Bioengineered Enzymes and Precision Fermentation in the Food Industry
Abstract
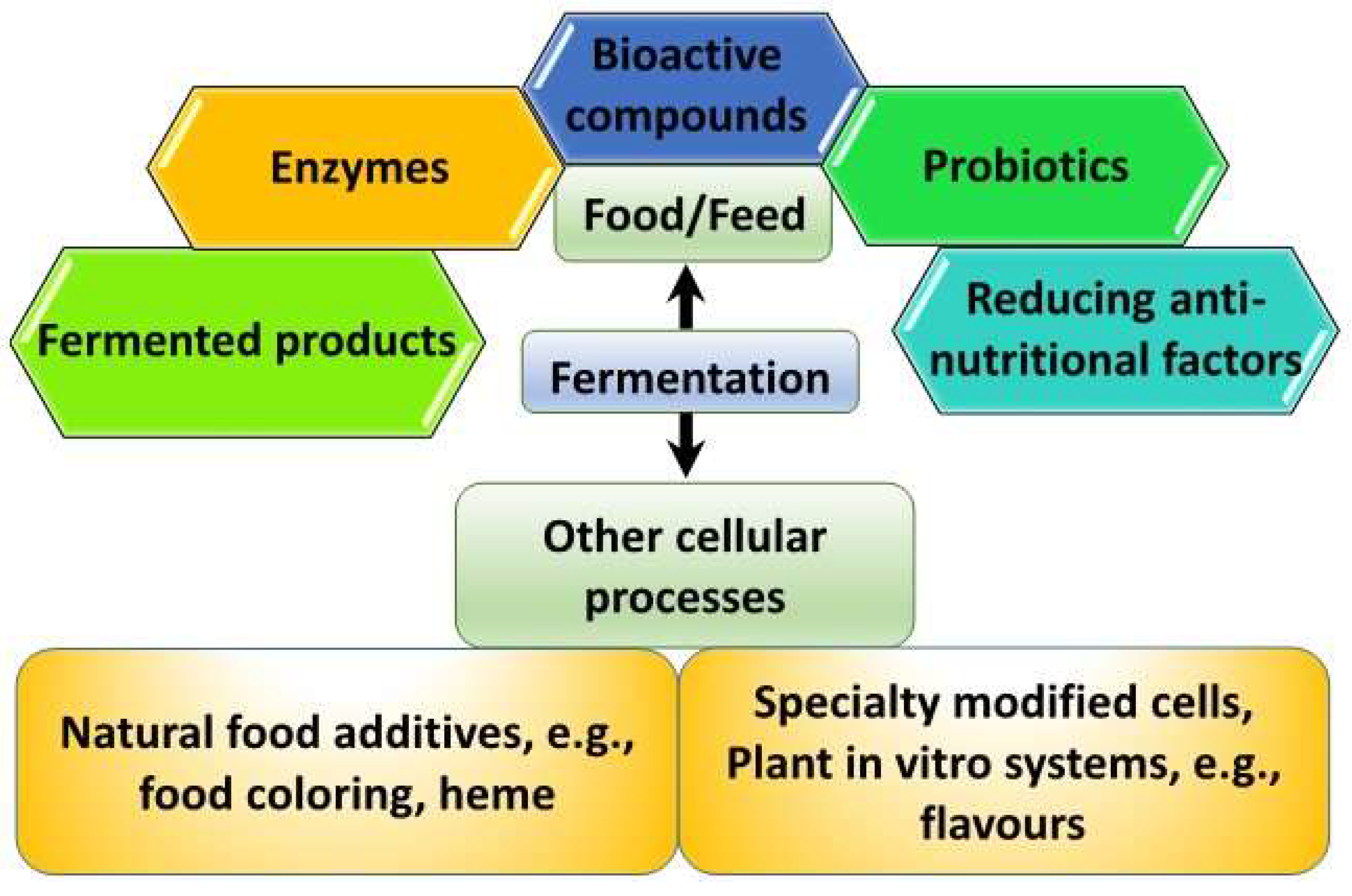
1. Introduction
2. Strategies and Challenges for the Development of Engineered Enzymes in the Food Industry
2.1. Directed Evolution
2.2. Rational Design
2.3. Semi-Rational Design
2.4. Gene Editing
2.5. Safety Challenges
2.6. Cell-Free Systems to Circumvent GMO Concerns
3. Engineered Enzymes for Improved Plant-Based Beverages and Meat Alternatives
4. Fermentation Scale-Up Challenges
4.1. Submerged Liquid Fermentation (SmF)
4.2. Solid-State Fermentation (SSF)
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hassoun, A.; Bekhit, A.E.; Jambrak, A.R.; Regenstein, J.M.; Chemat, F.; Morton, J.D.; Gudjónsdóttir, M.; Carpena, M.; Prieto, M.A.; Varela, P.; et al. The fourth industrial revolution in the food industry-part II: Emerging food trends. Crit. Rev. Food Sci. Nutr. 2022, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Teng, T.S.; Chin, Y.L.; Chai, K.F.; Chen, W.N. Fermentation for future food systems. EMBO Rep. 2021, 22, e52680. [Google Scholar] [CrossRef] [PubMed]
- Fasim, A.; More, V.S.; More, S.S. Large-scale production of enzymes for biotechnology uses. Curr. Opin. Biotechnol. 2021, 69, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Mesbah, N.M. Industrial Biotechnology Based on Enzymes from Extreme Environments. Front. Bioeng. Biotechnol. 2022, 10, 870083. [Google Scholar] [CrossRef]
- Al-Ghanayem, A.A.; Joseph, B. Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 2020, 104, 2871–2882. [Google Scholar] [CrossRef]
- Rocha, R.A.; Speight, R.E.; Scott, C. Engineering Enzyme Properties for Improved Biocatalytic Processes in Batch and Continuous Flow. Org. Process. Res. Dev. 2022, 26, 1914–1924. [Google Scholar] [CrossRef]
- Ali, M.; Ishqi, H.M.; Husain, Q. Enzyme engineering: Reshaping the biocatalytic functions. Biotechnol. Bioeng. 2020, 117, 1877–1894. [Google Scholar] [CrossRef]
- Jones, A.; Lamsa, M.; Frandsen, T.P.; Spendler, T.; Harris, P.; Sloma, A.; Xu, F.; Nielsen, J.B.; Cherry, J.R. Directed evolution of a maltogenic α-amylase from Bacillus sp. TS-25. J. Biotechnol. 2008, 134, 325–333. [Google Scholar] [CrossRef]
- Li, S.; Yang, Q.; Tang, B. Improving the thermostability and acid resistance of Rhizopus oryzae α-amylase by using multiple sequence alignment based site-directed mutagenesis. Biotechnol. Appl. Biochem. 2020, 67, 677–684. [Google Scholar] [CrossRef]
- da Silva Amatto, I.V.; da Rosa-Garzon, N.G.; de Oliveira Simões, F.A.; Santiago, F.; da Silva Leite, N.P.; Raspante Martins, J.; Cabral, H. Enzyme engineering and its industrial applications. Biotechnol. Appl. Biochem. 2022, 69, 389–409. [Google Scholar] [CrossRef]
- Seyedhosseini Ghaheh, H.; Sajjadi, S.; Shafiee, F.; Barzegari, E.; Moazen, F.; Sadeghi, H. Rational design of a new variant of Reteplase with optimized physicochemical profile and large-scale production in Escherichia coli. World J. Microbiol. Biotechnol. 2022, 38, 29. [Google Scholar] [CrossRef]
- Ruan, Y.; Zhang, R.; Xu, Y. Directed evolution of maltogenic amylase from Bacillus licheniformis R-53: Enhancing activity and thermostability improves bread quality and extends shelf life. Food Chem. 2022, 381, 132222. [Google Scholar] [CrossRef]
- Pouyan, S.; Lagzian, M.; Sangtarash, M.H. Enhancing thermostabilization of a newly discovered α-amylase from Bacillus cereus GL96 by combining computer-aided directed evolution and site-directed mutagenesis. Int. J. Biol. Macromol. 2022, 197, 12–22. [Google Scholar] [CrossRef]
- Xu, X.; Liu, M.-Q.; Huo, W.-K.; Dai, X.-J. Obtaining a mutant of Bacillus amyloliquefaciens xylanase A with improved catalytic activity by directed evolution. Enzym. Microb. Technol. 2016, 86, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-Q.; Li, J.-Y.; Rehman, A.U.; Xu, X.; Gu, Z.-J.; Wu, R.-C. Laboratory Evolution of GH11 Endoxylanase through DNA Shuffling: Effects of Distal Residue Substitution on Catalytic Activity and Active Site Architecture. Front. Bioeng. Biotechnol. 2019, 7, 350. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.; Huang, L.; Gui, S.; Zheng, D.; Jia, L.; Fu, Y.; Lu, F. Improvement in thermostability of an alkaline lipase I from Penicillium cyclopium by directed evolution. RSC Adv. 2017, 7, 38538–38548. [Google Scholar] [CrossRef]
