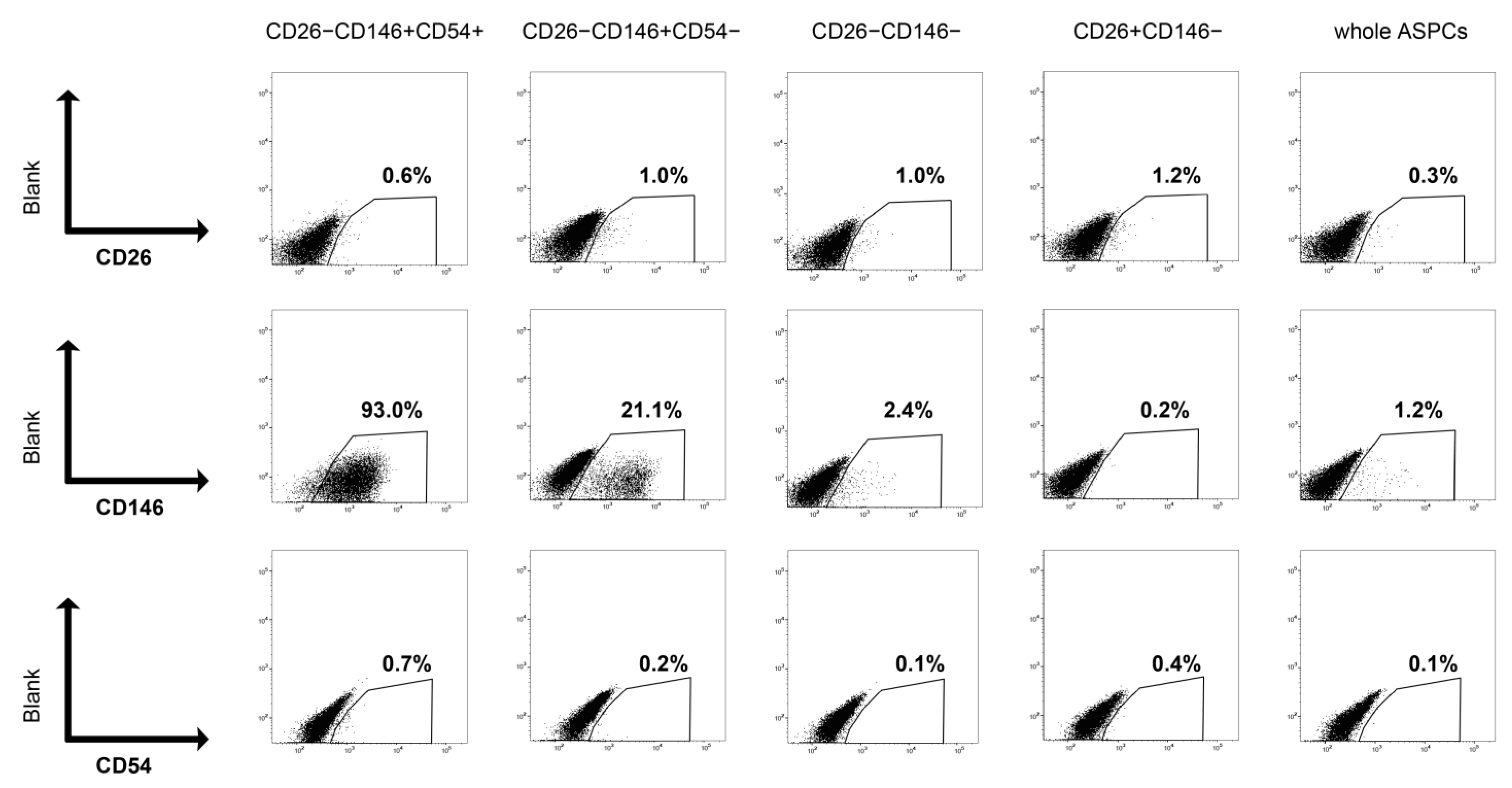
First, possible PreAs were isolated from bovine adipose as CD26−CD146+CD54+ or CD26−CD146+CD54−. CD146, also known as a MCAM, has been reported to be a marker for highly adipogenic progenitors in adipose tissue [
9,
10]. Furthermore, CD54, also known as intercellular adhesion molecule-1, is a marker of PreAs in human and mouse white adipose tissue [
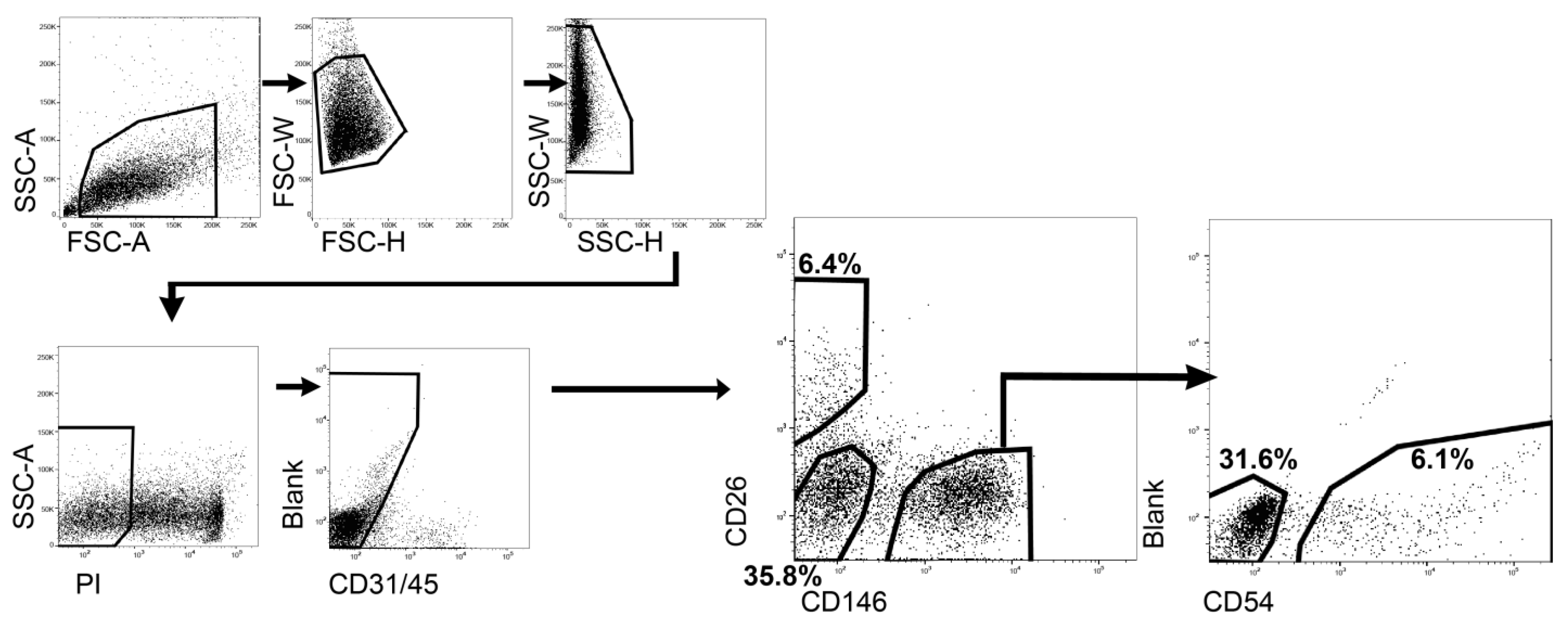
6]. This study shared these markers; however, the fractions of positive populations were lower than those reported in previous reports. For example, concerning CD54-positive populations, there was only 6.1% in this study compared to 40.8% reported by Merrick et al. [
6]. This difference probably indicates that anti-mouse CD54 (clone3E2) antibodies failed to bind to bovine CD54. Additionally, CD26−CD146+ populations, especially CD54+, from bovine fat showed slow proliferation and a high adipogenic capacity. These phenotypes are consistent with the characteristics of PreAs [
1]; however, CD26−CD146+ populations from bovine fat showed lower adipogenic capacities than PreAs in humans and mice described in a previous report [
6]. Specifically, possible PreAs from bovine fat cannot differentiate into adipocytes by insulin alone, but PreAs in humans and mice have been reported [
6]. This difference was possibly caused by the difference in the optimal adipogenic induction method between cattle and other experimental animals. To support this possibility, adipogenic protocols for humans and mice are known to be ineffective for cattle, and three-dimensional spheroid culture and endothelial basal medium facilitate bovine adipogenesis [
16].
