Establishment and Characterization of Free-Floating 3D Macrophage Programming Model in the Presence of Cancer Cell Spheroids
Abstract
1. Introduction
2. Results
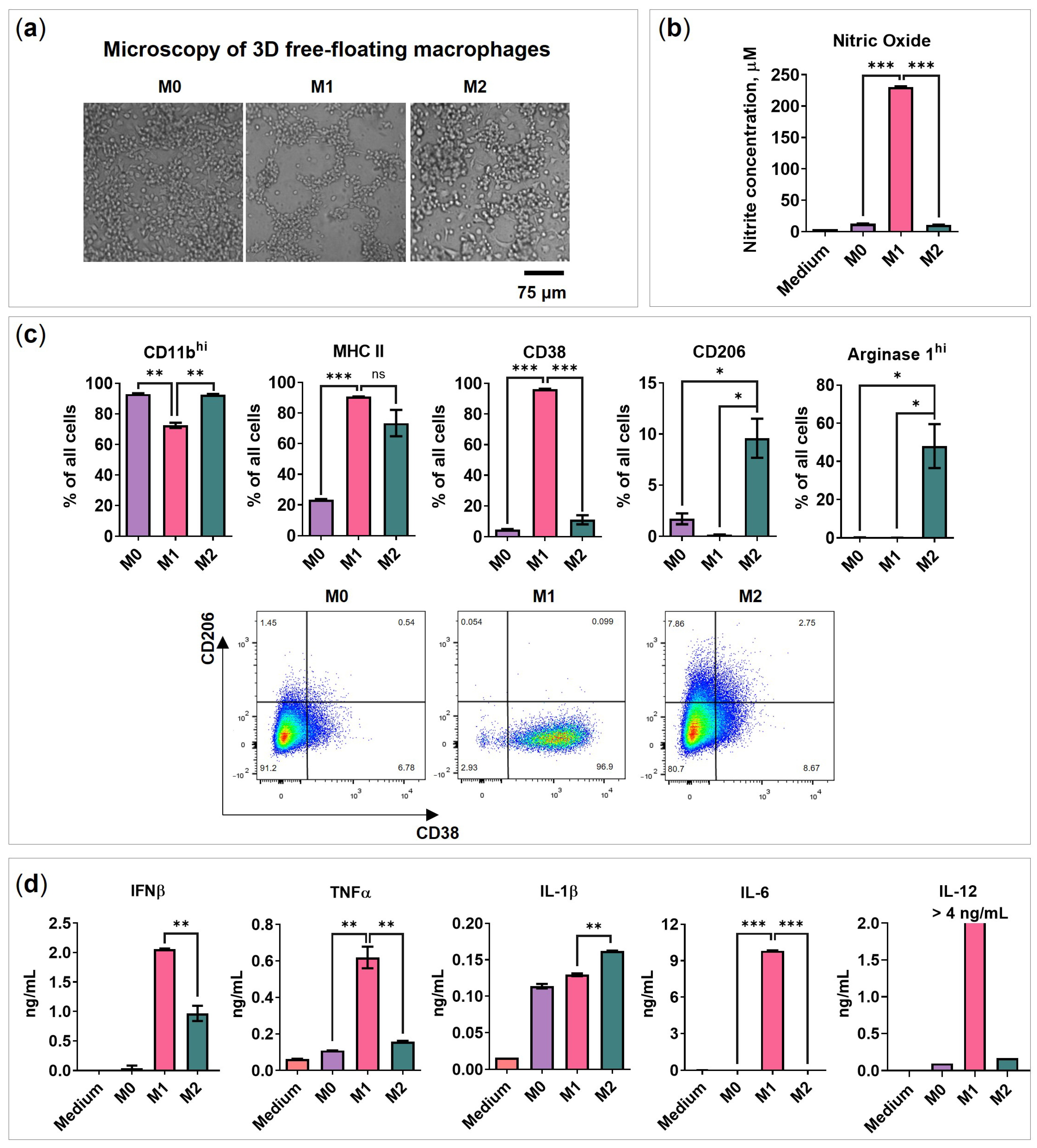
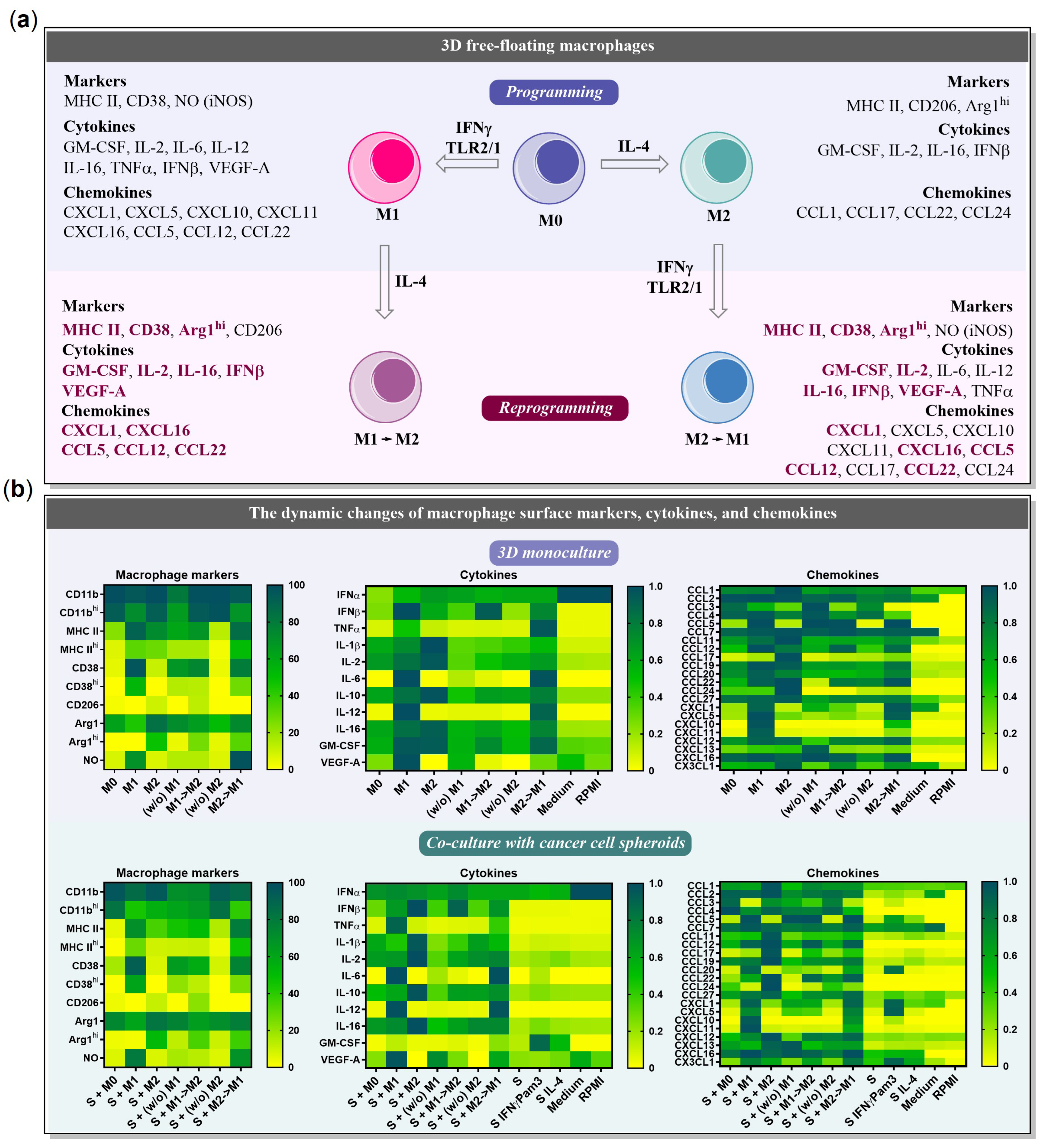
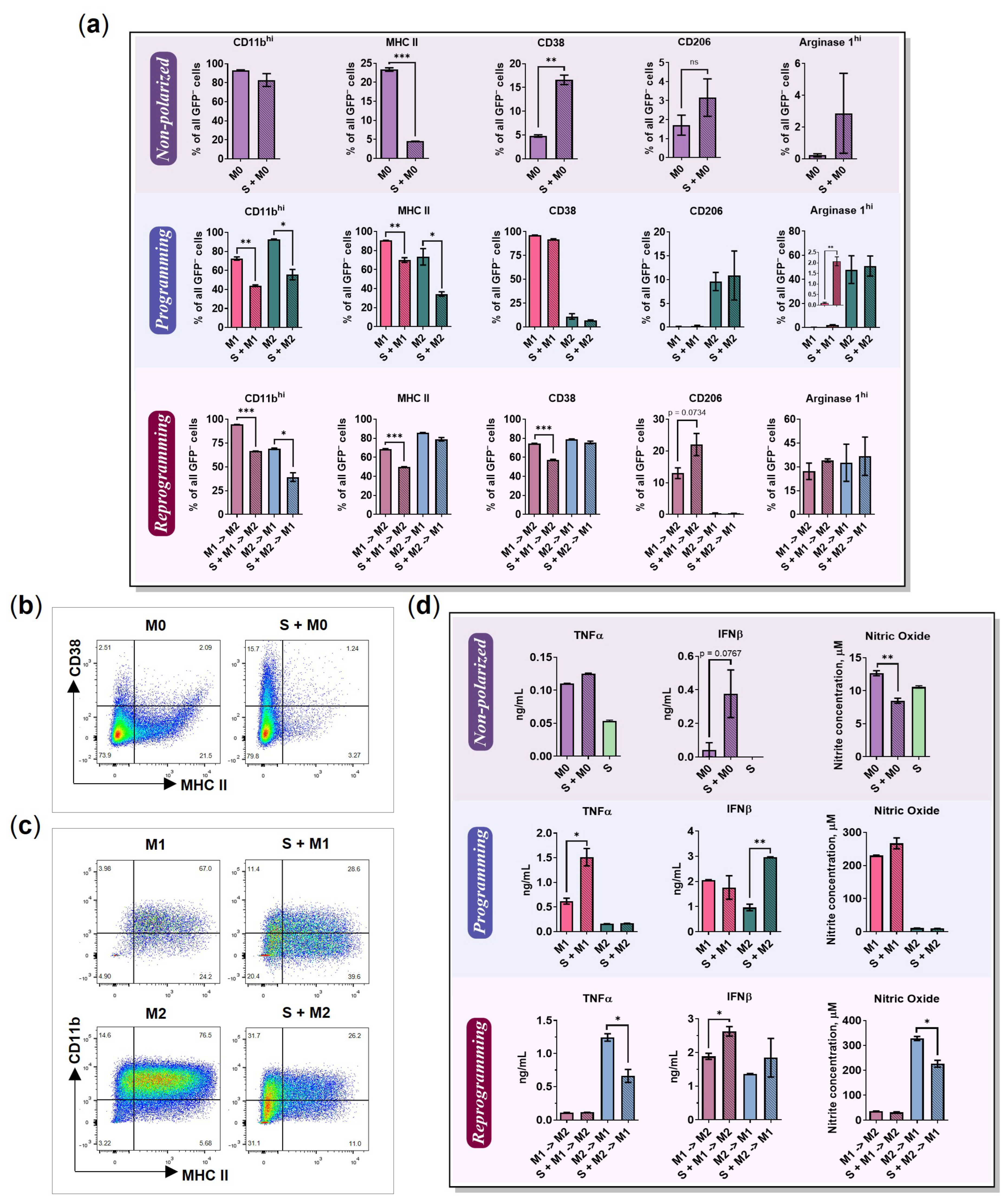
2.1. BMDMs Programming under Free-Floating 3D Conditions
2.2. BMDMs Reprogramming under Free-Floating 3D Conditions
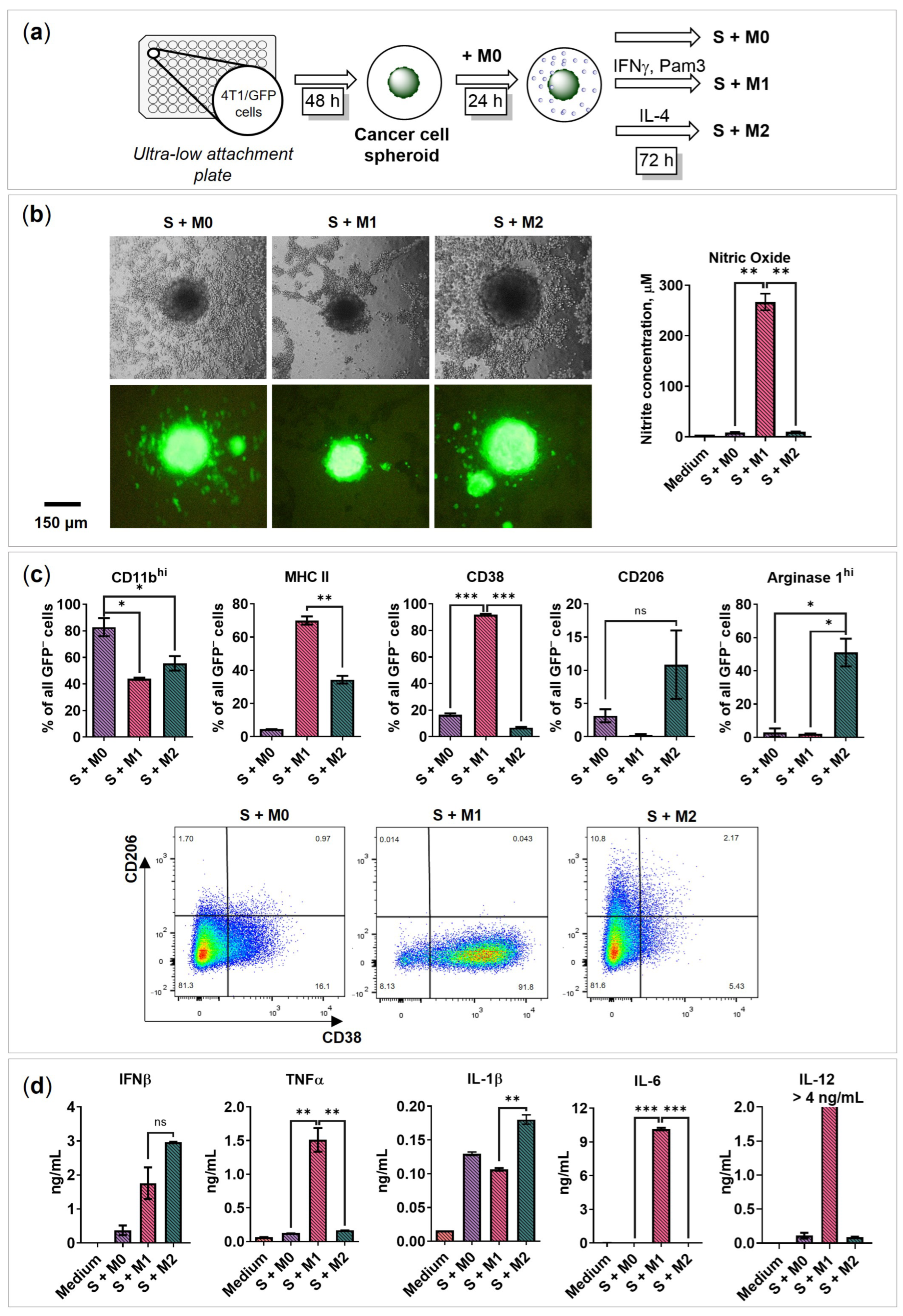
2.3. BMDMs Programming in the Presence of Cancer Cell Spheroids
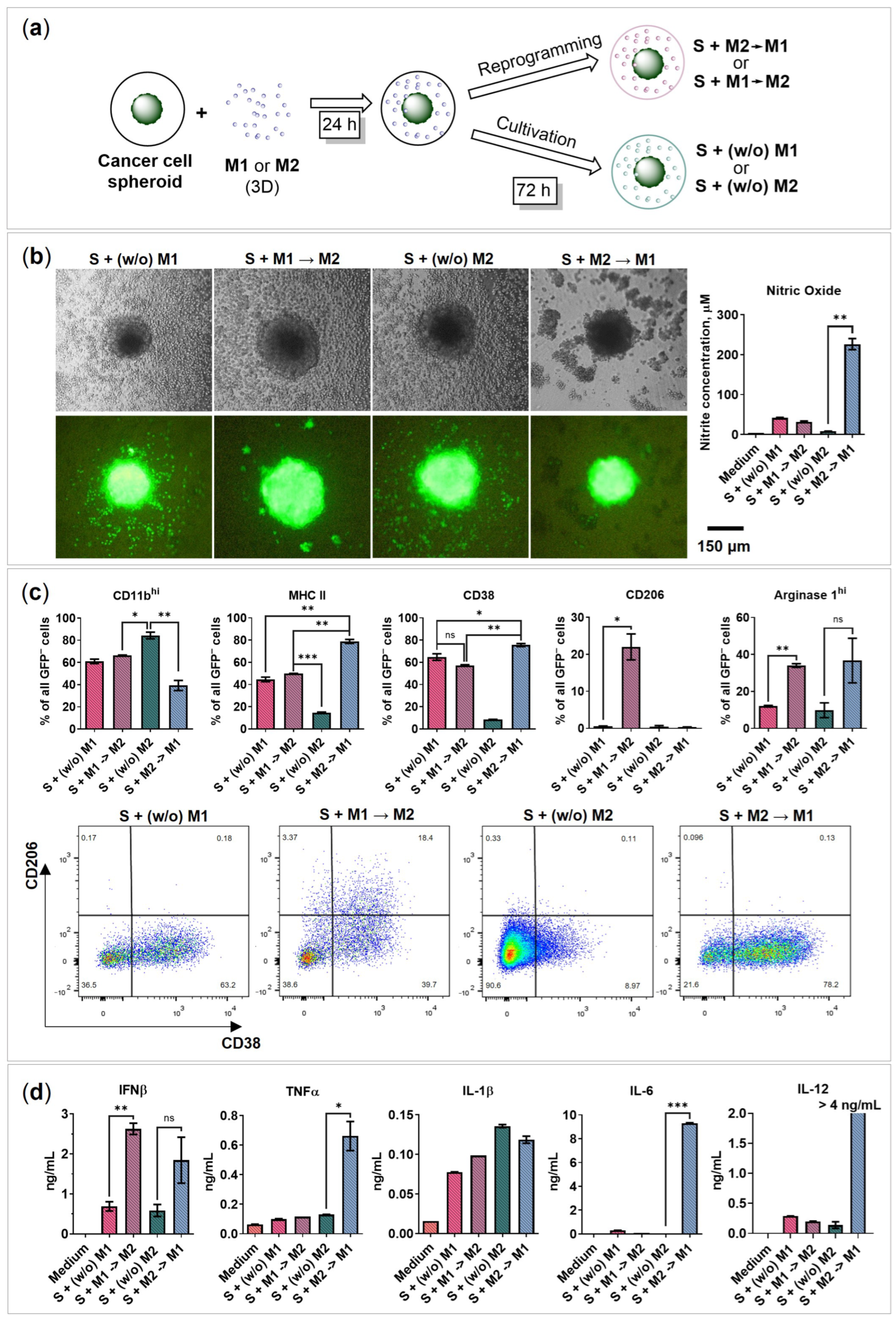
2.4. BMDMs Reprogramming in the Presence of Cancer Cell Spheroids
2.5. Chemokines Secreted by Macrophages and 4T1/GFP Spheroids
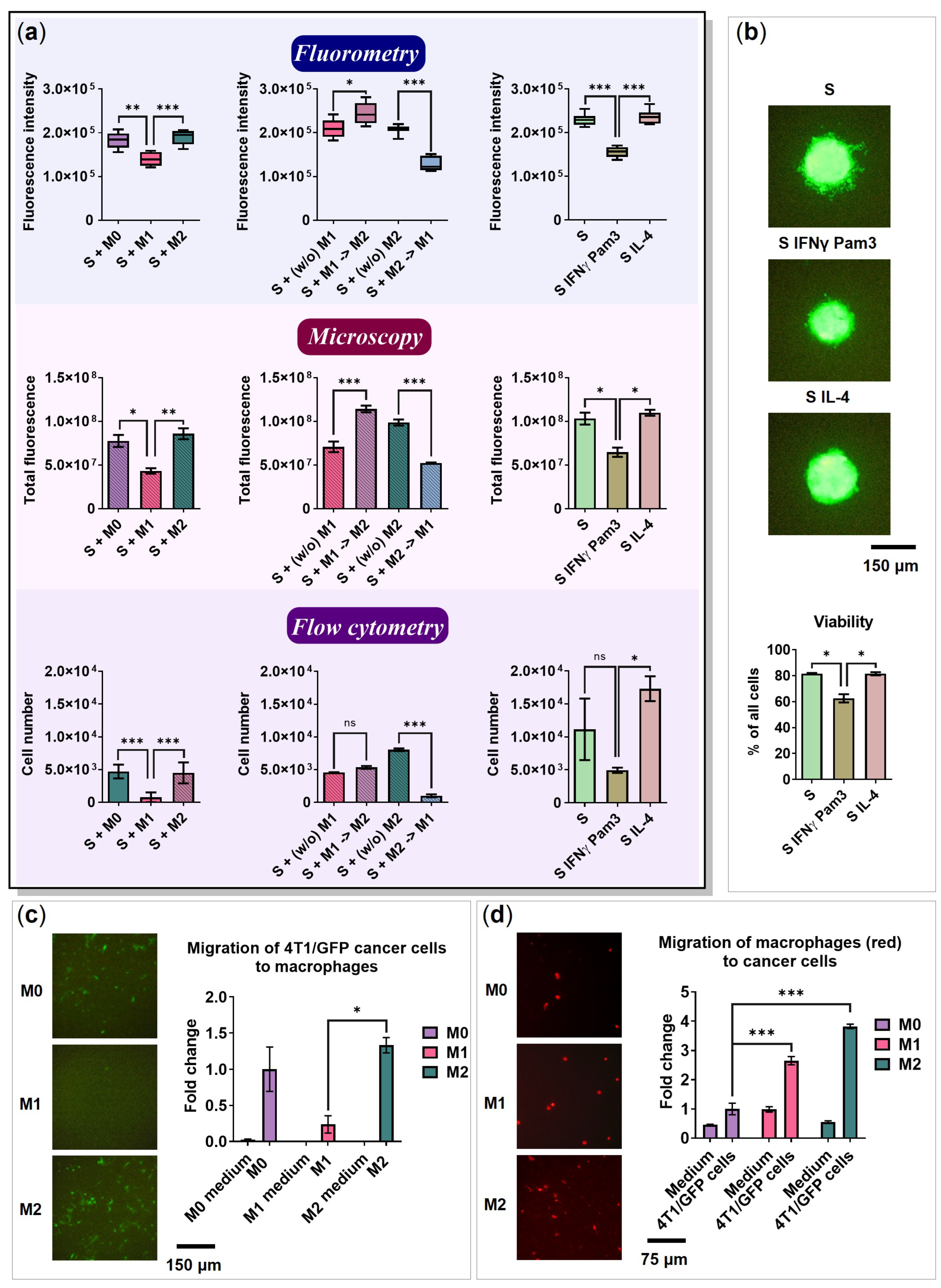
2.6. The Influence of Cancer Cell Spheroids on BMDMs
2.7. BMDMs Do Not Affect the Spheroid Size
2.8. 4T1 Cells and Macrophages Migrate toward Each Other
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Isolation and Generation of Bone Marrow-Derived Macrophages (BMDMs)
4.3. BMDMs Polarization into M0/M1/M2 Phenotype
4.4. Reprogramming of BMDMs
4.5. Formation of Spheroids
4.6. Co-Cultivation of Macrophages with the Cancer Cell Spheroid
4.7. Flow Cytometry
4.8. Analysis of Cytokines and Chemokines by ELISA and Luminex Assays
4.9. Nitric Oxide (NO) Assay
4.10. Migration Assay
4.11. Determination of Cancer Cell Spheroid Growth
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zhou, J.; Tang, Z.; Gao, S.; Li, C.; Feng, Y.; Zhou, X. Tumor-Associated Macrophages: Recent Insights and Therapies. Front. Oncol. 2020, 10, 188. [Google Scholar] [CrossRef]
- Mills, C. M1 and M2 Macrophages: Oracles of Health and Disease. Crit. Rev. Immunol. 2012, 32, 463–488. [Google Scholar] [CrossRef] [PubMed]
- Müller, E.; Speth, M.; Christopoulos, P.F.; Lunde, A.; Avdagic, A.; Øynebråten, I.; Corthay, A. Both Type I and Type II Interferons Can Activate Antitumor M1 Macrophages When Combined with TLR Stimulation. Front. Immunol. 2018, 9, 2520. [Google Scholar] [CrossRef]
- Müller, E.; Christopoulos, P.F.; Halder, S.; Lunde, A.; Beraki, K.; Speth, M.; Øynebråten, I.; Corthay, A. Toll-like Receptor Ligands and Interferon-γ Synergize for Induction of Antitumor M1 Macrophages. Front. Immunol. 2017, 8, 1383. [Google Scholar] [CrossRef]
- Jablonski, K.A.; Amici, S.A.; Webb, L.M.; De Dios Ruiz-Rosado, J.; Popovich, P.G.; Partida-Sanchez, S.; Guerau-de-Arellano, M. Novel Markers to Delineate Murine M1 and M2 Macrophages. PLoS ONE 2015, 10, e0145342. [Google Scholar] [CrossRef]
- Jaguin, M.; Houlbert, N.; Fardel, O.; Lecureur, V. Polarization Profiles of Human M-CSF-Generated Macrophages and Comparison of M1-Markers in Classically Activated Macrophages from GM-CSF and M-CSF Origin. Cell. Immunol. 2013, 281, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Amici, S.A.; Young, N.A.; Narvaez-Miranda, J.; Jablonski, K.A.; Arcos, J.; Rosas, L.; Papenfuss, T.L.; Torrelles, J.B.; Jarjour, W.N.; Guerau-de-Arellano, M.; et al. CD38 Is Robustly Induced in Human Macrophages and Monocytes in Inflammatory Conditions. Front. Immunol. 2018, 9, 1593. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Sanson, M.; Distel, E.; Fisher, E.A. HDL Induces the Expression of the M2 Macrophage Markers Arginase 1 and Fizz-1 in a STAT6-Dependent Process. PLoS ONE 2013, 8, e74676. [Google Scholar] [CrossRef]