- Guan, L.; Gao, Y.; Li, J.; Wang, K.; Zhang, Z.; Yan, S.; Ji, N.; Zhou, Y.; Lu, S. Directed Evolution of Pseudomonas fluorescens Lipase Variants with Improved Thermostability Using Error-Prone PCR. Front. Bioeng. Biotechnol. 2020, 8, 1034. [Google Scholar] [CrossRef] [PubMed]
- Rentschler, E.; Schwarz, T.; Stressler, T.; Fischer, L. Development and validation of a screening system for a β-galactosidase with increased specific activity produced by directed evolution. Eur. Food Res. Technol. 2016, 242, 2129–2138. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, T.; Zhang, Z.; Sun, T.; Wang, J.; Lu, F. Improvement of cold adaptation of Bacillus alcalophilus alkaline protease by directed evolution. J. Mol. Catal. B Enzym. 2014, 106, 117–123. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, L.; Shan, M.; Sang, J.; Li, Y.; Jia, L.; Wang, N.; Wang, S.; Shao, S.; Liu, F.; et al. Enhancing the activity and thermostability of Streptomyces mobaraensis transglutaminase by directed evolution and molecular dynamics simulation. Biochem. Eng. J. 2019, 151, 107333. [Google Scholar] [CrossRef]
- Ashraf, N.; Krishnagopal, A.; Hussain, A.; Kastner, D.; Sayed, A.; Mok, Y.K.; Swaminathan, K.; Zeeshan, N. Engineering of serine protease for improved thermostability and catalytic activity using rational design. Int. J. Biol. Macromol. 2018, 126, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Xiong, K.; Xiong, S.; Gao, S.; Li, Q.; Sun, B.; Li, X. Improving Hydrolysis Characteristics of Xylanases by Site-Directed Mutagenesis in Binding-Site Subsites from Streptomyces L10608. Int. J. Mol. Sci. 2018, 19, 834. [Google Scholar] [CrossRef]
- Niu, C.; Zhu, L.; Zhu, P.; Li, Q. Lysine-Based Site-Directed Mutagenesis Increased Rigid β-Sheet Structure and Thermostability of Mesophilic 1,3–1,4-β-Glucanase. J. Agric. Food Chem. 2015, 63, 5249–5256. [Google Scholar] [CrossRef]
- Lee, J.M.; Moon, S.Y.; Kim, Y.-R.; Kim, K.W.; Lee, B.-J.; Kong, I.-S. Improvement of thermostability and halostability of β-1,3-1,4-glucanase by substituting hydrophobic residue for Lys48. Int. J. Biol. Macromol. 2017, 94, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-W.; Huang, M.; Hsieh, Y.-H.; Luo, Y.-T.; Wu, T.-T.; Tsai, C.-W.; Chen, C.-S.; Shaw, J.-F. Simultaneous production of fatty acid methyl esters and diglycerides by four recombinant Candida rugosa lipase’s isozymes. Food Chem. 2014, 155, 140–145. [Google Scholar] [CrossRef]
- Costa, M.G.S.; Silva, Y.F.; Batista, P.R. Computational engineering of cellulase Cel9A-68 functional motions through mutations in its linker region. Phys. Chem. Chem. Phys. 2018, 20, 7643–7652. [Google Scholar] [CrossRef]
- Elatico, A.J.J.; Nellas, R.B. Computational reverse engineering of the lipase from Pseudomonas aeruginosa PAO1: α-helices. J. Mol. Graph. Model. 2020, 100, 107657. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Miao, H.; Ding, J.; Li, J.; Mu, Y.; Zhou, J.; Huang, Z. Improving the thermostability of a fungal GH11 xylanase via site-directed mutagenesis guided by sequence and structural analysis. Biotechnol. Biofuels 2017, 10, 133. [Google Scholar] [CrossRef]
- Takita, T.; Nakatani, K.; Katano, Y.; Suzuki, M.; Kojima, K.; Saka, N.; Mikami, B.; Yatsunami, R.; Nakamura, S.; Yasukawa, K. Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis. Enzym. Microb. Technol. 2019, 130, 109363. [Google Scholar] [CrossRef]
- Wu, X.; Tian, Z.; Jiang, X.; Zhang, Q.; Wang, L. Enhancement in catalytic activity of Aspergillus niger XynB by selective site-directed mutagenesis of active site amino acids. Appl. Microbiol. Biotechnol. 2018, 102, 249–260. [Google Scholar] [CrossRef]