Next, a bovine CD26-positive cell population was isolated as a candidate ASC against possible PreAs. This population comprised 6.4% of whole ASPCs (CD31−CD45− live singlets), and the percentage was comparable with a previous report [
6]. In addition, the bovine CD26+CD146− cell population is highly proliferative, and this phenotype is characteristic of ASCs [
1]. In contrast, bovine CD26+CD146− cells did not differentiate into adipocytes after complete adipogenic induction for 2 weeks. The human CD26-positive cell population has adipogenic capacity via complete induction. The optimization of adipogenic induction protocol for bovine CD26-positive cells may overcome this difference. Alternatively, the bovine CD26-positive population may not have an adipogenic capacity. To confirm the possibilities, it is required to try other induction methods optimized for cattle and other ruminants, such as the addition of PPAR-γ agonist [
17,
18] and the removal of dexamethasone and isobutylmethylxanthine [
19].
We were unable to isolate two fractions from the bovine fat: CD142+ adipogenesis regulators (Aregs) and CD26+CD146+ (double positive, DP). Aregs are commonly recognized as adipogenesis suppressors in a paracrine manner; however, there is no consensus on whether Aregs can differentiate into adipocytes [
5,
6]. In this study, we could not use markers such as CD142 or substitutes for CD142; therefore, populations other than CD54 positive or CD26 positive were candidates for Aregs. However, CD26−CD146+CD54− or CD26−CD146− were still heterogeneous in this study. Further classification and assays are required to confirm the presence of Aregs, with a focus on adipogenesis suppression. In addition, DP was reported as the transition state between ASCs and PreAs in vivo [
6]; however, we did not observe DP in prospective isolation or after in vitro culture. These observations are probably due to the poverty caused by cross-reactivity. Another possibility is that bovine CD26 positive and CD54 positive are highly independent.
In this study, we could not observe each population repeatedly and multilaterally. The position of bovine fat was in bovine cheek meat only and the biological replicate was poor. Furthermore, gene-level analyses and histochemical analyses were not carried out.
In the future, each ASPC fraction of this study can make a contribution to the evaluation of adipogenesis and lipogenesis in vitro regarding a variety of species. For example, CD26−CD146+ will be useful to validate adipogenesis and lipogenesis more accurately, as the fraction is homogeneous and high adipogenic progenitors. Furthermore, the fraction may facilitate the analysis of the fatty-acid metabolism in vitro, which is helpful in developing the livestock industry. Also, the sorting strategy and characteristics of bovine ASPCs are expected to be applied to other livestock animals as they are more closely related to cattle than humans and mice, which traces back to Boreoeutheria. For example, in order of closest relatives, bison and buffalo of a common tribe (Bovini), goats and sheep of a common family (Bovidae), and pigs of a common order (Cetartiodactyla) may share the characteristics of ASPCs in this study. ASPCs of other livestock animals can make the in vitro assay described above more comprehensive and acceptable to different cultures in terms of ethics, religion, and environment.
4. Materials and Methods
4.1. Preparation of SVF Cells from Livestock Tissue
Bovine fat was derived from bovine cheek meat after slaughter. The meat was obtained from Tokyo Shibaura Zoki Co., Ltd. (Tokyo, Japan) and transported to our laboratory on ice. Fat around the fascia of the cheek meat was harvested and stored in Hanks’ Balanced Salt Solution (HBSS) with a penicillin–streptomycin–amphotericin B Suspension (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan; 161-23181) at 4 °C until the following procedures were conducted.
