Adipose Tissue in Breast Cancer Microphysiological Models to Capture Human Diversity in Preclinical Models
Abstract
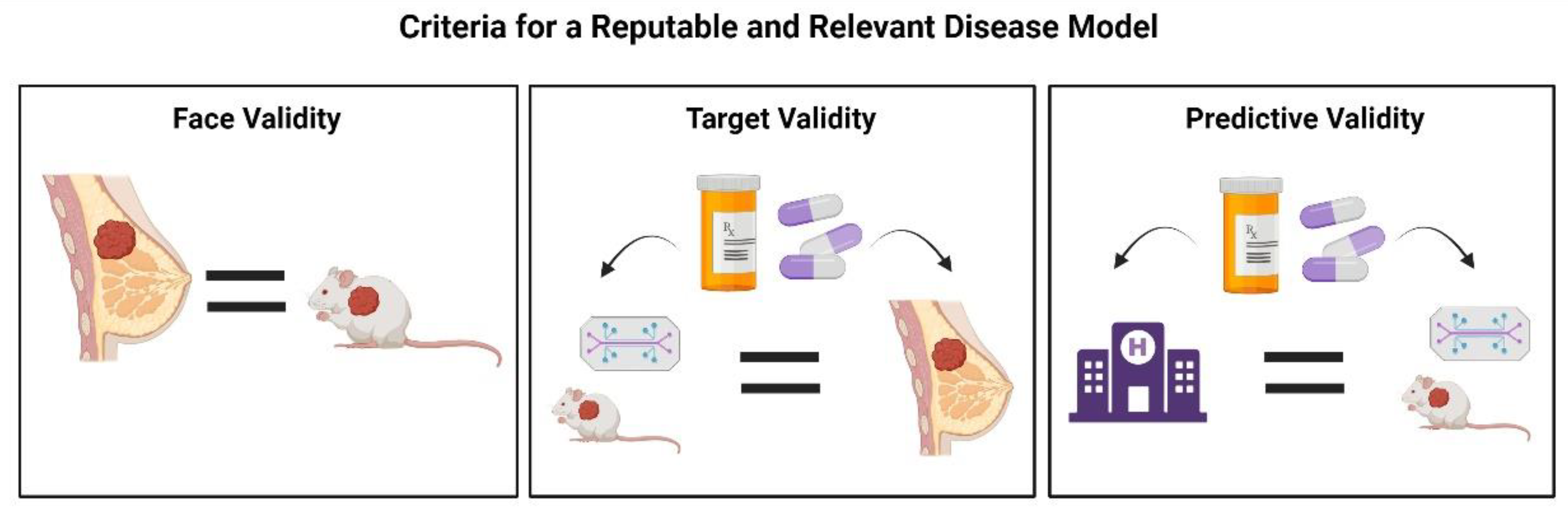
1. Introduction
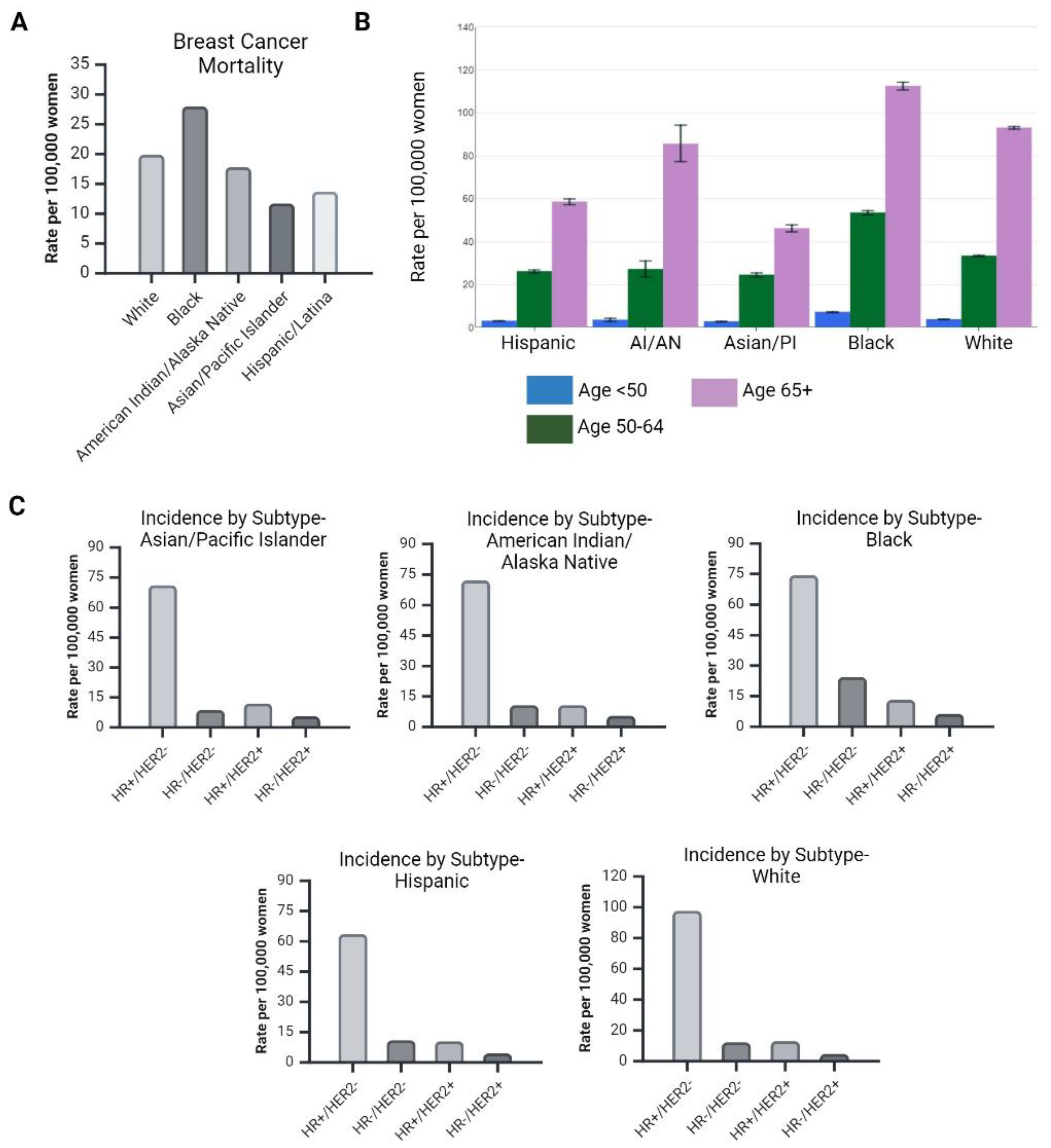
1.1. Diversity in Breast Cancer

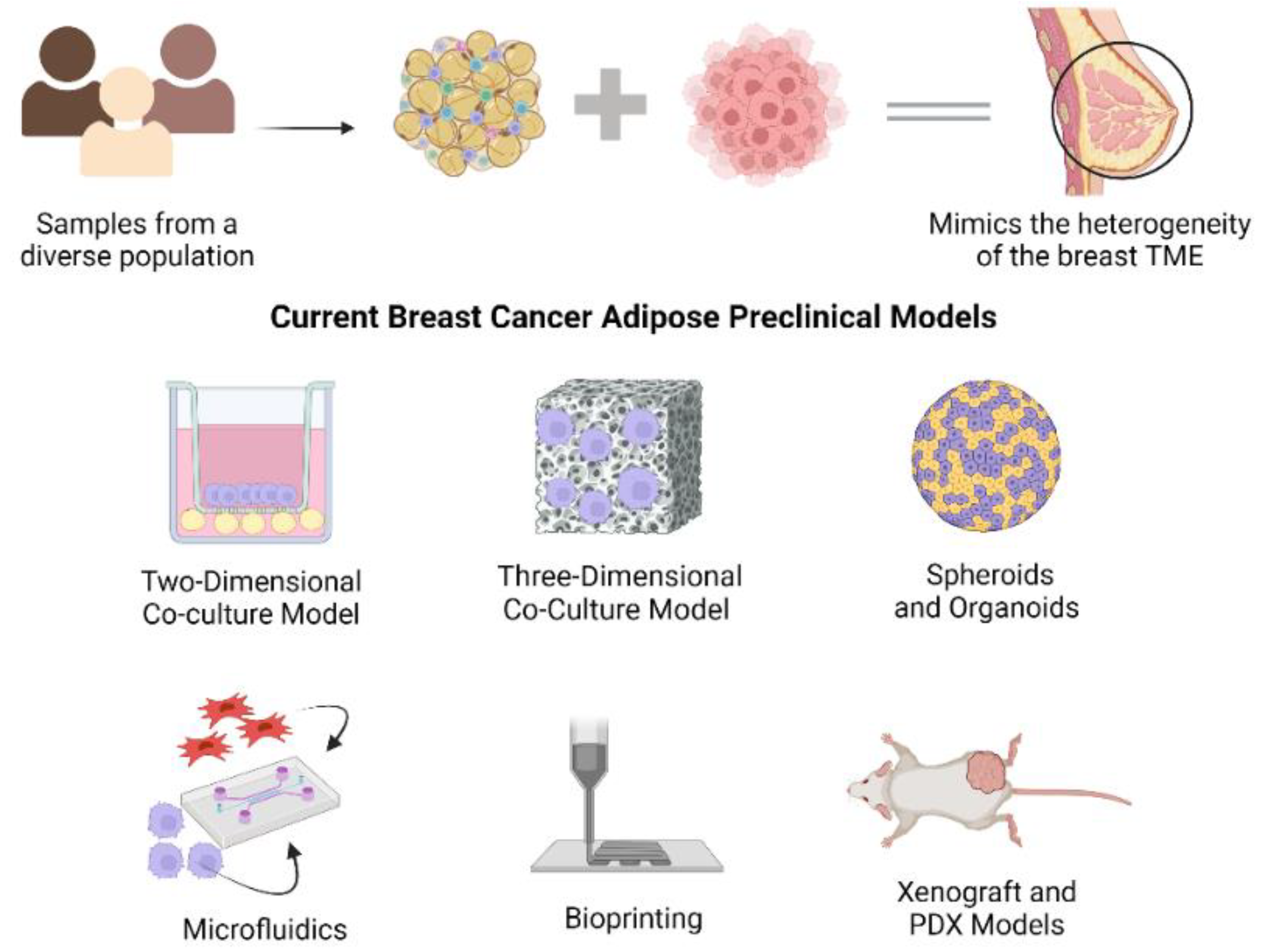
1.2. Current Breast Cancer-Adipose Preclinical Models
1.2.1. Co-Culture Models
| Platform | Cell Type(s) | Human-Derived (Y/N) | Patient-Derived Adipocytes or Adipose-Derived Stromal/Stem Cells (ASCs) (Y/N) | Multi Patient Adipocytes or ASCs (Y/N) | Key Findings | Strengths and/or Weaknesses | Reference |
|---|---|---|---|---|---|---|---|
| 2D Co-Culture | THP-1 Macrophages, HUVECs, and Mammary Preadipocytes | Yes | Yes | No | Macrophage vascular endothelial growth factor A (VEGFA) expression increased post co-culture. Macrophage expression of pro-angiogenic and pro-metastatic genes significantly increased post co-culture. Conditioned media derived from co-cultures promoted HUVEC endothelial tube formation | No breast cancer cells were used | [48] |
| MSCs, U937, MCF-7, and MDA-MB-231 | Yes | Yes | No | Macrophage paracrine activity intensifies breast cancer cell-adipocyte crosstalk | 2D co-culture | [27] | |
| ASCs, MCF-7, T47D, and ZR75 | Yes | Yes | Yes | Obesity-altered ASCs contribute to the radiation resistance observed in ER+ breast cancer | 2D co-culture | [49] | |
| ASCs, MCF-7, and MDA-MB-231 | Yes | Yes | Yes | ASCs increase breast cancer cell proliferation. ASC paracrine activity increases breast tumor cell proliferation | 2D co-culture | [26] | |
| MDA-MB-231 and Adipocytes | No | Yes | No | MDA-MB-231 pro-inflammatory gene expression was upregulated in the presence of obese murine adipose tissue. MDA-MB-231 impacted adipocyte biosynthesis pathways | 2D co-culture | [25] | |
| HS578Bst and ASCs | Yes | Yes | Yes | ASCs disrupted the expression of ECM maintenance-related genes and increased leptin and inflammatory marker gene expression | 2D co-culture | [24] | |
| ASCs, MDA-MB-231, and MCF-7 | Yes | Yes | Yes | ASCs do not impact breast cancer cell proliferation via direct cell contact or paracrine activity. ASCs do not significantly increase breast cancer cell EMT-related gene expression via direct cell contact | 2D co-culture | [33] | |
| 3D Co-Culture | 3T3-F442A, ZR75, SUM159PT, MCF-7, T47D, and MDA-MB-231 | Yes-Breast Cancer Cell Lines No-Preadipocytes | No | No | Adipocytes promote the invasion of MDA-MB-231, MCF-7, and other breast cancer cell lines | No human-derived preadipocytes used | [45] |
| ASCs and breast cancer cell lines | Yes | Yes | No | Demonstrates breast cancer cell-primary preadipocyte crosstalk in vitro | Sandwich white adipose tissue-breast cancer model | [59] | |
| Breast Adipose Tissue, ASCs, and TU-BcX-41C | Yes | Yes | Yes | Increase in cancer stem cell population when TU-BcX-41C cells are cultured in breast cancer-adipose MPS | Modified sandwich MPS model | [60] | |