- Zhou, Y.; Jiao, L.; Shen, J.; Chi, H.; Lu, Z.; Liu, H.; Lu, F.; Zhu, P. Enhancing the Catalytic Activity of Type II L-Asparaginase from Bacillus licheniformis through Semi-Rational Design. Int. J. Mol. Sci. 2022, 23, 9663. [Google Scholar] [CrossRef] [PubMed]
- Verhaeghe, T.; De Winter, K.; Berland, M.; De Vreese, R.; D’Hooghe, M.; Offmann, B.; Desmet, T. Converting bulk sugars into prebiotics: Semi-rational design of a transglucosylase with controlled selectivity. Chem. Commun. 2016, 52, 3687–3689. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Xiao, Y.; Shakhnovich, E.I.; Zhang, W.; Mu, W. Semi-rational design and molecular dynamics simulations study of the thermostability enhancement of cellobiose 2-epimerases. Int. J. Biol. Macromol. 2020, 154, 1356–1365. [Google Scholar] [CrossRef]
- Iqbal, Z.; Sadaf, S. A patent-based consideration of latest platforms in the art of directed evolution: A decade long untold story. Biotechnol. Genet. Eng. Rev. 2022, 38, 133–246. [Google Scholar] [CrossRef] [PubMed]
- Ghahremani Nezhad, N.; Rahman, R.; Normi, Y.; Oslan, S.; Mohd Shariff, F.; Leow, T. Thermostability engineering of industrial enzymes through structure modification. Appl. Microbiol. Biotechnol. 2022, 106, 4845–4866. [Google Scholar] [CrossRef]
- Jang, W.D.; Kim, G.B.; Kim, Y.; Lee, S.Y. Applications of artificial intelligence to enzyme and pathway design for metabolic engineering. Curr. Opin. Biotechnol. 2022, 73, 101–107. [Google Scholar] [CrossRef]
- Freschlin, C.R.; Fahlberg, S.A.; Romero, P.A. Machine learning to navigate fitness landscapes for protein engineering. Curr. Opin. Biotechnol. 2022, 75, 102713. [Google Scholar] [CrossRef]
- Saito, Y.; Oikawa, M.; Sato, T.; Nakazawa, H.; Ito, T.; Kameda, T.; Tsuda, K.; Umetsu, M. Machine-Learning-Guided Library Design Cycle for Directed Evolution of Enzymes: The Effects of Training Data Composition on Sequence Space Exploration. ACS Catal. 2021, 11, 14615–14624. [Google Scholar] [CrossRef]
- Siedhoff, N.E.; Schwaneberg, U.; Davari, M.D. Machine learning-assisted enzyme engineering. In Methods in Enzymology; Tawfik, D.S., Ed.; Academic Press: Cambridge, MA, USA, 2020; Volume 643, pp. 281–315. [Google Scholar]
- Siedhoff, N.E.; Illig, A.-M.; Schwaneberg, U.; Davari, M.D. PyPEF—An Integrated Framework for Data-Driven Protein Engineering. J. Chem. Inf. Model. 2021, 61, 3463–3476. [Google Scholar] [CrossRef]
- Yan, Q.; Zhang, X.; Chen, Y.; Guo, B.; Zhou, P.; Chen, B.; Huang, Q.; Wang, J.-B. From Semirational to Rational Design: Developing a Substrate-Coupled System of Glucose Dehydrogenase for Asymmetric Synthesis. ACS Catal. 2022, 12, 6746–6755. [Google Scholar] [CrossRef]
- Savino, S.; Desmet, T.; Franceus, J. Insertions and deletions in protein evolution and engineering. Biotechnol. Adv. 2022, 60, 108010. [Google Scholar] [CrossRef]
- Chi, H.; Wang, Y.; Xia, B.; Zhou, Y.; Lu, Z.; Lu, F.; Zhu, P. Enhanced Thermostability and Molecular Insights for L-Asparaginase from Bacillus licheniformis via Structure- and Computation-Based Rational Design. J. Agric. Food Chem. 2022, 70, 14499–14509. [Google Scholar] [CrossRef]
- Suzuki, M.; Date, M.; Kashiwagi, T.; Suzuki, E.; Yokoyama, K. Rational design of a disulfide bridge increases the thermostability of microbial transglutaminase. Appl. Microbiol. Biotechnol. 2022, 106, 4553–4562. [Google Scholar] [CrossRef]
- Leys, S.; Pauly, A.; Delcour, J.A.; Courtin, C.M. Modification of the Secondary Binding Site of Xylanases Illustrates the Impact of Substrate Selectivity on Bread Making. J. Agric. Food Chem. 2016, 64, 5400–5409. [Google Scholar] [CrossRef]
- Wang, X.; Du, J.; Zhao, B.; Wang, H.; Rao, S.; Du, G.; Zhou, J.; Chen, J.; Liu, S. Significantly Improving the Thermostability and Catalytic Efficiency of Streptomyces mobaraenesis Transglutaminase through Combined Rational Design. J. Agric. Food Chem. 2021, 69, 15268–15278. [Google Scholar] [CrossRef]