SVF cells were prepared from bovine fat via mechanical and enzymatic dissociation as described previously [
13]. Briefly, harvested tissues were mechanically dissected with scissors for approximately 10 min and digested with a collagenase solution. The collagenase solution consisted of 2 mg/mL collagenase (FUJIFILM; 032-22364), 25 Units/mL DNase 1 (Sigma-Aldrich, St. Louis, MO, USA; D5025), HEPES 10 mM (Nacalai Tesque, Kyoto, Japan; 17557-94), and penicillin–streptomycin–amphotericin B Suspension in Dulbecco’s modified Eagle medium (DMEM) (Gibco; 10567-014). Tissues cut into tiny pieces were rotated in collagenase solution for 1 h at 37 °C. After rotation, the tissue fragments were removed using a drain net for the kitchen, and the filtrate was centrifuged at 490×
g for 5 min in HBSS. The cell pellet was resuspended in HBSS and centrifuged at 490×
g for 5 min. Finally, the cell pellet was frozen in CELLBANKER 1plus (Nippon Zenyaku Kogyo Co., Ltd., Fukushima, Japan; 11912). All procedures until cell freezing were performed within 48 h of slaughter, and the cells were stored in liquid nitrogen.
4.2. Isolation of Each ASPC Population with Flow Cytometry
The cells were rapidly thawed and filtered in HBSS through a 100 µm cell strainer (Corning, Durham, NC, USA; 352360). Then, the filtrate was centrifuged at 490× g for 5 min and resuspended in basic staining buffer, which comprised 1 mM ethylenediaminetetraacetic acid (EDTA) (Invitrogen, Carlsbad, CA, USA; 15575-038), 10 mM HEPES, 2% fetal bovine serum (FBS) (Gibco; 10270-106), and a penicillin–streptomycin–amphotericin B Suspension in HBSS. Finally, the cell suspensions were stained with the following antibodies for analysis and sorting: The primary antibody set was fluorescein isothiocyanate (FITC)-conjugated anti-sheep CD31 (clone: CO.3E1D4) antibody (1:50; Bio-Rad Laboratories, Inc., Hercules, CA, USA; MCA1097F), FITC-conjugated anti-sheep CD45 (clone:1.11.32) antibody (1:200; Bio-Rad; MCA2220F), phycoerythrin (PE)-conjugated anti-human CD146 (clone: P1H12) antibody (1:200; BD Biosciences, Franklin Lakes, NJ, USA; 550315), non-conjugated anti-bovine CD26 (clone: CACT114A) antibody (1:200; Monoclonal Antibody Center, Washington State University, Pullman, WA, USA; BOV2078), and Brilliant Violet 510 (BV510)-conjugated anti-mouse CD54 (clone:3E2) antibody (1:200; BD Biosciences; 563628). The secondary antibody was Alexa Fluor 647-conjugated anti-mouse IgG2b antibody (1:1000; Jackson ImmunoResearch Inc., West Grove, PA, USA; 115-605-207) and binding anti-CD26 antibody. The cell suspension was incubated with primary and secondary antibodies on ice for 30 min each. The cell suspension was washed once after the primary response. Finally, a 200 ng/mL propidium iodide (PI) solution (Sigma-Aldrich; P4864) was used to remove the dead cells. Flow cytometry and cell sorting were performed using an FACSAria II Cell Sorter (BD Biosciences).
4.3. Cell Culture, Proliferation Assay, and Cell Surface Marker Analysis
Isolated cells were cultured on plastic dishes (Thermo Fisher Scientific Inc., Waltham, MA, USA; 150464, 150468) in DMEM (Gibco; 10567-014), which contained 10% FBS, 10 mM HEPES, and penicillin–streptomycin–amphotericin B Suspension in 5% CO2 at 37 °C. The medium was changed twice weekly and cultured cells were passaged with 0.05% trypsin-EDTA (Gibco; 25300-620) until confluence. Proliferation assays were performed from passages 1 to 7, and CPD was evaluated. The number of passaged cells was accordingly adjusted not to cause contact inhibition.
For cell surface marker analysis, the cultured cells were detached using trypsin-EDTA. The cells were then stained with antibodies and analyzed via flow cytometry in the same manner as for cell isolation.