- Georgoudaki, A.-M.; Prokopec, K.E.; Boura, V.F.; Hellqvist, E.; Sohn, S.; Östling, J.; Dahan, R.; Harris, R.A.; Rantalainen, M.; Klevebring, D.; et al. Reprogramming Tumor-Associated Macrophages by Antibody Targeting Inhibits Cancer Progression and Metastasis. Cell Rep. 2016, 15, 2000–2011. [Google Scholar] [CrossRef]
- Watkins, S.K.; Egilmez, N.K.; Suttles, J.; Stout, R.D. IL-12 Rapidly Alters the Functional Profile of Tumor-Associated and Tumor-Infiltrating Macrophages In Vitro and In Vivo. J. Immunol. 2007, 178, 1357–1362. [Google Scholar] [CrossRef]
- Johansson, A.; Hamzah, J.; Payne, C.J.; Ganss, R. Tumor-Targeted TNFα Stabilizes Tumor Vessels and Enhances Active Immunotherapy. Proc. Natl. Acad. Sci. USA 2012, 109, 7841–7846. [Google Scholar] [CrossRef] [PubMed]
- Spiller, K.L.; Nassiri, S.; Witherel, C.E.; Anfang, R.R.; Ng, J.; Nakazawa, K.R.; Yu, T.; Vunjak-Novakovic, G. Sequential Delivery of Immunomodulatory Cytokines to Facilitate the M1-to-M2 Transition of Macrophages and Enhance Vascularization of Bone Scaffolds. Biomaterials 2015, 37, 194–207. [Google Scholar] [CrossRef]
- Kurena, B.; Müller, E.; Christopoulos, P.F.; Johnsen, I.B.; Stankovic, B.; Øynebråten, I.; Corthay, A.; Zajakina, A. Generation and Functional In Vitro Analysis of Semliki Forest Virus Vectors Encoding TNF-α and IFN-γ. Front. Immunol. 2017, 8, 1667. [Google Scholar] [CrossRef] [PubMed]
- Trofimova, O.; Korotkaja, K.; Skrastina, D.; Jansons, J.; Spunde, K.; Isaguliants, M.; Zajakina, A. Alphavirus-Driven Interferon Gamma (IFNg) Expression Inhibits Tumor Growth in Orthotopic 4T1 Breast Cancer Model. Vaccines 2021, 9, 1247. [Google Scholar] [CrossRef] [PubMed]
- Sherman, H.; Gitschier, H.J.; Rossi, A.E. A Novel Three-Dimensional Immune Oncology Model for High-Throughput Testing of Tumoricidal Activity. Front. Immunol. 2018, 9, 857. [Google Scholar] [CrossRef]
- Boutin, M.E.; Voss, T.C.; Titus, S.A.; Cruz-Gutierrez, K.; Michael, S.; Ferrer, M. A High-Throughput Imaging and Nuclear Segmentation Analysis Protocol for Cleared 3D Culture Models. Sci. Rep. 2018, 8, 11135. [Google Scholar] [CrossRef]
- Nyga, A.; Neves, J.; Stamati, K.; Loizidou, M.; Emberton, M.; Cheema, U. The next Level of 3D Tumour Models: Immunocompetence. Drug Discov. Today 2016, 21, 1421–1428. [Google Scholar] [CrossRef]
- Klinder, A.; Markhoff, J.; Jonitz-Heincke, A.; Sterna, P.; Salamon, A.; Bader, R. Comparison of Different Cell Culture Plates for the Enrichment of Non-adherent Human Mononuclear Cells. Exp. Ther. Med. 2019, 17, 2004–2012. [Google Scholar] [CrossRef]
- Rostam, H.M.; Reynolds, P.M.; Alexander, M.R.; Gadegaard, N.; Ghaemmaghami, A.M. Image Based Machine Learning for Identification of Macrophage Subsets OPEN. Sci. Rep. 2017, 7, 3521. [Google Scholar] [CrossRef]