| 3T3-L1 pre-adipocytes, MDA-MB-231, MCF-7, SUM159, and HS578t | Yes | No | No | Breast cancer cell interactions with the ECM and adipocytes alter breast cancer cell MET, potentially contributing to secondary tumor formation | Used 3T3-L1 preadipocytes | [53] | |
| MSCs and MDA-MB-231 | Yes | Yes | No | MDA-MB-231 invasion is enhanced in the presence of adipocytes and collagen matrix | Integrated a collagen plug into traditional Boyden chamber | [55] | |
| 3T3-L1 pre-adipocytes, MDA-MB-453, MDA-MB-435S, MDA-MB-231, and MDA-MB-468 | Yes | No | No | Adipocytes induce migration and invasion of breast cancer cells. Adipocytes stimulate breast cancer cells to adopt an aggressive tumor phenotype by inducing EMT-associated traits | Used 3T3-L1 preadipocytes | [28] | |
| Stromal vascular fraction (SVF), ASCs, and MDA-MB-231 | Yes | Yes | Yes | Direct and indirect contact with adipocytes induces similar invasive behaviors in the MDA-MB-231 TNBC cell line | Enhanced cellular heterogeneity by including SVF | [29] | |
| Mammary adipocytes and MDA-MB-231 | Yes | Yes | Yes | Breast cancer-adipocyte crosstalk is amplified by obesity. Supports the study of mammary adipocyte lipid secretion on tumor secretions and overall tumor aggressiveness in lean and obese conditions | Used a fibrin matrix in Boyden chamber | [56] | |
| ASCs, Fibroblasts, and MDA-MB-231 | Yes | No | No | Collagen VI, a highly oncogenic collagen isoform linked to breast cancer, was decreased in the irradiated cancer co-culture. Irradiation not only makes cells ablative but also may influence the oncogenic potential of the microenvironment | Used decellularized scaffold | [57] | |
| Murine peri-uterine and inguinal white adipose tissue (WAT) and MDA-MB-231 | Yes | No | No | Adipose tissue paracrine activity induces MET-like changes in the MDA-MB-231 TNBC cell line | Used murine WAT | [54] | |
| Microfluidics | ASCs and MDA-MB-231 | Yes | Yes | No | Statistically significant increase in MDA-MB-231 proliferation in the presence of ASCs. MDA-MB-231 cells adopt more of a mesenchymal phenotype in the presence of ASCs. Paclitaxel has reduced effectiveness in inhibiting MDA-MB-231 replication in the presence of ASCs | One ASC donor used (female, Caucasian, normal BMI) | [30] |
| MCF-7 and ASCs | Yes | Yes | Yes | Predicts anastrazole sensitivity with respect to ASC BMI better than a 2D co-culture system. Primary mammary adipose stromal cells derived from obese patients exhibited increased aromatase mRNA compared to lean controls | Multiple ASC donors used | [32] | |
| Spheroids | ASCs, MDA-MB-231, MCF-7, DT28, and HMLER3 CSC | Yes | Yes | Yes | The proportion of mammosphere-forming cells and cells expressing stem-like markers increases when in direct or indirect contact with adipocytes | Multiple ASC donors used | [62] |
| Multipotent adipose-derived stem cells (MADS)-adipocytes, breast adipocytes, MCF-7, and MDA-MB-231 | Yes | Yes | No | Adipocyte lipid droplet size decreases in the presence of mammospheres. UCP1 expression is dependent on adipocyte-mammosphere distance. Mammospheres produce adrenomedullin, which is critical in the interactions between adipocytes and breast cancer cells | hMADS cells isolated from young donors [63] | [64] | |