4.4. Adipogenic Differentiation
Adipogenic differentiation was assessed via ORO staining and ICC analysis for each population at early passages. The three methods used for adipogenic differentiation were complete, middle, and minimal induction. In complete induction, DMEM (Gibco; 10567-014) was supplemented with 1 µg/mL of human recombinant insulin (FUJIFILM; 093-06471), 400 µM of oleic acid (Tokyo Chemical Industry Co., Ltd., Tokyo, Japan; O0180) conjugated with bovine serum albumin (Nacalai Tesque; 19361-84), as previously described [
20], and penicillin–streptomycin–amphotericin B suspension. Furthermore, 1 µM dexamethasone (FUJIFILM; 041-18861) and 500 µM isobutylmethylxanthine (FUJIFILM; 095-03413) were added to the induction medium for the first 3 d. Complete induction was performed over 2 weeks. In contrast, middle and minimal inductions were performed for 3 d. The middle induction was identical to the first 3 d of complete induction, and the minimal induction lacked dexamethasone and isobutylmethylxanthine. Each fraction was passaged on laminin (FUJIFILM; 120-05751)-coated plastic plates (Greiner Bio-One, Kremsmünster, Oberösterreich, Austria; 665180) for ORO or glass chambers (Matsunami Glass Ind., Ltd., Osaka, Japan; SCS-N08) for ICC and induced after confluency.
4.5. ORO Staining
Induced cells were fixed in 4% paraformaldehyde (FUJIFILM, 163-20145) for 10 min and washed with phosphate-buffered saline (PBS) twice. Then, the cells were acclimated with 60% 2-propanol (Nacalai Tesque; 29112-63) for 3 min and stained with ORO working solution for 10–20 min, which comprised ORO solution (Muto Pure Chemicals Co., Ltd., Tokyo, Japan; 40491) and water in the ratio of 6:4. After washing with 60% 2-propanol for a short period, the cells were further washed with water thrice. Stained cells were observed using a BZ-X710 microscope (Osaka, Japan). Scanned images were then analyzed to evaluate the ORO positive area using ImageJ 1.53q software (
Supplementary Material Figure S1) [
21,
22].
4.6. ICC
Induced cells were fixed, washed as described above, and blocked with 2% bovine serum albumin (BSA) in PBS for 1 h at room temperature. Then, the cells were stained with the following primary antibody in 2% BSA/PBS overnight at 4 °C: PE-conjugated anti-human CD146 (clone: P1H12) antibody (1:100; BD Biosciences; 550315). After two washes, the cells were stained with Hoechst 33258 (1:1000; FUJIFILM; 343-07961), 1 µg/mL Bodipy 493/503 (Invitrogen; D3922), and Alexa Fluor 555-conjugated anti-mouse IgG1 secondary Antibody (1:1000; Invitrogen; A-21127) for 1 h at room temperature. The stained cells were washed twice and mounted using Fluoremount g (Southern Biotech, Birmingham, AL, USA; 0100-01). The prepared samples were observed under an LSM 700 confocal microscope (Zeiss, Oberkochen, Germany).
4.7. Confirmation of Cross-Reactivity between Bovine and Other Species for Immunogen
Thirty antigens (58 clones) were analyzed using flow cytometry and a homology search. For flow cytometry analyses, targeted SVF cells were isolated from bovine muscle and analyzed using a FACSAria II cell sorter (BD Biosciences). SVF cells were pre-gated as CD29 (Ha2/5)-positive singlets (BD Biosciences; 562153 or 562154) to concentrate colony-forming cells, as described previously [
13]. Then, the cells were validated for reactivity of each antibody in terms of “%pos” and “Staining index.” “%pos” was defined as the fraction of positive events to total CD29+ singlets. Additionally, the “Staining index” was commonly defined as the formula below, showing the degree of separation between the positive and the negative populations.
In the homology search, the p-distance was measured as the evolutionary distance between the cattle and antigen species. The amino acid sequence of each protein was obtained from the National Center for Biotechnology Information. Notably, the information on the focal site targeted by each antibody was often close to the public; therefore, we analyzed the full length of the total amino acids. Multiple sequence alignment (ClustalW) and measurements of the p-distance by pairwise deletion were performed using the Molecular Evolutionary Genetics Analysis software [
23,
24,
25].