- Feng, L.; Song, P.; Zhou, H.; Li, A.; Ma, Y.; Zhang, X.; Liu, H.; Xu, G.; Zhou, Y.; Wu, X.; et al. Pentamethoxyflavanone Regulates Macrophage Polarization and Ameliorates Sepsis in Mice. Biochem. Pharmacol. 2014, 89, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, H.; Mao, C.; Sun, M.; Dominah, G.; Chen, L.; Zhuang, Z. Fatty Acid Oxidation Contributes to IL-1β Secretion in M2 Macrophages and Promotes Macrophage-Mediated Tumor Cell Migration. Mol. Immunol. 2019, 94, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zheng, S.; Liu, T.; Liu, Q.; Chen, Y.; Tan, D.; Ma, R.; Lu, X. IL-1β from M2 Macrophages Promotes Migration and Invasion of ESCC Cells Enhancing Epithelial-Mesenchymal Transition and Activating NF-ΚB Signaling Pathway. J. Cell. Biochem. 2018, 119, 7040–7052. [Google Scholar] [CrossRef] [PubMed]
- Jetten, N.; Roumans, N.; Gijbels, M.J.; Romano, A.; Post, M.J.; de Winther, M.P.J.; Van Der Hulst, R.R.W.J.; Xanthoulea, S. Wound Administration of M2-Polarized Macrophages Does Not Improve Murine Cutaneous Healing Responses. PLoS ONE 2014, 9, e102994. [Google Scholar] [CrossRef]
- Ogasawara, S.; Yano, H.; Momosaki, S.; Jun, A.; Nishida, N.; Kojiro, S.; Moriya, F.; Ishizaki, H.; Kuratomi, K.; Kojiro, M. Growth Inhibitory Effects of IFN-β on Human Liver Cancer Cells in Vitro and in Vivo. J. Interferon Cytokine Res. 2007, 27, 507–516. [Google Scholar] [CrossRef]
- Kumaran Satyanarayanan, S.; El Kebir, D.; Soboh, S.; Butenko, S.; Sekheri, M.; Saadi, J.; Peled, N.; Assi, S.; Othman, A.; Schif-Zuck, S.; et al. IFN-β Is a Macrophage-Derived Effector Cytokine Facilitating the Resolution of Bacterial Inflammation. Nat. Commun. 2019, 10, 3471. [Google Scholar] [CrossRef]
- Vogel, D.Y.S.; Heijnen, P.D.A.M.; Breur, M.; de Vries, H.E.; Tool, A.T.J.; Amor, S.; Dijkstra, C.D. Macrophages Migrate in an Activation-Dependent Manner to Chemokines Involved in Neuroinflammation. J. Neuroinflamm. 2014, 11, 23. [Google Scholar] [CrossRef]
- Jenkins, S.J.; Ruckerl, D.; Thomas, G.D.; Hewitson, J.P.; Duncan, S.; Brombacher, F.; Maizels, R.M.; Hume, D.A.; Allen, J.E. IL-4 Directly Signals Tissue-Resident Macrophages to Proliferate beyond Homeostatic Levels Controlled by CSF-1. J. Exp. Med. 2013, 210, 2477–2491. [Google Scholar] [CrossRef]
- Goldberg, M.; Belkowski, L.S.; Bloom, B.R. Regulation of Macrophage Growth and Antiviral Activity by Interferon-γ. J. Cell Biol. 1989, 109, 1331–1340. [Google Scholar] [CrossRef]
- Stout, R.D.; Jiang, C.; Matta, B.; Tietzel, I.; Watkins, S.K.; Suttles, J. Macrophages Sequentially Change Their Functional Phenotype in Response to Changes in Microenvironmental Influences. J. Immunol. 2005, 175, 342–349. [Google Scholar] [CrossRef]