| ASCs, MDA_MB-231, and MCF-7 | Yes | No | No | ASC C-C motif chemokine ligand 5 (CCL5) expression was elevated when co-cultured with the MDA-MB-231 TNBC cell line | One ASC donor used | [65] | |
| 3T3-L1 pre-adipocytes, SKBR-3, MDA-MB-231, and MDA-MB-468 | Yes | No | No | Adipocytes increase the invasiveness of breast cancer cells | Used 3T3-L1 preadipocytes | [43] | |
| ASCs, MCF10AT, MCF10A, MCF10DCIS.com, MCF10CA1a | Yes | Yes | No | ASCs promote premalignant breast cell invasions via direct cell contact. Obese ASCs have a pro-invasive effect on premalignant and malignant breast cell lines | Combination of models established in study | [66] | |
| Bioprinting | ASCs and MDA-MB-231 | Yes | No | No | MDA-MB-231 TNBC cell line induces adipose tissue ECM remodeling and lipid content modulation | Used hyaluronic acid-based bioink and extrusion-based bioprinting | [67] |
| ASCs and MDA-MB-231 | Yes | Yes | Yes | Adipose cells hasten the invasion and escape of tumor cells via soluble factor secretion. Tumor invasion and escape are more strongly induced by ASCs than adipocytes | Multiple demographics represented in ASC donor selection | [31] |
1.2.2. Spheroids
1.2.3. Microfluidics
1.2.4. Bioprinting
1.2.5. Xenograft and PDX Models
| Platform | Cell Type(s) | Human-Derived (Y/N) | Patient-Derived Adipocytes or ASCs (Y/N) | Multi Patient Adipocytes or ASCs (Y/N) | Key Findings | Reference |
|---|---|---|---|---|---|---|
| Xenograft | ASCs, BT20, MDA-MB-231, MDA-MB-468, MCF-7, and HCC1806 | Yes | Yes | No | ASCs derived from obese donors promote a pro-metastatic phenotype by upregulating epithelial–mesenchymal transition (EMT)-associated genes and promoting migration in vitro | [91] |
| ASCs, MCF-7, T47D, and ZR-75 | Yes | Yes | Yes | MCF-7 co-cultured with obese ASCs and irradiated prior to injection had increased tumor growth compared to cells that were not co-cultured before radiation | [49] | |
| PDX | MDA-MB-231, TU-BCX-41C PDX, and TU-BCX-41C PDX derived cells | Yes | No | No | Provided a detailed characterization of a PDX model for metastatic breast cancer (MBC). The established PDX model maintained consistent matrix architecture and stiffness after multiple serial passages | [60] |
| ASCs, human breast cancer PDX cells | Yes | Yes | Yes | Adipsin secreted from mammary ASCs promotes cancer stem cell-like properties and proliferation of human breast cancer PDX cells in vitro and in vivo | [92] | |
| TU-BCX-2 K1 PDX, ASCs | Yes | Yes | Yes | ASCs derived from obese donors promote a pro-metastatic phenotype by upregulating EMT-associated genes and promoting migration in vitro | [91] |
2. Discussion
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cancer of the Breast (Female)-Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/breast.html (accessed on 30 October 2023).