- Ferreira, P.; Fernandes, P.A.; Ramos, M.J. Modern computational methods for rational enzyme engineering. Chem. Catal. 2022, 2, 2481–2498. [Google Scholar] [CrossRef]
- Mehta, A.; Guleria, S.; Sharma, R.; Gupta, R. The lipases and their applications with emphasis on food industry. In Microbial Biotechnology in Food and Health; Ray, R.C., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 143–164. [Google Scholar]
- Anishchenko, I.; Pellock, S.J.; Chidyausiku, T.M.; Ramelot, T.A.; Ovchinnikov, S.; Hao, J.; Bafna, K.; Norn, C.; Kang, A.; Bera, A.K.; et al. De novo protein design by deep network hallucination. Nature 2021, 600, 547–552. [Google Scholar] [CrossRef]
- Schmitt, L.T.; Paszkowski-Rogacz, M.; Jug, F.; Buchholz, F. Prediction of designer-recombinases for DNA editing with generative deep learning. Nat. Commun. 2022, 13, 7966. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, J.Z.H. DenseCPD: Improving the Accuracy of Neural-Network-Based Computational Protein Sequence Design with DenseNet. J. Chem. Inf. Model. 2020, 60, 1245–1252. [Google Scholar] [CrossRef]
- Li, S.; Cao, L.; Yang, X.; Wu, X.; Xu, S.; Liu, Y. Simultaneously optimizing multiple properties of β-glucosidase Bgl6 using combined (semi-)rational design strategies and investigation of the underlying mechanisms. Bioresour. Technol. 2023, 374, 128792. [Google Scholar] [CrossRef]
- Pang, C.; Liu, S.; Zhang, G.; Zhou, J.; Du, G.; Li, J. Improving the catalytic efficiency of Pseudomonas aeruginosa lipoxygenase by semi-rational design. Enzym. Microb. Technol. 2023, 162, 110120. [Google Scholar] [CrossRef]
- Qu, G.; Sun, Z.; Reetz, M.T. Iterative Saturation Mutagenesis for Semi-rational Enzyme Design. In Protein Engineering; Wiley-VCH GmbH: Weinheim, Germany, 2021; pp. 105–132. [Google Scholar]
- Chen, L.; Jiang, K.; Zhou, Y.; Zhu, L.; Chen, X. Improving the Thermostability of α-Glucosidase from Xanthomonas campestris through Proline Substitutions Guided by Semi-rational Design. Biotechnol. Bioprocess. Eng. 2022, 27, 631–639. [Google Scholar] [CrossRef]
- Jia, R.; Wan, X.; Geng, X.; Xue, D.; Xie, Z.; Chen, C. Microbial L-asparaginase for Application in Acrylamide Mitigation from Food: Current Research Status and Future Perspectives. Microorganisms 2021, 9, 1659. [Google Scholar] [CrossRef]
- Beerens, K.; De Winter, K.; Van de Walle, D.; Grootaert, C.; Kamiloglu, S.; Miclotte, L.; Van de Wiele, T.; Van Camp, J.; Dewettinck, K.; Desmet, T. Biocatalytic Synthesis of the Rare Sugar Kojibiose: Process Scale-Up and Application Testing. J. Agric. Food Chem. 2017, 65, 6030–6041. [Google Scholar] [CrossRef]
- Klug, A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q. Rev. Biophys. 2010, 43, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Eleonora, I.I.; Matthew, T.N.Y.; Cian, S.-U.; Rohan, N.K.; Justin, L.; Lukas, V.; Wenyuan, Z.; Kaiyi, J.; Nathaniel, R.; Liyang, Z.; et al. Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lopes, T.S.; Klootwijk, J.; Veenstra, A.E.; van der Aar, P.C.; van Heerikhuizen, H.; Raué, H.A.; Planta, R.J. High-copy-number integration into the ribosomal DNA of Saccharomyces cerevisiae: A new vector for high-level expression. Gene 1989, 79, 199–206. [Google Scholar] [CrossRef]
- Deckers, M.; Deforce, D.; Fraiture, M.A.; Roosens, N.H.C. Genetically Modified Micro-Organisms for Industrial Food Enzyme Production: An Overview. Foods 2020, 9, 326. [Google Scholar] [CrossRef]
- Zhang, K.; Su, L.; Wu, J. Enhanced extracellular pullulanase production in Bacillus subtilis using protease-deficient strains and optimal feeding. Appl. Microbiol. Biotechnol. 2018, 102, 5089–5103. [Google Scholar] [CrossRef]
- Salazar-Cerezo, S.; Kun, R.S.; de Vries, R.P.; Garrigues, S. CRISPR/Cas9 technology enables the development of the filamentous ascomycete fungus Penicillium subrubescens as a new industrial enzyme producer. Enzym. Microb. Technol. 2020, 133, 109463. [Google Scholar] [CrossRef]