- Stout, R.D.; Suttles, J. Functional Plasticity of Macrophages: Reversible Adaptation to Changing Microenvironments. J. Leukoc. Biol. 2004, 76, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Tarique, A.A.; Logan, J.; Thomas, E.; Holt, P.G.; Sly, P.D.; Fantino, E. Phenotypic, Functional, and Plasticity Features of Classical and Alternatively Activated Human Macrophages. Am. J. Respir. Cell Mol. Biol. 2015, 53, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, T.; Imamichi, T.; Weiss, J.M.; Sato, M.; Li, L.; Matsukawa, A.; Wang, J.M. Induction of Monocyte Chemoattractant Proteins in Macrophages via the Production of Granulocyte/Macrophage Colony-Stimulating Factor by Breast Cancer Cells. Front. Immunol. 2016, 7, 2. [Google Scholar] [CrossRef]
- Aldinucci, D.; Borghese, C.; Casagrande, N. The CCL5/CCR5 Axis in Cancer Progression. Cancers 2020, 12, 1765. [Google Scholar] [CrossRef]
- Hsu, Y.-L.; Hou, M.-F.; Kuo, P.-L.; Huang, Y.-F.; Tsai, E.-M. Breast Tumor-Associated Osteoblast-Derived CXCL5 Increases Cancer Progression by ERK/MSK1/Elk-1/Snail Signaling Pathway. Oncogene 2013, 32, 4436–4447. [Google Scholar] [CrossRef] [PubMed]
- Rego, S.L.; Helms, R.S.; Dréau, D. Breast Tumor Cell TACE-Shed MCSF Promotes pro-Angiogenic Macrophages through NF-ΚB Signaling. Angiogenesis 2014, 17, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Fluhr, H.; Seitz, T.; Zygmunt, M. Heparins Modulate the IFN-γ-Induced Production of Chemokines in Human Breast Cancer Cells. Breast Cancer Res. Treat. 2013, 137, 109–118. [Google Scholar] [CrossRef]
- Spiller, K.L.; Anfang, R.R.; Spiller, K.J.; Ng, J.; Nakazawa, K.R.; Daulton, J.W.; Vunjak-Novakovic, G. The Role of Macrophage Phenotype in Vascularization of Tissue Engineering Scaffolds. Biomaterials 2014, 35, 4477–4488. [Google Scholar] [CrossRef]
- Movahedi, K.; Laoui, D.; Gysemans, C.; Baeten, M.; Stangé, G.; Van den Bossche, J.; Mack, M.; Pipeleers, D.; Veld, P.I.; De Baetselier, P.; et al. Different Tumor Microenvironments Contain Functionally Distinct Subsets of Macrophages Derived from Ly6C(High) Monocytes. Cancer Res. 2010, 70, 5728–5739. [Google Scholar] [CrossRef]
- Gobert, M.; Treilleux, I.; Bendriss-Vermare, N.; Bachelot, T.; Goddard-Leon, S.; Arfl, V.; Biota, C.; Doffin, A.C.; Durand, I.; Olive, D.; et al. Regulatory T Cells Recruited through CCL22/CCR4 Are Selectively Activated in Lymphoid Infiltrates Surrounding Primary Breast Tumors and Lead to an Adverse Clinical Utcome. Cancer Res. 2009, 69, 2000–2009. [Google Scholar] [CrossRef]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific Recruitment of Regulatory T Cells in Ovarian Carcinoma Fosters Immune Privilege and Predicts Reduced Survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef]
- Mantovani, A.; Savino, B.; Locati, M.; Zammataro, L.; Allavena, P.; Bonecchi, R. The Chemokine System in Cancer Biology and Therapy. Cytokine Growth Factor Rev. 2010, 21, 27–39. [Google Scholar] [CrossRef]
- Konen, J.M.; Fradette, J.J.; Gibbons, D.L. The Good, the Bad and the Unknown of CD38 in the Metabolic Microenvironment and Immune Cell Functionality of Solid Tumors. Cells 2019, 9, 52. [Google Scholar] [CrossRef]