- Fröhlich, E. The Variety of 3D Breast Cancer Models for the Study of Tumor Physiology and Drug Screening. Int. J. Mol. Sci. 2023, 24, 7116. [Google Scholar] [CrossRef] [PubMed]
- Mak, I.W.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114–118. [Google Scholar] [PubMed]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef]
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef]
- Hirko, K.A.; Rocque, G.; Reasor, E.; Taye, A.; Daly, A.; Cutress, R.I.; Copson, E.R.; Lee, D.W.; Lee, K.H.; Im, S.A.; et al. The impact of race and ethnicity in breast cancer-disparities and implications for precision oncology. BMC Med. 2022, 20, 72. [Google Scholar] [CrossRef]
- Guo, L.; Kong, D.; Liu, J.; Zhan, L.; Luo, L.; Zheng, W.; Zheng, Q.; Chen, C.; Sun, S. Breast cancer heterogeneity and its implication in personalized precision therapy. Exp. Hematol. Oncol. 2023, 12, 3. [Google Scholar] [CrossRef]
- Kim, G.; Pastoriza, J.M.; Condeelis, J.S.; Sparano, J.A.; Filippou, P.S.; Karagiannis, G.S.; Oktay, M.H. The Contribution of Race to Breast Tumor Microenvironment Composition and Disease Progression. Front. Oncol. 2020, 10, 1022. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.E.; Hunziker, R.; Wikswo, J.P. Fitting tissue chips and microphysiological systems into the grand scheme of medicine, biology, pharmacology, and toxicology. Exp. Biol. Med. 2017, 242, 1559–1572. [Google Scholar] [CrossRef]
- Schexnayder, C.; Broussard, K.; Onuaguluchi, D.; Poché, A.; Ismail, M.; McAtee, L.; Llopis, S.; Keizerweerd, A.; McFerrin, H.; Williams, C. Metformin Inhibits Migration and Invasion by Suppressing ROS Production and COX2 Expression in MDA-MB-231 Breast Cancer Cells. Int. J. Mol. Sci. 2018, 19, 3692. [Google Scholar] [CrossRef]
- Rowan, B.G.; Lacayo, E.A.; Sheng, M.; Anbalagan, M.; Gimble, J.M.; Jones, R.K.; Joseph, W.J.; Friedlander, P.L.; Chiu, E.S. Human Adipose Tissue-Derived Stromal/Stem Cells Promote Migration and Early Metastasis of Head and Neck Cancer Xenografts. Aesthetic Surg. J. 2015, 36, 93–104. [Google Scholar] [CrossRef]
- Hoang, V.T.; Matossian, M.D.; Ucar, D.A.; Elliott, S.; La, J.; Wright, M.K.; Burks, H.E.; Perles, A.; Hossain, F.; King, C.T.; et al. ERK5 Is Required for Tumor Growth and Maintenance Through Regulation of the Extracellular Matrix in Triple Negative Breast Cancer. Front. Oncol. 2020, 10, 1164. [Google Scholar] [CrossRef]
- Frazier, T.; Williams, C.; Henderson, M.; Duplessis, T.; Rogers, E.; Wu, X.; Hamel, K.; Martin, E.C.; Mohiuddin, O.; Shaik, S.; et al. Breast Cancer Reconstruction: Design Criteria for a Humanized Microphysiological System. Tissue Eng. Part A 2021, 27, 479–488. [Google Scholar] [CrossRef]
- Brock, C.K.; Hebert, K.L.; Artiles, M.; Wright, M.K.; Cheng, T.; Windsor, G.O.; Nguyen, K.; Alzoubi, M.S.; Collins-Burow, B.M.; Martin, E.C.; et al. A Role for Adipocytes and Adipose Stem Cells in the Breast Tumor Microenvironment and Regenerative Medicine. Front. Physiol. 2021, 12, 751239. [Google Scholar] [CrossRef]
- Bunnell, B.A.; Martin, E.C.; Matossian, M.D.; Brock, C.K.; Nguyen, K.; Collins-Burow, B.; Burow, M.E. The effect of obesity on adipose-derived stromal cells and adipose tissue and their impact on cancer. Cancer Metastasis Rev. 2022, 41, 549–573. [Google Scholar] [CrossRef]
- Strong, A.L.; Burow, M.E.; Gimble, J.M.; Bunnell, B.A. Concise review: The obesity cancer paradigm: Exploration of the interactions and crosstalk with adipose stem cells. Stem Cells 2015, 33, 318–326. [Google Scholar] [CrossRef]
- Strong, A.L.; Strong, T.A.; Rhodes, L.V.; Semon, J.A.; Zhang, X.; Shi, Z.; Zhang, S.; Gimble, J.M.; Burow, M.E.; Bunnell, B.A. Obesity associated alterations in the biology of adipose stem cells mediate enhanced tumorigenesis by estrogen dependent pathways. Breast Cancer Res. 2013, 15, R102. [Google Scholar] [CrossRef] [PubMed]
- Matossian, M.D.; Giardina, A.A.; Wright, M.K.; Elliott, S.; Loch, M.M.; Nguyen, K.; Zea, A.H.; Lau, F.H.; Moroz, K.; Riker, A.I.; et al. Patient-Derived Xenografts as an Innovative Surrogate Tumor Model for the Investigation of Health Disparities in Triple Negative Breast Cancer. Womens Health Rep. 2020, 1, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Fane, M.; Weeraratna, A.T. How the ageing microenvironment influences tumour progression. Nat. Rev. Cancer 2020, 20, 89–106. [Google Scholar] [CrossRef] [PubMed]
- Bandera, E.V.; Chandran, U.; Hong, C.C.; Troester, M.A.; Bethea, T.N.; Adams-Campbell, L.L.; Haiman, C.A.; Park, S.Y.; Olshan, A.F.; Ambrosone, C.B.; et al. Obesity, body fat distribution, and risk of breast cancer subtypes in African American women participating in the AMBER Consortium. Breast Cancer Res. Treat. 2015, 150, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Mertz, D.; Sentosa, J.; Luker, G.; Takayama, S. Studying Adipose Tissue in the Breast Tumor Microenvironment In Vitro: Progress and Opportunities. Tissue Eng. Regen. Med. 2020, 17, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Cottrill, A.; Samadi, Y.; Marra, K. Chapter 9—Adipose stem cells and donor demographics: Impact of anatomic location, donor sex, race, BMI, and health. In Scientific Principles of Adipose Stem Cells; Kokai, L., Marra, K., Rubin, J.P., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 149–163. [Google Scholar]
- Rezaee, R.; Abdollahi, M. The importance of translatability in drug discovery. Expert Opin. Drug Discov. 2017, 12, 237–239. [Google Scholar] [CrossRef] [PubMed]
- Delort, L.; Cholet, J.; Decombat, C.; Vermerie, M.; Dumontet, C.; Castelli, F.A.; Fenaille, F.; Auxenfans, C.; Rossary, A.; Caldefie-Chezet, F. The Adipose Microenvironment Dysregulates the Mammary Myoepithelial Cells and Could Participate to the Progression of Breast Cancer. Front. Cell Dev. Biol. 2021, 8, 571948. [Google Scholar] [CrossRef] [PubMed]
- Blücher, C.; Iberl, S.; Schwagarus, N.; Müller, S.; Liebisch, G.; Höring, M.; Hidrobo, M.S.; Ecker, J.; Spindler, N.; Dietrich, A.; et al. Secreted Factors from Adipose Tissue Reprogram Tumor Lipid Metabolism and Induce Motility by Modulating PPARα/ANGPTL4 and FAK. Mol. Cancer Res. 2020, 18, 1849–1862. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, X.; Zhao, H.; Wang, J.; Zhang, Q. CXCL5 secreted from adipose tissue-derived stem cells promotes cancer cell proliferation. Oncol. Lett. 2018, 15, 1403–1410. [Google Scholar] [PubMed]