- Park, J.; Yu, B.J.; Choi, J.-I.; Woo, H.M. Heterologous Production of Squalene from Glucose in Engineered Corynebacterium glutamicum Using Multiplex CRISPR Interference and High-Throughput Fermentation. J. Agric. Food Chem. 2019, 67, 308–319. [Google Scholar] [CrossRef]
- Dong, L.; Yu, D.; Lin, X.; Wang, B.; Pan, L. Improving expression of thermostable trehalase from Myceliophthora sepedonium in Aspergillus niger mediated by the CRISPR/Cas9 tool and its purification, characterization. Protein Expr. Purif. 2020, 165, 105482. [Google Scholar] [CrossRef]
- Waltz, E. GABA-enriched tomato is first CRISPR-edited food to enter market. Nat. Biotechnol. 2022, 40, 9–11. [Google Scholar] [CrossRef]
- González, M.N.; Massa, G.A.; Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Storani, L.; Décima Oneto, C.A.; Hofvander, P.; Feingold, S.E. Reduced Enzymatic Browning in Potato Tubers by Specific Editing of a Polyphenol Oxidase Gene via Ribonucleoprotein Complexes Delivery of the CRISPR/Cas9 System. Front. Plant. Sci. 2020, 10, 1649. [Google Scholar] [CrossRef]
- EFSA Panel on Food Contact Materials, Enzymes, Processing Aids (CEP); Silano, V.; Barat Baviera, J.M.; Bolognesi, C.; Brüschweiler, B.J.; Cocconcelli, P.S.; Crebelli, R.; Gott, D.M.; Grob, K.; Lampi, E.; et al. Safety evaluation of the food enzyme alpha-amylase from a genetically modified Bacillus subtilis (strain NBA). EFSA J. 2019, 17, e05681. [Google Scholar] [CrossRef]
- Budnik, L.T.; Scheer, E.; Burge, P.S.; Baur, X. Sensitising effects of genetically modified enzymes used in flavour, fragrance, detergence and pharmaceutical production: Cross-sectional study. Occup. Environ. Med. 2017, 74, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Kuske, M.; Berndt, K.; Spornraft-Ragaller, P.; Neumeister, V.; Raulf, M.; Sander, I.; Koschel, D.; Bickhardt, J.; Beissert, S.; Bauer, A. Occupational allergy to phytase: Case series of eight production workers exposed to animal feed additives. J. Dtsch. Dermatol. Ges. 2020, 18, 859–865. [Google Scholar] [CrossRef]
- Sewalt, V.; Shanahan, D.; Gregg, L.; La Marta, J.; Carrillo, R. The Generally Recognized as Safe (GRAS) Process for Industrial Microbial Enzymes. Ind. Biotechnol. 2016, 12, 295–302. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards; Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; de Cesare, A.; Hilbert, F.; Lindqvist, R.; et al. Update of the list of qualified presumption of safety (QPS) recommended microorganisms intentionally added to food or feed as notified to EFSA. EFSA J. 2023, 21, e07747. [Google Scholar] [CrossRef]
- Seo, S.-O.; Jin, Y.-S. Next-Generation Genetic and Fermentation Technologies for Safe and Sustainable Production of Food Ingredients: Colors and Flavorings. Annu. Rev. Food Sci. Technol. 2022, 13, 463–488. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); Paraskevopoulos, K.; Federici, S. Overview of EFSA and European national authorities’ scientific opinions on the risk assessment of plants developed through New Genomic Techniques. EFSA J. 2021, 19, e06314. [Google Scholar] [CrossRef] [PubMed]
- Rostoks, N. Implications of the EFSA Scientific Opinion on Site Directed Nucleases 1 and 2 for Risk Assessment of Genome-Edited Plants in the EU. Agronomy 2021, 11, 572. [Google Scholar] [CrossRef]
- Brookwell, A.; Oza, J.P.; Caschera, F. Biotechnology Applications of Cell-Free Expression Systems. Life 2021, 11, 1367. [Google Scholar] [CrossRef] [PubMed]
- Rasor, B.J.; Vögeli, B.; Landwehr, G.M.; Bogart, J.W.; Karim, A.S.; Jewett, M.C. Toward sustainable, cell-free biomanufacturing. Curr. Opin. Biotechnol. 2021, 69, 136–144. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. The fourth wave of biocatalysis is approaching. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2017, 376, 20170063. [Google Scholar] [CrossRef]
- Poppe, L.; Vértessy, B.G. The Fourth Wave of Biocatalysis Emerges—The 13th International Symposium on Biocatalysis and Biotransformations. ChemBioChem 2018, 19, 284–287. [Google Scholar] [CrossRef]