- Madera, L.; Greenshields, A.; Power Coombs, M.R.; Hoskin, D.W. 4T1 Murine Mammary Carcinoma Cells Enhance Macrophage-Mediated Innate Inflammatory Responses. PLoS ONE 2015, 10, e0133385. [Google Scholar] [CrossRef] [PubMed]
- Hollmén, M.; Roudnicky, F.; Karaman, S.; Detmar, M. Characterization of Macrophage-Cancer Cell Crosstalk in Estrogen Receptor Positive and Triple-Negative Breast Cancer. Sci. Rep. 2015, 5, 9188. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, W.; Zheng, Y.; Wang, S.; Yang, B.; Li, M.; Song, J.; Zhang, F.; Zhang, X.; Wang, Q.; et al. CXCL1 Derived from Tumor-Associated Macrophages Promotes Breast Cancer Metastasis via Activating NF-ΚB/SOX4 Signaling. Cell Death Dis. 2018, 9, 880. [Google Scholar] [CrossRef]
- Hagemann, T.; Wilson, J.; Burke, F.; Kulbe, H.; Li, N.F.; Plüddemann, A.; Charles, K.; Gordon, S.; Balkwill, F.R. Ovarian Cancer Cells Polarize Macrophages Toward A Tumor-Associated Phenotype. J. Immunol. 2006, 176, 5023–5032. [Google Scholar] [CrossRef]
- Olkhanud, P.B.; Baatar, D.; Bodogai, M.; Hakim, F.; Gress, R.; Anderson, R.L.; Deng, J.; Xu, M.; Briest, S.; Biragyn, A. Breast Cancer Lung Metastasis Requires Expression of Chemokine Receptor CCR4 and Regulatory T Cells. Cancer Res 2009, 69, 5996–6004. [Google Scholar] [CrossRef]
- Yu, P.F.; Huang, Y.; Xu, C.L.; Lin, L.Y.; Han, Y.Y.; Sun, W.H.; Hu, G.H.; Rabson, A.B.; Wang, Y.; Shi, Y.F. Downregulation of CXCL12 in Mesenchymal Stromal Cells by TGFβ Promotes Breast Cancer Metastasis. Oncogene 2017, 36, 840–849. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, N.; Li, Q.; Zhang, W.; Ke, F.; Leng, Q.; Wang, H.; Chen, J.; Wang, H. Tumor-Associated Macrophages Recruit CCR6+ Regulatory T Cells and Promote the Development of Colorectal Cancer via Enhancing CCL20 Production in Mice. PLoS ONE 2011, 6, e19495. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Lu, J. Interferon Gamma in Cancer Immunotherapy. Cancer Med. 2018, 7, 4509–4516. [Google Scholar] [CrossRef]
- Lee, C.-C.; Lin, J.-C.; Hwang, W.-L.; Kuo, Y.-J.; Chen, H.-K.; Tai, S.-K.; Lin, C.-C.; Yang, M.-H. Macrophage-Secreted Interleukin-35 Regulates Cancer Cell Plasticity to Facilitate Metastatic Colonization. Nat. Commun. 2018, 9, 3763. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Qu, Q.; Zheng, B.; Xiong, S.; Fan, G.-H. The Chemotaxis of M1 and M2 Macrophages Is Regulated by Different Chemokines. J. Leukoc. Biol. 2015, 97, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Gerrick, K.Y.; Gerrick, E.R.; Gupta, A.; Wheelan, S.J.; Yegnasubramanian, S.; Jaffee, E.M. Transcriptional Profiling Identifies Novel Regulators of Macrophage Polarization. PLoS ONE 2018, 13, e0208602. [Google Scholar] [CrossRef] [PubMed]
- Genin, M.; Clement, F.; Fattaccioli, A.; Raes, M.; Michiels, C. M1 and M2 Macrophages Derived from THP-1 Cells Differentially Modulate the Response of Cancer Cells to Etoposide. BMC Cancer 2015, 15, 577. [Google Scholar] [CrossRef]