- Vallega, K.A.; Bosco, D.B.; Ren, Y.; Sang, Q.-X.A. Macrophage-Conditioned Media Promotes Adipocyte Cancer Association, Which in Turn Stimulates Breast Cancer Proliferation and Migration. Biomolecules 2022, 12, 1757. [Google Scholar] [CrossRef]
- Lee, Y.; Jung, W.H.; Koo, J.S. Adipocytes can induce epithelial-mesenchymal transition in breast cancer cells. Breast Cancer Res. Treat. 2015, 153, 323–335. [Google Scholar] [CrossRef]
- D’Esposito, V.; Liguoro, D.; Ambrosio, M.R.; Collina, F.; Cantile, M.; Spinelli, R.; Raciti, G.A.; Miele, C.; Valentino, R.; Campiglia, P.; et al. Adipose microenvironment promotes triple negative breast cancer cell invasiveness and dissemination by producing CCL5. Oncotarget 2016, 7, 24495–24509. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.M.; Campbell, J.M.; Coates, R.N.; Render, K.M.; Byrne, C.E.; Martin, E.C.; Melvin, A.T. Evaluation of intercellular communication between breast cancer cells and adipose-derived stem cells via passive diffusion in a two-layer microfluidic device. Lab Chip 2020, 20, 2009–2019. [Google Scholar] [CrossRef]
- Dance, Y.W.; Meshulam, T.; Seibel, A.J.; Obenreder, M.C.; Layne, M.D.; Nelson, C.M.; Tien, J. Adipose Stroma Accelerates the Invasion and Escape of Human Breast Cancer Cells from an Engineered Microtumor. Cell. Mol. Bioeng. 2022, 15, 15–29. [Google Scholar] [CrossRef]
- Morgan, M.M.; Arendt, L.M.; Alarid, E.T.; Beebe, D.J.; Johnson, B.P. Mammary adipose stromal cells derived from obese women reduce sensitivity to the aromatase inhibitor anastrazole in an organotypic breast model. FASEB J. 2019, 33, 8623–8633. [Google Scholar] [CrossRef]
- Ejaz, A.; Yang, K.S.; Venkatesh, K.P.; Chinnapaka, S.; Kokai, L.E.; Rubin, J.P. The Impact of Human Lipoaspirate and Adipose Tissue-Derived Stem Cells Contact Culture on Breast Cancer Cells: Implications in Breast Reconstruction. Int. J. Mol. Sci. 2020, 21, 9171. [Google Scholar] [CrossRef]
- Vis, M.A.M.; Ito, K.; Hofmann, S. Impact of Culture Medium on Cellular Interactions in in vitro Co-culture Systems. Front. Bioeng. Biotechnol. 2020, 8, 911. [Google Scholar] [CrossRef]
- Mason, J.; Öhlund, D. Key aspects for conception and construction of co-culture models of tumor-stroma interactions. Front. Bioeng. Biotechnol. 2023, 11, 1150764. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.A.; Song, A.; Chen, W.; Schwalie, P.C.; Zhang, F.; Vishvanath, L.; Jiang, L.; Ye, R.; Shao, M.; Tao, C.; et al. Reversible De-differentiation of Mature White Adipocytes into Preadipocyte-like Precursors during Lactation. Cell Metab. 2018, 28, 282–288.e283. [Google Scholar] [CrossRef] [PubMed]
- Zwick, R.K.; Rudolph, M.C.; Shook, B.A.; Holtrup, B.; Roth, E.; Lei, V.; Van Keymeulen, A.; Seewaldt, V.; Kwei, S.; Wysolmerski, J.; et al. Adipocyte hypertrophy and lipid dynamics underlie mammary gland remodeling after lactation. Nat. Commun. 2018, 9, 3592. [Google Scholar] [CrossRef] [PubMed]
- Kothari, C.; Diorio, C.; Durocher, F. The Importance of Breast Adipose Tissue in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 5760. [Google Scholar] [CrossRef] [PubMed]
- D’Esposito, V.; Ambrosio, M.R.; Giuliano, M.; Cabaro, S.; Miele, C.; Beguinot, F.; Formisano, P. Mammary Adipose Tissue Control of Breast Cancer Progression: Impact of Obesity and Diabetes. Front. Oncol. 2020, 10, 1554. [Google Scholar] [CrossRef] [PubMed]
- Cozzo, A.J.; Fuller, A.M.; Makowski, L. Contribution of Adipose Tissue to Development of Cancer. Compr. Physiol. 2017, 8, 237–282. [Google Scholar] [CrossRef]
- Iyengar, P.; Combs, T.P.; Shah, S.J.; Gouon-Evans, V.; Pollard, J.W.; Albanese, C.; Flanagan, L.; Tenniswood, M.P.; Guha, C.; Lisanti, M.P.; et al. Adipocyte-secreted factors synergistically promote mammary tumorigenesis through induction of anti-apoptotic transcriptional programs and proto-oncogene stabilization. Oncogene 2003, 22, 6408–6423. [Google Scholar] [CrossRef]
- Wu, Q.; Li, B.; Li, Z.; Li, J.; Sun, S.; Sun, S. Cancer-associated adipocytes: Key players in breast cancer progression. J. Hematol. Oncol. 2019, 12, 95. [Google Scholar] [CrossRef]
- He, J.Y.; Wei, X.H.; Li, S.J.; Liu, Y.; Hu, H.L.; Li, Z.Z.; Kuang, X.H.; Wang, L.; Shi, X.; Yuan, S.T.; et al. Adipocyte-derived IL-6 and leptin promote breast Cancer metastasis via upregulation of Lysyl Hydroxylase-2 expression. Cell Commun. Signal 2018, 16, 100. [Google Scholar] [CrossRef]
- Park, J.Y.; Kang, S.E.; Ahn, K.S.; Um, J.Y.; Yang, W.M.; Yun, M.; Lee, S.G. Inhibition of the PI3K-AKT-mTOR pathway suppresses the adipocyte-mediated proliferation and migration of breast cancer cells. J. Cancer 2020, 11, 2552–2559. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Attané, C.; Milhas, D.; Dirat, B.; Dauvillier, S.; Guerard, A.; Gilhodes, J.; Lazar, I.; Alet, N.; Laurent, V.; et al. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI Insight 2017, 2, e87489. [Google Scholar] [CrossRef] [PubMed]
- Fujisaki, K.; Fujimoto, H.; Sangai, T.; Nagashima, T.; Sakakibara, M.; Shiina, N.; Kuroda, M.; Aoyagi, Y.; Miyazaki, M. Cancer-mediated adipose reversion promotes cancer cell migration via IL-6 and MCP-1. Breast Cancer Res. Treat. 2015, 150, 255–263. [Google Scholar] [CrossRef] [PubMed]
- D’Esposito, V.; Passaretti, F.; Hammarstedt, A.; Liguoro, D.; Terracciano, D.; Molea, G.; Canta, L.; Miele, C.; Smith, U.; Beguinot, F.; et al. Adipocyte-released insulin-like growth factor-1 is regulated by glucose and fatty acids and controls breast cancer cell growth in vitro. Diabetologia 2012, 55, 2811–2822. [Google Scholar] [CrossRef] [PubMed]
- Yadav, N.V.S.; Barcikowski, A.; Uehana, Y.; Jacobs, A.T.; Connelly, L. Breast Adipocyte Co-culture Increases the Expression of Pro-angiogenic Factors in Macrophages. Front. Oncol. 2020, 10, 454. [Google Scholar] [CrossRef] [PubMed]
- Sabol, R.A.; Villela, V.A.; Denys, A.; Freeman, B.T.; Hartono, A.B.; Wise, R.M.; Harrison, M.A.A.; Sandler, M.B.; Hossain, F.; Miele, L.; et al. Obesity-Altered Adipose Stem Cells Promote Radiation Resistance of Estrogen Receptor Positive Breast Cancer through Paracrine Signaling. Int. J. Mol. Sci. 2020, 21, 2722. [Google Scholar] [CrossRef] [PubMed]
