Genome-Wide Identification and Expression Analysis of Senescence-Associated Genes in Grapevine (Vitis vinifera L.) Reveal Their Potential Functions in Leaf Senescence Order
Abstract
1. Introduction
2. Results
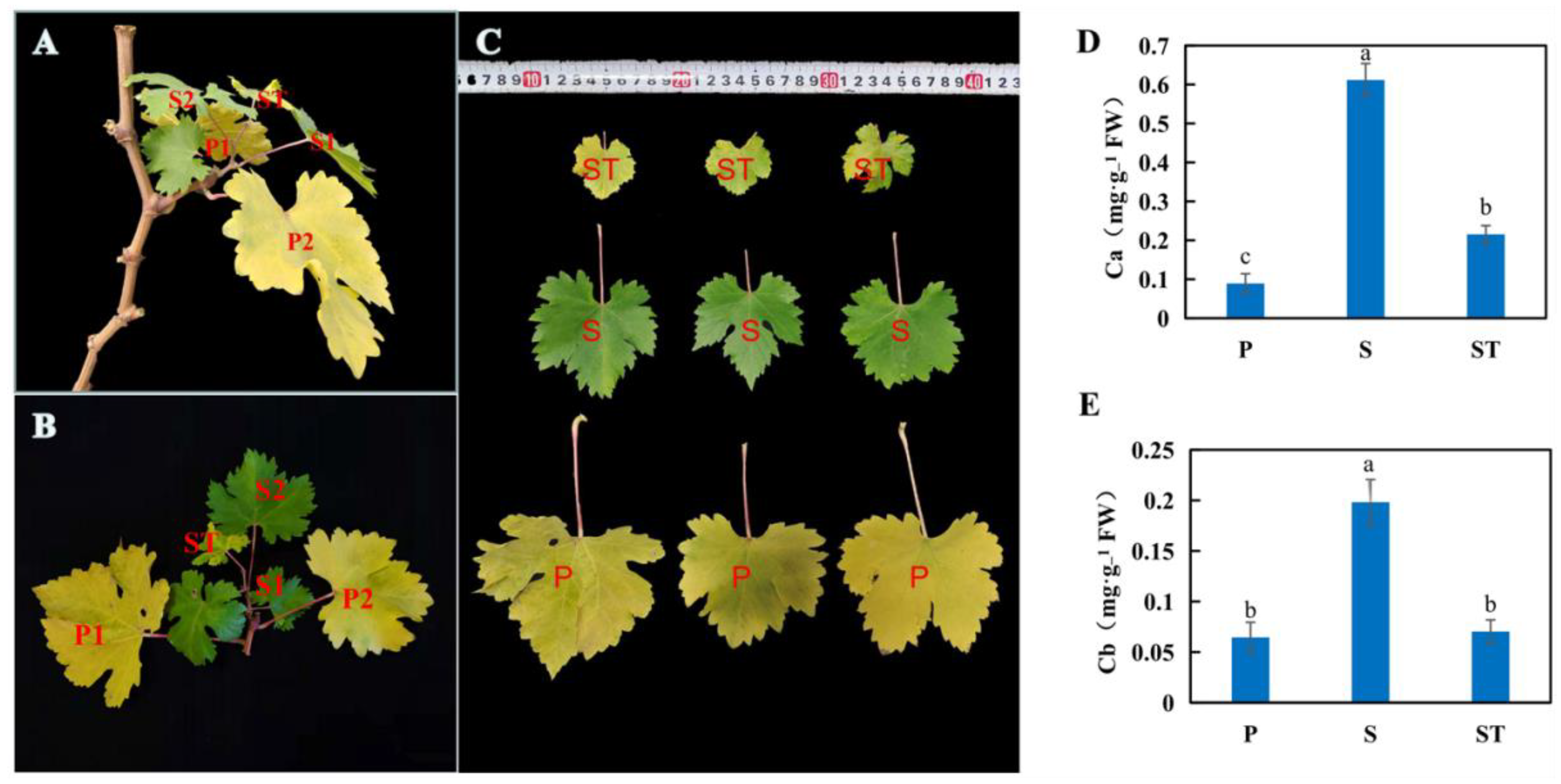
2.1. The Impact of Leaf Position on Senescence of Grapevine Leaves
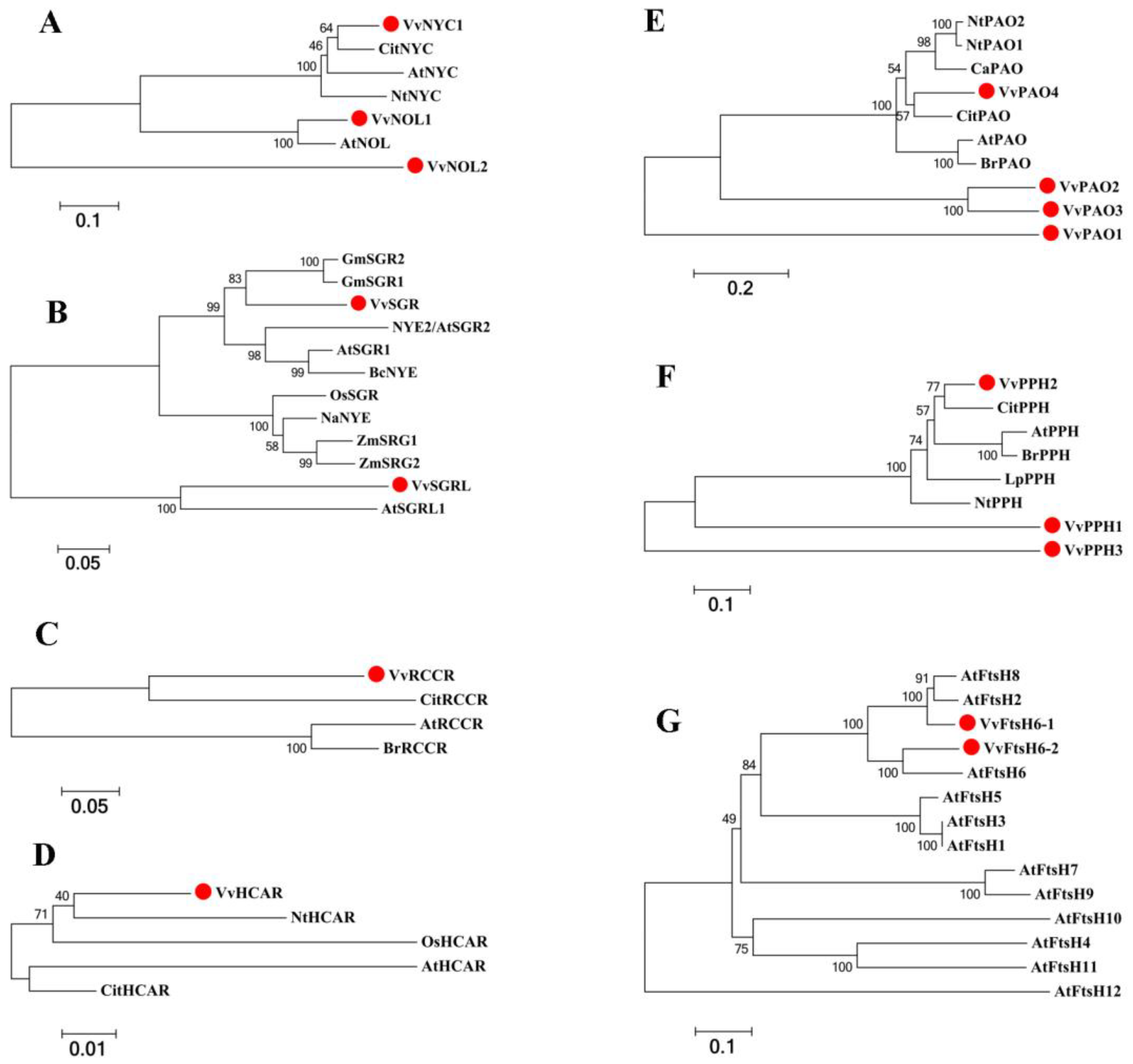
2.2. Genome-Wide Identification and Characterization of Chl Degradation-Related SAGs in Grapevine Leaves
2.3. Cis-Acting Regulatory Element Analysis of SAG Promoters
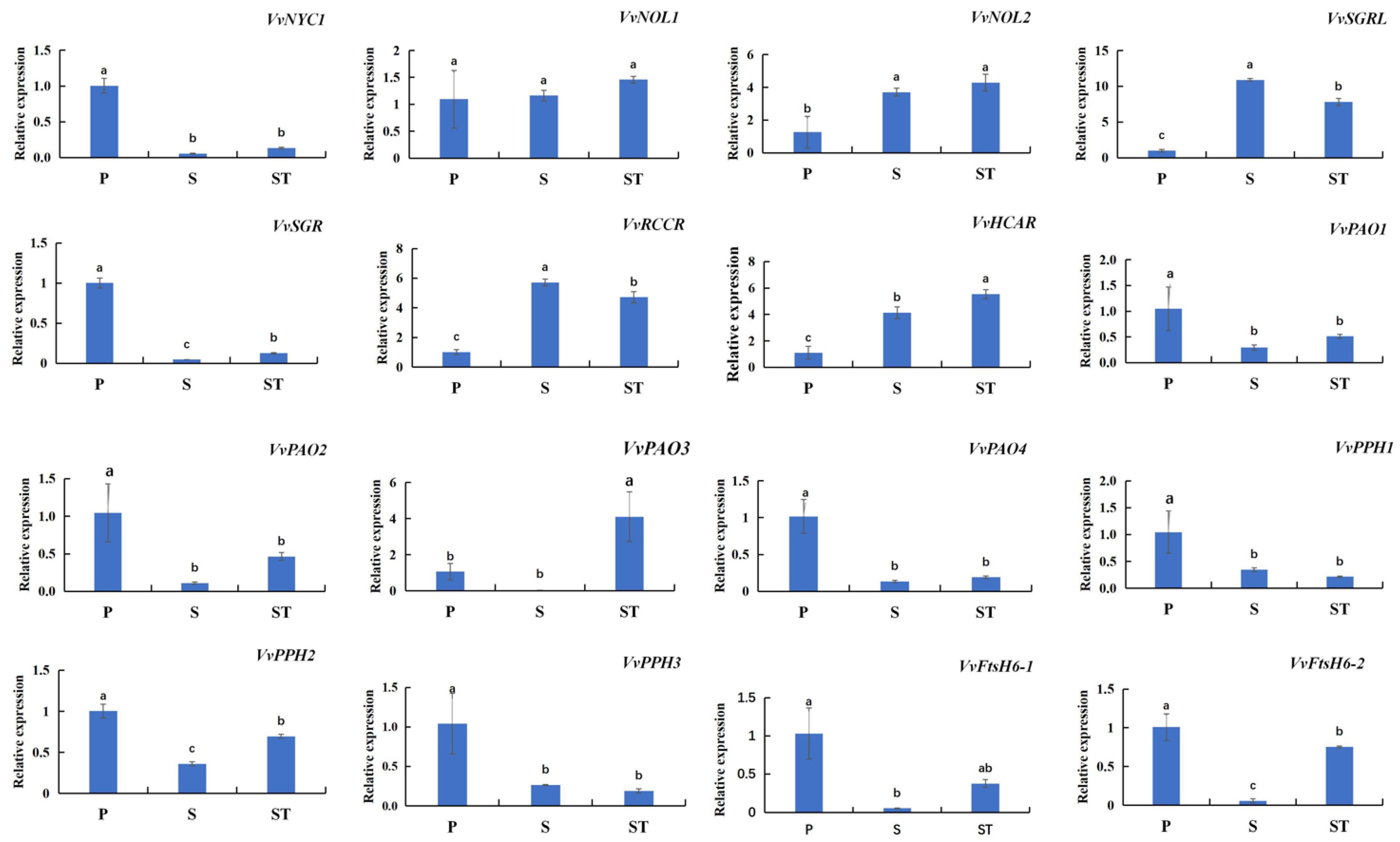
2.4. Chl Degradation-Related SAGs Are Characterized by Diverse Expression Profiles in Different Leaf Positions of Grape Shoots
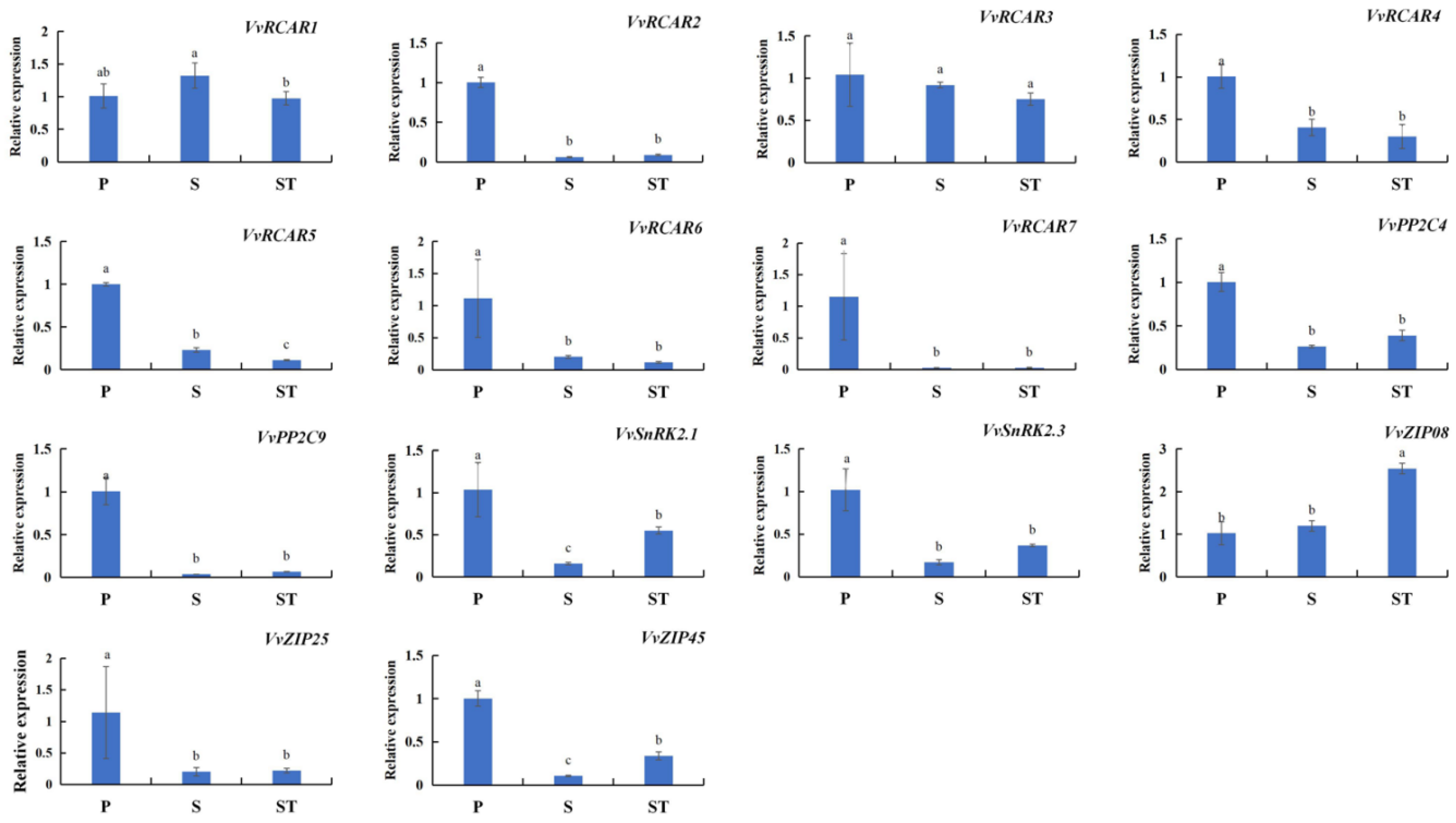
2.5. ABA-Related SAGs Are Characterized by Consistent Expression Profiles with Chl Degradation-Related SAGs in Different Leaf Positions of Grape Shoots
3. Discussion
4. Materials and Methods
4.1. Plant Material and Sampling
4.2. Measurement of Chl Content
4.3. Identification of Chl Degradation-Related SAGs in Grapevine Leaves
4.4. Phylogenetic Analysis, Gene Structure Analysis, and Promoter Analysis
4.5. Expression Analysis by Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schippers, J.H.; Schmidt, R.; Wagstaff, C.; Jing, H.C. Living to Die and Dying to Live: The Survival Strategy behind Leaf Senescence. Plant Physiol. 2015, 169, 914–930. [Google Scholar] [CrossRef] [PubMed]