- Delprat, V.; Tellier, C.; Demazy, C.; Raes, M.; Feron, O.; Michiels, C. Cycling Hypoxia Promotes a Pro-Inflammatory Phenotype in Macrophages via JNK/P65 Signaling Pathway. Sci. Rep. 2020, 10, 882. [Google Scholar] [CrossRef]
- Müller, J.; Von Bernstorff, W.; Heidecke, C.D.; Schulze, T. Differential S1P Receptor Profiles on M1- and M2-Polarized Macrophages Affect Macrophage Cytokine Production and Migration. BioMed Res. Int. 2017, 2017, 7584621. [Google Scholar] [CrossRef]
- Rolny, C.; Mazzone, M.; Tugues, S.; Laoui, D.; Johansson, I.; Coulon, C.; Squadrito, M.L.; Segura, I.; Li, X.; Knevels, E.; et al. HRG Inhibits Tumor Growth and Metastasis by Inducing Macrophage Polarization and Vessel Normalization through Downregulation of PlGF. Cancer Cell 2011, 19, 31–44. [Google Scholar] [CrossRef]
- Li, C.; Levin, M.; Kaplan, D.L. Bioelectric Modulation of Macrophage Polarization. Sci. Rep. 2016, 6, 21044. [Google Scholar] [CrossRef]
- Geraghty, T.; Rajagopalan, A.; Aslam, R.; Pohlman, A.; Venkatesh, I.; Zloza, A.; Cimbaluk, D.; DeNardo, D.G.; Gupta, V. Positive Allosteric Modulation of CD11b as a Novel Therapeutic Strategy Against Lung Cancer. Front. Oncol. 2020, 10, 748. [Google Scholar] [CrossRef]
- Lizotte, P.H.; Baird, J.R.; Stevens, C.A.; Lauer, P.; Green, W.R.; Brockstedt, D.G.; Fiering, S.N. Attenuated Listeria monocytogenes Reprograms M2-Polarized Tumor-Associated Macrophages in Ovarian Cancer Leading to INOS-Mediated Tumor Cell Lysis. Oncoimmunology 2014, 3, e28926. [Google Scholar] [CrossRef]
- Bonaventura, P.; Shekarian, T.; Alcazer, V.; Valladeau-Guilemond, J.; Valsesia-Wittmann, S.; Amigorena, S.; Caux, C.; Depil, S. Cold Tumors: A Therapeutic Challenge for Immunotherapy. Front. Immunol. 2019, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Spunde, K.; Korotkaja, K.; Zajakina, A. Recombinant Viral Vectors for Therapeutic Programming of Tumour Microenvironment: Advantages and Limitations. Biomedicines 2022, 10, 2142. [Google Scholar] [CrossRef] [PubMed]
- Arnaout, M.A. Biology and Structure of Leukocyte Β2 Integrins and Their Role in Inflammation. F1000Research 2016, 5, 2433. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Jin, X.; Liao, Q.; Chen, Z.; Peng, H.; Zhou, Y. CD38: A Significant Regulator of Macrophage Function. Front. Oncol. 2022, 12, 775649. [Google Scholar] [CrossRef]
- Karakasheva, T.A.; Waldron, T.J.; Eruslanov, E.; Kim, S.-B.; Lee, J.-S.; O’Brien, S.; Hicks, P.D.; Basu, D.; Singhal, S.; Malavasi, F.; et al. Microenvironment and Immunology CD38-Expressing Myeloid-Derived Suppressor Cells Promote Tumor Growth in a Murine Model of Esophageal Cancer. Cancer Res. 2015, 75, 4074–4085. [Google Scholar] [CrossRef]
- La Fleur, L.; Botling, J.; He, F.; Pelicano, C.; Zhou, C.; He, C.; Palano, G.; Mezheyeuski, A.; Micke, P.; Ravetch, J.V.; et al. Targeting MARCO and IL37R on Immunosuppressive Macrophages in Lung Cancer Blocks Regulatory T Cells and Supports Cytotoxic Lymphocyte Function. Cancer Res. 2021, 81, 956–967. [Google Scholar] [CrossRef]
- Wu, T.; Yang, W.; Sun, A.; Wei, Z.; Lin, Q. The Role of CXC Chemokines in Cancer Progression. Cancers 2022, 15, 167. [Google Scholar] [CrossRef] [PubMed]