- Passaniti, A.; Kleinman, H.K.; Martin, G.R. Matrigel: History/background, uses, and future applications. J. Cell Commun. Signal 2022, 16, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.M.; Doyle, A.D.; Lu, J. Cell-3D matrix interactions: Recent advances and opportunities. Trends Cell Biol. 2022, 32, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Köhrmann, A.; Kammerer, U.; Kapp, M.; Dietl, J.; Anacker, J. Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: New findings and review of the literature. BMC Cancer 2009, 9, 188. [Google Scholar] [CrossRef]
- Pallegar, N.K.; Garland, C.J.; Mahendralingam, M.; Viloria-Petit, A.M.; Christian, S.L. A Novel 3-Dimensional Co-culture Method Reveals a Partial Mesenchymal to Epithelial Transition in Breast Cancer Cells Induced by Adipocytes. J. Mammary Gland. Biol. Neoplasia 2019, 24, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Asante, E.C.; Pallegar, N.K.; Hoffmann, A.J.; Viloria-Petit, A.M.; Christian, S.L. Adipose Tissue from Lean and Obese Mice Induces a Mesenchymal to Epithelial Transition-Like Effect in Triple Negative Breast Cancers Cells Grown in 3-Dimensional Culture. Int. J. Mol. Sci. 2020, 21, 6439. [Google Scholar] [CrossRef]
- Hume, R.D.; Berry, L.; Reichelt, S.; D’Angelo, M.; Gomm, J.; Cameron, R.E.; Watson, C.J. An Engineered Human Adipose/Collagen Model for In Vitro Breast Cancer Cell Migration Studies. Tissue Eng. Part A 2018, 24, 1309–1319. [Google Scholar] [CrossRef]
- Rebeaud, M.; Bouche, C.; Dauvillier, S.; Attané, C.; Arellano, C.; Vaysse, C.; Fallone, F.; Muller, C. A novel 3D culture model for human primary mammary adipocytes to study their metabolic crosstalk with breast cancer in lean and obese conditions. Sci. Rep. 2023, 13, 4707. [Google Scholar] [CrossRef]
- Brett, E.; Rosemann, M.; Azimzadeh, O.; Pagani, A.; Prahm, C.; Daigeler, A.; Duscher, D.; Kolbenschlag, J. Irradiated Triple-Negative Breast Cancer Co-Culture Produces a Less Oncogenic Extracellular Matrix. Int. J. Mol. Sci. 2022, 23, 8265. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, L.; Maffini, M.V.; Soto, A.; Sonnenschein, C.; Kaplan, D.L. A complex 3D human tissue culture system based on mammary stromal cells and silk scaffolds for modeling breast morphogenesis and function. Biomaterials 2010, 31, 3920–3929. [Google Scholar] [CrossRef] [PubMed]
- Au-Brown, L.M.; Au-Hebert, K.L.; Au-Gurrala, R.R.; Au-Byrne, C.E.; Au-Burow, M.; Au-Martin, E.C.; Au-Lau, F.H. Modeling Breast Cancer in Human Breast Tissue using a Microphysiological System. JoVE (J. Vis. Exp.) 2021, 170, e62009. [Google Scholar] [CrossRef]
- Matossian, M.D.; Chang, T.; Wright, M.K.; Burks, H.E.; Elliott, S.; Sabol, R.A.; Wathieu, H.; Windsor, G.O.; Alzoubi, M.S.; King, C.T.; et al. In-depth characterization of a new patient-derived xenograft model for metaplastic breast carcinoma to identify viable biologic targets and patterns of matrix evolution within rare tumor types. Clin. Transl. Oncol. 2022, 24, 127–144. [Google Scholar] [CrossRef]
- Lau, F.H.; Vogel, K.; Luckett, J.P.; Hunt, M.; Meyer, A.; Rogers, C.L.; Tessler, O.; Dupin, C.L.; St Hilaire, H.; Islam, K.N.; et al. Sandwiched White Adipose Tissue: A Microphysiological System of Primary Human Adipose Tissue. Tissue Eng. Part C Methods 2018, 24, 135–145. [Google Scholar] [CrossRef]
- Picon-Ruiz, M.; Pan, C.; Drews-Elger, K.; Jang, K.; Besser, A.H.; Zhao, D.; Morata-Tarifa, C.; Kim, M.; Ince, T.A.; Azzam, D.J.; et al. Interactions between Adipocytes and Breast Cancer Cells Stimulate Cytokine Production and Drive Src/Sox2/miR-302b–Mediated Malignant Progression. Cancer Res. 2016, 76, 491–504. [Google Scholar] [CrossRef]
- Rodriguez, A.M.; Pisani, D.; Dechesne, C.A.; Turc-Carel, C.; Kurzenne, J.Y.; Wdziekonski, B.; Villageois, A.; Bagnis, C.; Breittmayer, J.P.; Groux, H.; et al. Transplantation of a multipotent cell population from human adipose tissue induces dystrophin expression in the immunocompetent mdx mouse. J. Exp. Med. 2005, 201, 1397–1405. [Google Scholar] [CrossRef]
- Paré, M.; Darini, C.Y.; Yao, X.; Chignon-Sicard, B.; Rekima, S.; Lachambre, S.; Virolle, V.; Aguilar-Mahecha, A.; Basik, M.; Dani, C.; et al. Breast cancer mammospheres secrete Adrenomedullin to induce lipolysis and browning of adjacent adipocytes. BMC Cancer 2020, 20, 784. [Google Scholar] [CrossRef]
- Watzling, M.; Klaus, L.; Weidemeier, T.; Horder, H.; Ebert, R.; Blunk, T.; Bauer-Kreisel, P. Three-Dimensional Breast Cancer Model to Investigate CCL5/CCR1 Expression Mediated by Direct Contact between Breast Cancer Cells and Adipose-Derived Stromal Cells or Adipocytes. Cancers 2023, 15, 3501. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Mulligan, J.A.; Ouyang, Y.; Shimpi, A.A.; Williams, R.M.; Beeghly, G.F.; Hopkins, B.D.; Spector, J.A.; Adie, S.G.; Fischbach, C. Obesity-Associated Adipose Stromal Cells Promote Breast Cancer Invasion through Direct Cell Contact and ECM Remodeling. Adv. Funct. Mater. 2020, 30, 1910650. [Google Scholar] [CrossRef] [PubMed]
- Horder, H.; Guaza Lasheras, M.; Grummel, N.; Nadernezhad, A.; Herbig, J.; Ergün, S.; Teßmar, J.; Groll, J.; Fabry, B.; Bauer-Kreisel, P.; et al. Bioprinting and Differentiation of Adipose-Derived Stromal Cell Spheroids for a 3D Breast Cancer-Adipose Tissue Model. Cells 2021, 10, 803. [Google Scholar] [CrossRef] [PubMed]
- Hutmacher, D.W. Scaffolds in tissue engineering bone and cartilage. Biomaterials 2000, 21, 2529–2543. [Google Scholar] [CrossRef] [PubMed]
- Kapur, S.K.; Wang, X.; Shang, H.; Yun, S.; Li, X.; Feng, G.; Khurgel, M.; Katz, A.J. Human adipose stem cells maintain proliferative, synthetic and multipotential properties when suspension cultured as self-assembling spheroids. Biofabrication 2012, 4, 025004. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Cohen-Harazi, R.; Maizels, Y.; Koman, I. Patient-derived tumor spheroid cultures as a promising tool to assist personalized therapeutic decisions in breast cancer. Transl. Cancer Res. 2022, 11, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, D.S.; Tevis, K.M.; Blessing, W.A.; Colson, Y.L.; Zaman, M.H.; Grinstaff, M.W. Breast Cancer Spheroids Reveal a Differential Cancer Stem Cell Response to Chemotherapeutic Treatment. Sci. Rep. 2017, 7, 10382. [Google Scholar] [CrossRef]
- Huang, Z.; Yu, P.; Tang, J. Characterization of Triple-Negative Breast Cancer MDA-MB-231 Cell Spheroid Model. Onco Targets Ther. 2020, 13, 5395–5405. [Google Scholar] [CrossRef]