- Nirenberg, M.W.; Matthaei, J.H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA 1961, 47, 1588–1602. [Google Scholar] [CrossRef] [PubMed]
- Silverman, A.D.; Karim, A.S.; Jewett, M.C. Cell-free gene expression: An expanded repertoire of applications. Nat. Rev. Genet. 2020, 21, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Hershewe, J.; Kightlinger, W.; Jewett, M.C. Cell-free systems for accelerating glycoprotein expression and biomanufacturing. J. Ind. Microbiol. Biotechnol. 2020, 47, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Zawada, J.F.; Burgenson, D.; Yin, G.; Hallam, T.J.; Swartz, J.R.; Kiss, R.D. Cell-free technologies for biopharmaceutical research and production. Curr. Opin. Biotechnol. 2022, 76, 102719. [Google Scholar] [CrossRef]
- You, C.; Shi, T.; Li, Y.; Han, P.; Zhou, X.; Zhang, Y.-H.P. An in vitro synthetic biology platform for the industrial biomanufacturing of myo-inositol from starch. Biotechnol. Bioeng. 2017, 114, 1855–1864. [Google Scholar] [CrossRef]
- Zhong, C.; You, C.; Wei, P.; Zhang, Y.-H.P. Thermal Cycling Cascade Biocatalysis of myo-Inositol Synthesis from Sucrose. ACS Catal. 2017, 7, 5992–5999. [Google Scholar] [CrossRef]
- Cheng, K.; Zheng, W.; Chen, H.; Zhang, Y.-H.P.J. Upgrade of wood sugar d-xylose to a value-added nutraceutical by in vitro metabolic engineering. Metab. Eng. 2019, 52, 1–8. [Google Scholar] [CrossRef]
- Buntru, M.; Hahnengress, N.; Croon, A.; Schillberg, S. Plant-Derived Cell-Free Biofactories for the Production of Secondary Metabolites. Front. Plant. Sci. 2022, 12, 794999. [Google Scholar] [CrossRef]
- Schlegel, K.; Sontheimer, K.; Hickisch, A.; Wani, A.A.; Eisner, P.; Schweiggert-Weisz, U. Enzymatic hydrolysis of lupin protein isolates—Changes in the molecular weight distribution, technofunctional characteristics, and sensory attributes. Food Sci. Nutr. 2019, 7, 2747–2759. [Google Scholar] [CrossRef] [PubMed]
- Akbarian, M.; Khani, A.; Eghbalpour, S.; Uversky, V.N. Bioactive Peptides: Synthesis, Sources, Applications, and Proposed Mechanisms of Action. Int. J. Mol. Sci. 2022, 23, 1445. [Google Scholar] [CrossRef]
- Prongjit, D.; Lekakarn, H.; Bunterngsook, B.; Aiewviriyasakul, K.; Sritusnee, W.; Champreda, V. Functional Characterization of Recombinant Raw Starch Degrading α-Amylase from Roseateles terrae HL11 and Its Application on Cassava Pulp Saccharification. Catalysts 2022, 12, 647. [Google Scholar] [CrossRef]
- Peng, H.; Li, R.; Li, F.; Zhai, L.; Zhang, X.; Xiao, Y.; Gao, Y. Extensive hydrolysis of raw rice starch by a chimeric α-amylase engineered with α-amylase (AmyP) and a starch-binding domain from Cryptococcus sp. S-2. Appl. Microbiol. Biotechnol. 2018, 102, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, Y.; Guan, R.; Jia, G.; Ma, Y.; Zhang, Y. Advances in research on calf rennet substitutes and their effects on cheese quality. Food Res. Int. 2021, 149, 110704. [Google Scholar] [CrossRef] [PubMed]
- Nicosia, F.D.; Puglisi, I.; Pino, A.; Caggia, C.; Randazzo, C.L. Plant Milk-Clotting Enzymes for Cheesemaking. Foods 2022, 11, 871. [Google Scholar] [CrossRef]
- Wen, Y.; Kim, H.W.; Park, H.J. Effects of transglutaminase and cooking method on the physicochemical characteristics of 3D-printable meat analogs. Innov. Food Sci. Emerg. Technol. 2022, 81, 103114. [Google Scholar] [CrossRef]
- Schlangen, M.; Ribberink, M.A.; Taghian Dinani, S.; Sagis, L.M.C.; van der Goot, A.J. Mechanical and rheological effects of transglutaminase treatment on dense plant protein blends. Food Hydrocoll. 2023, 136, 108261. [Google Scholar] [CrossRef]
- Boukid, F.; Hassoun, A.; Zouari, A.; Tülbek, M.Ç.; Mefleh, M.; Aït-Kaddour, A.; Castellari, M. Fermentation for Designing Innovative Plant-Based Meat and Dairy Alternatives. Foods 2023, 12, 1005. [Google Scholar] [CrossRef]