- Schippers, J.H. Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 2015, 27, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Noodén, L.D. Plant Cell Death Processes; Academic Press: London, UK, 2004. [Google Scholar]
- Tamary, E.; Nevo, R.; Naveh, L.; Levin-Zaidman, S.; Kiss, V.; Savidor, A.; Levin, Y.; Eyal, Y.; Reich, Z.; Adam, Z. Chlorophyll catabolism precedes changes in chloroplast structure and proteome during leaf senescence. Plant Direct 2019, 3, e00127. [Google Scholar] [CrossRef] [PubMed]
- Horie, Y.; Ito, H.; Kusaba, M.; Tanaka, R.; Tanaka, A. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 2009, 284, 17449–17456. [Google Scholar] [CrossRef] [PubMed]
- Meguro, M.; Ito, H.; Takabayashi, A.; Tanaka, R.; Tanaka, A. Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 2011, 23, 3442–3453. [Google Scholar] [CrossRef]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef]
- Morita, R.; Sato, Y.; Masuda, Y.; Nishimura, M.; Kusaba, M. Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef]
- Pruzinska, A.; Anders, I.; Aubry, S.; Schenk, N.; Tapernoux-Luthi, E.; Muller, T.; Krautler, B.; Hortensteiner, S. In Vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 2007, 19, 369–387. [Google Scholar] [CrossRef]
- Piao, W.; Han, S.H.; Sakuraba, Y.; Paek, N.C. Rice 7-Hydroxymethyl Chlorophyll a Reductase Is Involved in the Promotion of Chlorophyll Degradation and Modulates Cell Death Signaling. Mol. Cell 2017, 40, 773–786. [Google Scholar]