- Cavaco, M.; Fraga, P.; Valle, J.; Andreu, D.; Castanho, M.; Neves, V. Development of Breast Cancer Spheroids to Evaluate Cytotoxic Response to an Anticancer Peptide. Pharmaceutics 2021, 13, 1863. [Google Scholar] [CrossRef]
- Chen, G.; Liu, W.; Yan, B. Breast Cancer MCF-7 Cell Spheroid Culture for Drug Discovery and Development. J. Cancer Ther. 2022, 13, 117–130. [Google Scholar] [CrossRef]
- Tevis, K.M.; Colson, Y.L.; Grinstaff, M.W. Embedded Spheroids as Models of the Cancer Microenvironment. Adv. Biosyst. 2017, 1, 1700083. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.P.; Tsuchida, C.; Zheng, Y.; Himmelfarb, J.; Akilesh, S. A 3D Human Renal Cell Carcinoma-on-a-Chip for the Study of Tumor Angiogenesis. Neoplasia 2018, 20, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Nieskens, T.T.G.; Persson, M.; Kelly, E.J.; Sjögren, A.K. A Multicompartment Human Kidney Proximal Tubule-on-a-Chip Replicates Cell Polarization-Dependent Cisplatin Toxicity. Drug Metab. Dispos. 2020, 48, 1303–1311. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, D.; Wang, Y.; Lin, S.; Jiang, Y. A novel 3D breast-cancer-on-chip platform for therapeutic evaluation of drug delivery systems. Anal. Chim. Acta 2018, 1036, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Gioiella, F.; Urciuolo, F.; Imparato, G.; Brancato, V.; Netti, P.A. An Engineered Breast Cancer Model on a Chip to Replicate ECM-Activation In Vitro during Tumor Progression. Adv. Healthc. Mater. 2016, 5, 3074–3084. [Google Scholar] [CrossRef] [PubMed]
- Sung, K.E.; Yang, N.; Pehlke, C.; Keely, P.J.; Eliceiri, K.W.; Friedl, A.; Beebe, D.J. Transition to invasion in breast cancer: A microfluidic in vitro model enables examination of spatial and temporal effects. Integr. Biol. 2010, 3, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Belgodere, J.A.; King, C.T.; Bursavich, J.B.; Burow, M.E.; Martin, E.C.; Jung, J.P. Engineering Breast Cancer Microenvironments and 3D Bioprinting. Front. Bioeng. Biotechnol. 2018, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shi, W.; Kuss, M.; Mirza, S.; Qi, D.; Krasnoslobodtsev, A.; Zeng, J.; Band, H.; Band, V.; Duan, B. 3D Bioprinting of Breast Cancer Models for Drug Resistance Study. ACS Biomater. Sci. Eng. 2018, 4, 4401–4411. [Google Scholar] [CrossRef]
- Heinrich, M.A.; Bansal, R.; Lammers, T.; Zhang, Y.S.; Michel Schiffelers, R.; Prakash, J. 3D-Bioprinted Mini-Brain: A Glioblastoma Model to Study Cellular Interactions and Therapeutics. Adv. Mater. 2019, 31, e1806590. [Google Scholar] [CrossRef] [PubMed]
- Baka, Z.; Godier, C.; Lamy, L.; Mallick, A.; Gribova, V.; Figarol, A.; Bezdetnaya, L.; Chateau, A.; Magne, Z.; Stiefel, M.; et al. A Coculture Based, 3D Bioprinted Ovarian Tumor Model Combining Cancer Cells and Cancer Associated Fibroblasts. Macromol. Biosci. 2023, 23, e2200434. [Google Scholar] [CrossRef] [PubMed]
- Browning, J.R.; Derr, P.; Derr, K.; Doudican, N.; Michael, S.; Lish, S.R.; Taylor, N.A.; Krueger, J.G.; Ferrer, M.; Carucci, J.A.; et al. A 3D biofabricated cutaneous squamous cell carcinoma tissue model with multi-channel confocal microscopy imaging biomarkers to quantify antitumor effects of chemotherapeutics in tissue. Oncotarget 2020, 11, 2587–2596. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Hu, B.; Zhang, W.; Xie, W.; Mo, J.; Sun, H.; Shang, J. Robotic-assisted automated in situ bioprinting. Int. J. Bioprint 2023, 9, 629. [Google Scholar] [CrossRef] [PubMed]
- Santoni, S.; Gugliandolo, S.G.; Sponchioni, M.; Moscatelli, D.; Colosimo, B.M. 3D bioprinting: Current status and trends—A guide to the literature and industrial practice. Bio-Des. Manuf. 2022, 5, 14–42. [Google Scholar] [CrossRef]
- Whittle, J.R.; Lewis, M.T.; Lindeman, G.J.; Visvader, J.E. Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Res. 2015, 17, 17. [Google Scholar] [CrossRef] [PubMed]
- Abdolahi, S.; Ghazvinian, Z.; Muhammadnejad, S.; Saleh, M.; Asadzadeh Aghdaei, H.; Baghaei, K. Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J. Transl. Med. 2022, 20, 206. [Google Scholar] [CrossRef]
- Guillen, K.P.; Fujita, M.; Butterfield, A.J.; Scherer, S.D.; Bailey, M.H.; Chu, Z.; DeRose, Y.S.; Zhao, L.; Cortes-Sanchez, E.; Yang, C.-H.; et al. A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology. Nat. Cancer 2022, 3, 232–250. [Google Scholar] [CrossRef]
- Sabol, R.A.; Bowles, A.C.; Côté, A.; Wise, R.; O’Donnell, B.; Matossian, M.D.; Hossain, F.M.; Burks, H.E.; Del Valle, L.; Miele, L.; et al. Leptin produced by obesity-altered adipose stem cells promotes metastasis but not tumorigenesis of triple-negative breast cancer in orthotopic xenograft and patient-derived xenograft models. Breast Cancer Res. 2019, 21, 67. [Google Scholar] [CrossRef]
- Goto, H.; Shimono, Y.; Funakoshi, Y.; Imamura, Y.; Toyoda, M.; Kiyota, N.; Kono, S.; Takao, S.; Mukohara, T.; Minami, H. Adipose-derived stem cells enhance human breast cancer growth and cancer stem cell-like properties through adipsin. Oncogene 2019, 38, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Matossian, M.D.; Burks, H.E.; Bowles, A.C.; Elliott, S.; Hoang, V.T.; Sabol, R.A.; Pashos, N.C.; O’Donnell, B.; Miller, K.S.; Wahba, B.M.; et al. A novel patient-derived xenograft model for claudin-low triple-negative breast cancer. Breast Cancer Res. Treat. 2018, 169, 381–390. [Google Scholar] [CrossRef]
- Ginsburg, G.S.; Phillips, K.A. Precision Medicine: From Science To Value. Health Aff. 2018, 37, 694–701. [Google Scholar] [CrossRef]
- Xu, H.; Jiao, D.; Liu, A.; Wu, K. Tumor organoids: Applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 2022, 15, 58. [Google Scholar] [CrossRef]
- Bareche, Y.; Buisseret, L.; Gruosso, T.; Girard, E.; Venet, D.; Dupont, F.; Desmedt, C.; Larsimont, D.; Park, M.; Rothé, F.; et al. Unraveling Triple-Negative Breast Cancer Tumor Microenvironment Heterogeneity: Towards an Optimized Treatment Approach. JNCI J. Natl. Cancer Inst. 2019, 112, 708–719. [Google Scholar] [CrossRef]
- Neves Rebello Alves, L.; Dummer Meira, D.; Poppe Merigueti, L.; Correia Casotti, M.; do Prado Ventorim, D.; Ferreira Figueiredo Almeida, J.; Pereira de Sousa, V.; Cindra Sant’Ana, M.; Gonçalves Coutinho da Cruz, R.; Santos Louro, L.; et al. Biomarkers in Breast Cancer: An Old Story with a New End. Genes 2023, 14, 1364. [Google Scholar] [CrossRef]
- Ramani, P.K.; Sankaran, B.P. Tay-Sachs Disease. Available online: https://www.ncbi.nlm.nih.gov/books/NBK564432/ (accessed on 13 July 2023).