- Ganeshan, S.; Kim, S.H.; Vujanovic, V. Scaling-up production of plant endophytes in bioreactors: Concepts, challenges and perspectives. Bioresour. Bioprocess. 2021, 8, 63. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, G.; Ahmad, T.; Mansoor, S.; Kaur, B. Enzyme Engineering: Current Trends and Future Perspectives. Food Rev. Int. 2021, 37, 121–154. [Google Scholar] [CrossRef]
- Augustin, M.A.; Hartley, C.J.; Maloney, G.; Tyndall, S. Innovation in precision fermentation for food ingredients. Crit. Rev. Food Sci. Nutr. 2023, 1. [Google Scholar] [CrossRef]
- Nolan, D.P. Application of HAZOP and What-If Safety Reviews to the Petroleum, Petrochemical and Chemical Industries; Noyes Publications: Park Ridge, NJ, USA, 1994. [Google Scholar]
- Crater, J.; Lievense, J. Scale-up of Industrial Microbial Processes. FEMS Microbiol. Lett. 2018, 365, 1–5. [Google Scholar] [CrossRef]
- Al-Maqtari, Q.; Al-Ansi, W.; Mahdi, A. Microbial enzymes produced by fermentation and their applications in the food—A review. Int. J. Agric. Innov. Res. 2019, 8, 62–82. [Google Scholar]
- Mitchell, D.A.; Pitol, L.O.; Biz, A.; Finkler, A.T.J.; de Lima Luz, L.F.; Krieger, N. Design and Operation of a Pilot-Scale Packed-Bed Bioreactor for the Production of Enzymes by Solid-State Fermentation. In Solid. State Fermentation: Research and Industrial Applications; Steudler, S., Werner, A., Cheng, J.J., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 27–50. [Google Scholar]
- Teles, A.S.C.; Chávez, D.W.H.; Oliveira, R.A.; Bon, E.P.S.; Terzi, S.C.; Souza, E.F.; Gottschalk, L.M.F.; Tonon, R.V. Use of grape pomace for the production of hydrolytic enzymes by solid-state fermentation and recovery of its bioactive compounds. Food Res. Int. 2019, 120, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Benabda, O.; M’hir, S.; Kasmi, M.; Mnif, W.; Hamdi, M. Optimization of Protease and Amylase Production by Rhizopus oryzae Cultivated on Bread Waste Using Solid-State Fermentation. J. Chem. 2019, 2019, 3738181. [Google Scholar] [CrossRef]
- Balakrishnan, M.; Jeevarathinam, G.; Kumar, S.K.S.; Muniraj, I.; Uthandi, S. Optimization and scale-up of α-amylase production by Aspergillus oryzae using solid-state fermentation of edible oil cakes. BMC Biotechnol. 2021, 21, 33. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-G.; Kim, B.-Y.; Bae, J.-M.; Wang, Y.; Jin, Y.-S. Genome-edited Saccharomyces cerevisiae strains for improving quality, safety, and flavor of fermented foods. Food Microbiol. 2022, 104, 103971. [Google Scholar] [CrossRef]
- Singhania, R.R.; Patel, A.K.; Gottumukkala, L.D.; Rajasree, K.; Soccol, C.R.; Pandey, A. Solid-state fermentation—Current trends and future prospects. In Fermentation Microbiology and Biotechnology; El-Mansi, E.M.T., El-Mansi, M., Nielsen, J., Mousdale, D.M., Allman, T., Carlson, R., Eds.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Prajapati, B.P.; Kumar Suryawanshi, R.; Agrawal, S.; Ghosh, M.; Kango, N. Characterization of cellulase from Aspergillus tubingensis NKBP-55 for generation of fermentable sugars from agricultural residues. Bioresour. Technol. 2018, 250, 733–740. [Google Scholar] [CrossRef]

| Engineered Enzyme | Source | Method | Effect | Example of Application | References |
|---|---|---|---|---|---|
| Directed evolution | |||||
| α-amylase Novamyl | Bacillus sp. (TS-25) | Error-prone PCR | Increased thermostability at acidic pH | Bakery | [8] |
| α-amylase | Bacillus licheniformis | Error-prone PCR | Increased thermostability | Bakery | [12] |
| α-amylase | Rhizopus oryzae | Multiple sequence alignment-based site-directed mutagenesis | Improved the thermostability and acid resistance | Starch industry and brewery | [9] |
| α-amylase | Bacillus cereus GL96 | Combining computer-aided directed evolution and site-directed mutagenesis | Increased thermostability (70 °C) and stability over a range of pH from 4 to 11) | Bakery | [13] |
| Xylanase (reBaxA50) | Bacillus amyloliquefaciens | Error-prone touchdown PCR | Increased catalytic efficiency and stability under thermal and extreme pH | Biorefinery | [14] |