- Unver, N. Macrophage Chemoattractants Secreted by Cancer Cells: Sculptors of the Tumor Microenvironment and Another Crucial Piece of the Cancer Secretome as a Therapeutic Target. Cytokine Growth Factor Rev. 2019, 50, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Velázquez, M.; Pestell, R.G. The CCL5/CCR5 Axis Promotes Metastasis in Basal Breast Cancer. Oncoimmunology 2013, 2, e23660. [Google Scholar] [CrossRef]
- Qian, B.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Liam, R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 Recruits Inflammatory Monocytes to Facilitate Breast Tumor Metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Huang, K. CCL11 Increases the Proportion of CD4+CD25+Foxp3+ Treg Cells and the Production of IL-2 and TGF-β by CD4+ T Cells via the STAT5 Signaling Pathway. Mol. Med. Rep. 2020, 21, 2522–2532. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Ito, F.; Nakazawa, H.; Horita, S.; Osaka, Y.; Toma, H. High Expression of Chemokine Gene as a Favorable Prognostic Factor in Renal Cell Carcinoma. J. Urol. 2004, 171, 2171–2175. [Google Scholar] [CrossRef] [PubMed]
- Reschke, R.; Yu, J.; Flood, B.A.; Higgs, E.F.; Hatogai, K.; Gajewski, T.F. Immune Cell and Tumor Cell-Derived CXCL10 Is Indicative of Immunotherapy Response in Metastatic Melanoma. J. Immunother. Cancer 2021, 9, e003521. [Google Scholar] [CrossRef] [PubMed]
- Kadomoto, S.; Izumi, K.; Hiratsuka, K.; Nakano, T.; Naito, R.; Makino, T.; Iwamoto, H.; Yaegashi, H.; Shigehara, K.; Kadono, Y.; et al. Tumor-Associated Macrophages Induce Migration of Renal Cell Carcinoma Cells via Activation of the CCL20-CCR6 Axis. Cancers 2020, 12, 89. [Google Scholar] [CrossRef]
- Zou, A.; Lambert, D.; Yeh, H.; Yasukawa, K.; Behbod, F.; Fan, F.; Cheng, N. Elevated CXCL1 Expression in Breast Cancer Stroma Predicts Poor Prognosis and Is Inversely Associated with Expression of TGF-β Signaling Proteins. BMC Cancer 2014, 14, 781. [Google Scholar] [CrossRef] [PubMed]
- von Hundelshausen, P.; Agten, S.M.; Eckardt, V.; Blanchet, X.; Schmitt, M.M.; Ippel, H.; Neideck, C.; Bidzhekov, K.; Leberzammer, J.; Wichapong, K.; et al. Chemokine Interactome Mapping Enables Tailored Intervention in Acute and Chronic Inflammation. Sci. Transl. Med. 2017, 9, eaah6650. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and Tumor Progression: Signaling Pathways and Targeted Intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Gundra, U.M.; Girgis, N.M.; Ruckerl, D.; Jenkins, S.; Ward, L.N.; Kurtz, Z.D.; Wiens, K.E.; Tang, M.S.; Basu-Roy, U.; Mansukhani, A.; et al. Alternatively Activated Macrophages Derived from Monocytes and Tissue Macrophages Are Phenotypically and Functionally Distinct. Blood 2014, 123, e110–e122. [Google Scholar] [CrossRef]