- Myer, H.; Chupita, S.; Jnah, A. Cystic Fibrosis: Back to the Basics. Neonatal Netw. 2023, 42, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Sedrak, A.; Sickle, K.N. Cell Disease. Available online: https://www.ncbi.nlm.nih.gov/books/NBK482384/ (accessed on 13 July 2023).
- Hamel, K.M.; King, C.T.; Cavalier, M.B.; Liimatta, K.Q.; Rozanski, G.L.; King, T.A., Jr.; Lam, M.; Bingham, G.C.; Byrne, C.E.; Xing, D.; et al. Breast Cancer-Stromal Interactions: Adipose-Derived Stromal/Stem Cell Age and Cancer Subtype Mediated Remodeling. Stem Cells Dev. 2022, 31, 604–620. [Google Scholar] [CrossRef] [PubMed]
- Hamel, K.M.; Liimatta, K.Q.; Belgodere, J.A.; Bunnell, B.A.; Gimble, J.M.; Martin, E.C. Adipose-Derived Stromal/Stem Cell Response to Tumors and Wounds: Evaluation of Patient Age. Stem Cells Dev. 2022, 31, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Byrne, C.E.; Decombe, J.B.; Bingham, G.C.; Remont, J.; Miller, L.G.; Khalif, L.; King, C.T.; Hamel, K.; Bunnell, B.A.; Burow, M.E.; et al. Evaluation of Extracellular Matrix Composition to Improve Breast Cancer Modeling. Tissue Eng. Part A 2021, 27, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic alternatives to Matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Tamayo-Angorrilla, M.; López de Andrés, J.; Jiménez, G.; Marchal, J.A. The biomimetic extracellular matrix: A therapeutic tool for breast cancer research. Transl. Res. 2022, 247, 117–136. [Google Scholar] [CrossRef]
- Börgeson, E.; Boucher, J.; Hagberg, C.E. Of mice and men: Pinpointing species differences in adipose tissue biology. Front. Cell Dev. Biol. 2022, 10, 1003118. [Google Scholar] [CrossRef]
- Emont, M.P.; Jacobs, C.; Essene, A.L.; Pant, D.; Tenen, D.; Colleluori, G.; Di Vincenzo, A.; Jørgensen, A.M.; Dashti, H.; Stefek, A.; et al. A single-cell atlas of human and mouse white adipose tissue. Nature 2022, 603, 926–933. [Google Scholar] [CrossRef] [PubMed]
- Bourin, P.; Bunnell, B.A.; Casteilla, L.; Dominici, M.; Katz, A.J.; March, K.L.; Redl, H.; Rubin, J.P.; Yoshimura, K.; Gimble, J.M. Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: A joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT). Cytotherapy 2013, 15, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Peel, S.; Jackman, M. Imaging microphysiological systems: A review. Am. J. Physiol. Cell Physiol. 2021, 320, C669–C680. [Google Scholar] [CrossRef] [PubMed]
- Mou, L.; Mandal, K.; Mecwan, M.M.; Hernandez, A.L.; Maity, S.; Sharma, S.; Herculano, R.D.; Kawakita, S.; Jucaud, V.; Dokmeci, M.R.; et al. Integrated biosensors for monitoring microphysiological systems. Lab Chip 2022, 22, 3801–3816. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, E.; Palma, C.; Vesentini, S.; Occhetta, P.; Rasponi, M. Integrating Biosensors in Organs-on-Chip Devices: A Perspective on Current Strategies to Monitor Microphysiological Systems. Biosensors 2020, 10, 110. [Google Scholar] [CrossRef]
- Shakeri, A.; Wang, Y.; Zhao, Y.; Landau, S.; Perera, K.; Lee, J.; Radisic, M. Engineering Organ-on-a-Chip Systems for Vascular Diseases. Arterioscler. Thromb. Vasc. Biol. 2023, 43, 2241–2255. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, S.; Shin, K.; Kanamori, T. Perfusion culture of endothelial cells under shear stress on microporous membrane in a pressure-driven microphysiological system. J. Biosci. Bioeng. 2023, 135, 79–85. [Google Scholar] [CrossRef]
- Zhu, H.; Özkayar, G.; Lötters, J.; Tichem, M.; Ghatkesar, M.K. Portable and integrated microfluidic flow control system using off-the-shelf components towards organs-on-chip applications. Biomed. Microdevices 2023, 25, 19. [Google Scholar] [CrossRef]
- Parent, C.; Raj Melayil, K.; Zhou, Y.; Aubert, V.; Surdez, D.; Delattre, O.; Wilhelm, C.; Viovy, J.L. Simple droplet microfluidics platform for drug screening on cancer spheroids. Lab Chip 2023, 23, 5139–5150. [Google Scholar] [CrossRef] [PubMed]
- Hockney, S.; Parker, J.; Turner, J.E.; Todd, X.; Todryk, S.; Gieling, R.G.; Hilgen, G.; Simoes, D.C.M.; Pal, D. Next generation organoid engineering to replace animals in cancer drug testing. Biochem. Pharmacol. 2023, 213, 115586. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Li, C.; Li, H.; Yi, S.; Yang, N.; Miao, K.; Deng, C.; Jia, Y.; Mak, P.I.; Martins, R.P. Cancer drug screening with an on-chip multi-drug dispenser in digital microfluidics. Lab Chip 2021, 21, 4749–4759. [Google Scholar] [CrossRef] [PubMed]
- Marx, U.; Akabane, T.; Andersson, T.B.; Baker, E.; Beilmann, M.; Beken, S.; Brendler-Schwaab, S.; Cirit, M.; David, R.; Dehne, E.M.; et al. Biology-inspired microphysiological systems to advance patient benefit and animal welfare in drug development. Altex 2020, 37, 365–394. [Google Scholar] [CrossRef]
- Mansouri, M.; Lam, J.; Sung, K.E. Progress in developing microphysiological systems for biological product assessment. Lab Chip 2024. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamel, K.M.; Frazier, T.P.; Williams, C.; Duplessis, T.; Rowan, B.G.; Gimble, J.M.; Sanchez, C.G. Adipose Tissue in Breast Cancer Microphysiological Models to Capture Human Diversity in Preclinical Models. Int. J. Mol. Sci. 2024, 25, 2728. https://doi.org/10.3390/ijms25052728
Hamel KM, Frazier TP, Williams C, Duplessis T, Rowan BG, Gimble JM, Sanchez CG. Adipose Tissue in Breast Cancer Microphysiological Models to Capture Human Diversity in Preclinical Models. International Journal of Molecular Sciences. 2024; 25(5):2728. https://doi.org/10.3390/ijms25052728
Chicago/Turabian StyleHamel, Katie M., Trivia P. Frazier, Christopher Williams, Tamika Duplessis, Brian G. Rowan, Jeffrey M. Gimble, and Cecilia G. Sanchez. 2024. "Adipose Tissue in Breast Cancer Microphysiological Models to Capture Human Diversity in Preclinical Models" International Journal of Molecular Sciences 25, no. 5: 2728. https://doi.org/10.3390/ijms25052728
APA StyleHamel, K. M., Frazier, T. P., Williams, C., Duplessis, T., Rowan, B. G., Gimble, J. M., & Sanchez, C. G. (2024). Adipose Tissue in Breast Cancer Microphysiological Models to Capture Human Diversity in Preclinical Models. International Journal of Molecular Sciences, 25(5), 2728. https://doi.org/10.3390/ijms25052728