| Xylanase | Bacillus amyloliquefaciens xylanase A (BaxA) and Thermomonospora fusca | DNA shuffling | Increased specificity and catalytic efficiency | Production of prebiotic xylo-oligosaccharides | [15] |
| Lipase | Penicillium cyclopium | Error-prone PCR | Enhanced thermostability | Bakery and dairy | [16] |
| Lipase | Pseudomonas fluorescens | Error-prone PCR | Enhanced alkali stability | Bakery and dairy | [17] |
| β-galactosidase | Escherichia coli | Error-prone PCR | Increased activity | Milk processing | [18] |
| Alkaline protease | Bacillus alcalophilus | Error-prone PCR | Increased cold adaptation | Cold-temperature food processing | [19] |
| Transglutaminase | Streptomyces mobaraensis | Directed Evolution and Molecular Dynamics Simulation | Improved thermostability and specific activity | Bakery | [20] |
| Rational design | |||||
| Serine peptidase | Pseudomonas aeruginosa | Site-directed mutagenesis | Improved thermal stability and catalytic efficiency | Dairy | [21] |
| Xylanase | Streptomyces | Site-directed mutagenesis | Enhanced substrate specificity | Bread making | [22] |
| β-glucanase | Bacillus terquilensis | Site-directed mutagenesis | Enhanced thermostability | Cereal-based sector | [23] |
| β-glucanase | Bacillus sp. SJ-10 | Site-directed mutagenesis | Enhanced catalytic efficiency, halostability, and thermostability | Hemicelluloses hydrolysis | [24] |
| Lipase isozymes | Candida rugosa | Site-directed mutagenesis | Increased catalytic efficiency | Food emulsifiers | [25] |
| Cel9A-68 cellulase | Thermobifida fusca | Computer-aided enzyme simulation | Increased catalytic activity | Brewery and wine | [26] |
| Lipase | P. aeruginosa PAO1 | Computational “reverse engineering” | Increased activity and stability | Dairy products such as cheese | [27] |
| GH11 xylanase | Neocallimastix patriciarum | Site-directed mutagenesis guided by sequence and structural analysis | Improved thermostability and kinetic efficiency | Cereal processing | [28] |
| GH11 xylanase | Bacillus sp. strain (T82A) | Site-saturation mutagenesis | Increased catalytic activity | Cereal processing | [29] |
| GH11 xylanase | Aspergillus niger | Virtual mutation and molecular dynamics simulations | Increased catalytic activity and thermostability | Cereal processing | [30] |
| Semi-rational design | |||||
| Type II ASNase | Bacillus licheniformis | Structural alignment and molecular dynamic simulation | Increased catalytic efficiency, structure stability, and substrate binding | Fried potato products, bakery products, and coffee | [31] |
| Sucrose phosphorylase | Bacillus licheniformis | Semi-rational mutagenesis and low-throughput | Increased selectivity | Confectionery products | [32] |
| Cellobiose 2-epimerase | Caldicellulosiruptor saccharolyticus | Computational prediction performance and molecular dynamics simulation | Improved thermostability and catalytic efficiency | Production of lactose-based prebiotics | [33] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boukid, F.; Ganeshan, S.; Wang, Y.; Tülbek, M.Ç.; Nickerson, M.T. Bioengineered Enzymes and Precision Fermentation in the Food Industry. Int. J. Mol. Sci. 2023, 24, 10156. https://doi.org/10.3390/ijms241210156
Boukid F, Ganeshan S, Wang Y, Tülbek MÇ, Nickerson MT. Bioengineered Enzymes and Precision Fermentation in the Food Industry. International Journal of Molecular Sciences. 2023; 24(12):10156. https://doi.org/10.3390/ijms241210156
Chicago/Turabian StyleBoukid, Fatma, Seedhabadee Ganeshan, Yingxin Wang, Mehmet Çağlar Tülbek, and Michael T. Nickerson. 2023. "Bioengineered Enzymes and Precision Fermentation in the Food Industry" International Journal of Molecular Sciences 24, no. 12: 10156. https://doi.org/10.3390/ijms241210156
APA StyleBoukid, F., Ganeshan, S., Wang, Y., Tülbek, M. Ç., & Nickerson, M. T. (2023). Bioengineered Enzymes and Precision Fermentation in the Food Industry. International Journal of Molecular Sciences, 24(12), 10156. https://doi.org/10.3390/ijms241210156