- Sakuraba, Y.; Kim, Y.S.; Yoo, S.C.; Hortensteiner, S.; Paek, N.C. 7-Hydroxymethyl chlorophyll a reductase functions in metabolic channeling of chlorophyll breakdown intermediates during leaf senescence. Biochem. Biophys. Res. Commun. 2013, 430, 32–37. [Google Scholar] [CrossRef]
- Schelbert, S.; Aubry, S.; Burla, B.; Agne, B.; Kessler, F.; Krupinska, K.; Hortensteiner, S. Pheophytin Pheophorbide Hydrolase (Pheophytinase) Is Involved in Chlorophyll Breakdown during Leaf Senescence in Arabidopsis. Plant Cell 2009, 21, 767–785. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.H.; Xie, Z.I.; Zhang, J.; Lei, S.S.; Lin, W.J.; Xu, B.; Huang, B.R. NOL-mediated functional stay-green traits in perennial ryegrass (Lolium perenne L.) involving multifaceted molecular factors and metabolic pathways regulating leaf senescence. Plant J. 2021, 106, 1219–1232. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.D.; An, K.; Liao, Y.; Zhou, X.; Cao, Y.J.; Zhao, H.F.; Ge, X.C.; Kuai, B.K. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in arabidopsis. Plant Physiol. 2007, 144, 1429–1441. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Schelbert, S.; Park, S.Y.; Han, S.H.; Lee, B.D.; Andres, C.B.; Kessler, F.; Hortensteiner, S.; Paek, N.C. STAY-GREEN and Chlorophyll Catabolic Enzymes Interact at Light-Harvesting Complex II for Chlorophyll Detoxification during Leaf Senescence in Arabidopsis. Plant Cell 2012, 24, 507–518. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, D.; Kim, Y.S.; Hortensteiner, S.; Paek, N.C. Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett. 2014, 588, 3830–3837. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Park, S.Y.; Kim, Y.S.; Wang, S.H.; Yoo, S.C.; Hortensteiner, S.; Paek, N.C. Arabidopsis STAY-GREEN2 Is a Negative Regulator of Chlorophyll Degradation during Leaf Senescence. Mol. Plant 2014, 7, 1288–1302. [Google Scholar] [CrossRef]
- Wagner, R.; Aigner, H.; Funk, C. FtsH proteases located in the plant chloroplast. Physiol. Plant 2012, 145, 203–214. [Google Scholar] [CrossRef]
- Zelisko, A.; Garcia-Lorenzo, M.; Jackowski, G.; Jansson, S.; Funk, C. AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc. Natl. Acad. Sci. USA 2005, 102, 13699–13704. [Google Scholar] [CrossRef]
- Ueda, H.; Ito, T.; Inoue, R.; Masuda, Y.; Nagashima, Y.; Kozuka, T.; Kusaba, M. Genetic Interaction Among Phytochrome, Ethylene and Abscisic Acid Signaling During Dark-Induced Senescence in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 564. [Google Scholar] [CrossRef]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 Promote ABA-Mediated Chlorophyll Degradation and Leaf Senescence by Transcriptional Activation of Chlorophyll Catabolic Genes and Senescence-Associated Genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.Y.; Kuai, B.K.; Jia, J.Z.; Jing, H.C. Regulation of leaf senescence and crop genetic improvement. J. Integr. Plant Biol. 2012, 54, 936–952. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.G.; Li, G.R.; Yan, C.H.; Liu, L.; Zhang, Q.T.; Han, Z.; Li, B. DRL1, Encoding A NAC Transcription Factor, Is Involved in Leaf Senescence in Grapevine. Int. J. Mol. Sci. 2019, 20, 2678. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.R.; Xie, X.L.; Xia, X.J.; Yu, J.Q.; Ferguson, I.B.; Giovannoni, J.J.; Chen, K.S. Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening. Plant J. 2016, 86, 403–412. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Zou, D.; Zhao, Y.; Wang, H.L.; Zhang, Y.; Xia, X.; Luo, J.; Guo, H.; Zhang, Z. LSD 3.0: A comprehensive resource for the leaf senescence research community. Nucleic Acids Res. 2020, 48, D1069–D1075. [Google Scholar] [CrossRef]
- Tanaka, R.; Kobayashi, K.; Masuda, T. Tetrapyrrole metabolism in arabidopsis thaliana. Arab. Book 2011, 9, e0145. [Google Scholar] [CrossRef]
- Tanaka, R.; Hirashima, M.; Satoh, S.; Tanaka, A. The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: Inhibition of the pheophorbide a oxygenase activity does not lead to the“Stay-Green” phenotype in Arabidopsis. Plant Cell Physiol. 2003, 44, 1266–1274. [Google Scholar] [CrossRef]