- Jain, S.; Dash, P.; Minz, A.P.; Satpathi, S.; Samal, A.G.; Behera, P.K.; Satpathi, P.S.; Senapati, S. Lipopolysaccharide (LPS) Enhances Prostate Cancer Metastasis Potentially through NF-ΚB Activation and Recurrent Dexamethasone Administration Fails to Suppress It in Vivo. Prostate 2019, 79, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Qian, S.; Zhang, J.; Feng, J.; Luo, K.; Sun, L.L.; Zhao, L.; Ran, Y.; Sun, L.L.; Wang, J.; et al. Lipopolysaccharide Promotes Metastasis via Acceleration of Glycolysis by the Nuclear Factor-ΚB/Snail/Hexokinase3 Signaling Axis in Colorectal Cancer. Cancer Metab. 2021, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Horuluoglu, B.; Bayik, D.; Kayraklioglu, N.; Goguet, E.; Kaplan, M.J.; Klinman, D.M. PAM3 Supports the Generation of M2-like Macrophages from Lupus Patient Monocytes and Improves Disease Outcome in Murine Lupus. J. Autoimmun. 2019, 99, 24–32. [Google Scholar] [CrossRef] [PubMed]



| Group | Macrophage Markers | Cytokines | Chemokines | |
|---|---|---|---|---|
| S + M0 | upregulated | CD38 ↑ | IFNβ ↑ | CXCL1 ↑ |
| downregulated | MHC II ↓ | NO (iNOS) ↓ | CCL22↓, CXCL12↓, CXCL16 ↓ | |
| S + M1 | upregulated | Arg1hi ↑ | TNFα ↑ | CCL4 ↑, CCL12 ↑, CCL22 ↑ |
| downregulated | MHC II ↓, CD11bhi ↓ | IL-1β ↓ | CCL24 ↓, CXCL11 ↓ | |
| S + M2 | upregulated | ND | IFNβ ↑ | CCL3 ↑, CCL12 ↑, CXCL1 ↑ |
| downregulated | MHC II ↓, CD11bhi ↓ | IL-12 ↓ | CXCL16 ↓ | |
| S + (w/o) M1 | upregulated | ND | ND | CCL20 ↑, CXCL1 ↑ |
| downregulated | ND | ND | CCL3 ↓, CXCL13 ↓ | |
| S + M1→M2 | upregulated | CD206 ↑ | IFNβ ↑ | CCL17 ↑, CCL20 ↑, CCL22 ↑, CCL24 ↑, CXCL1 ↑ |
| downregulated | MHC II ↓, CD11bhi ↓, CD38 ↓ | ND | ND | |
| S + (w/o) M2 | upregulated | ND | IL-1β ↑, IFNβ ↑, TNFα ↑ | CCL24 ↑, CXCL1 ↑ |
| downregulated | ND | ND | ND | |
| S + M2→M1 | upregulated | ND | ND | CCL17 ↑, CCL24 ↑ |
| downregulated | CD11bhi ↓ | NO (iNOS) ↓, TNFα ↓ | ND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korotkaja, K.; Jansons, J.; Spunde, K.; Rudevica, Z.; Zajakina, A. Establishment and Characterization of Free-Floating 3D Macrophage Programming Model in the Presence of Cancer Cell Spheroids. Int. J. Mol. Sci. 2023, 24, 10763. https://doi.org/10.3390/ijms241310763
Korotkaja K, Jansons J, Spunde K, Rudevica Z, Zajakina A. Establishment and Characterization of Free-Floating 3D Macrophage Programming Model in the Presence of Cancer Cell Spheroids. International Journal of Molecular Sciences. 2023; 24(13):10763. https://doi.org/10.3390/ijms241310763
Chicago/Turabian StyleKorotkaja, Ksenija, Juris Jansons, Karina Spunde, Zhanna Rudevica, and Anna Zajakina. 2023. "Establishment and Characterization of Free-Floating 3D Macrophage Programming Model in the Presence of Cancer Cell Spheroids" International Journal of Molecular Sciences 24, no. 13: 10763. https://doi.org/10.3390/ijms241310763
APA StyleKorotkaja, K., Jansons, J., Spunde, K., Rudevica, Z., & Zajakina, A. (2023). Establishment and Characterization of Free-Floating 3D Macrophage Programming Model in the Presence of Cancer Cell Spheroids. International Journal of Molecular Sciences, 24(13), 10763. https://doi.org/10.3390/ijms241310763