- Mach, J.M.; Castillo, A.R.; Hoogstraten, R.; Greenberg, J.T. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. USA 2001, 98, 771–776. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, G.H.; Wen, W.W.; Ma, X.Q.; Xu, B.; Huang, B.R. Functional characterization and hormonal regulation of the PHEOPHYTINASE gene LpPPH controlling leaf senescence in perennial ryegrass. J. Exp. Bot. 2016, 67, 935–945. [Google Scholar] [CrossRef]
- Choi, H.I.; Hong, J.H.; Ha, J.O.; Kang, J.Y.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Boneh, U.; Biton, I.; Zheng, C.; Schwartz, A.; Ben-Ari, G. Characterization of potential ABA receptors in Vitis vinifera. Plant Cell Rep. 2012, 31, 311–321. [Google Scholar] [CrossRef]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [PubMed]
- Boneh, U.; Biton, I.; Schwartz, A.; Ben-Ari, G. Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant Sci. 2012, 187, 89–96. [Google Scholar] [CrossRef]
- Burnison, B.K. Modified dimethyl sulfoxide (dmso) extraction for chlorophyll analysis of phytoplankton. Can. J. Fish. Aquat. 2011, 37, 729–733. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463. [Google Scholar]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Gambino, G.; Perrone, I.; Gribaudo, I. A Rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Gene ID | Location | Peptide (aa) |
|---|---|---|---|
| VvNYC1 | VIT_11s0016g03890.2 | chr11:3174337..3179203− | 518 |
| VvNOL1 | VIT_01s0010g00590.1 | chr1:15587022..15619072− | 317 |
| VvNOL2 | VIT_12s0035g01780.1 | chr12:21916237..21924762− | 265 |
| VvSGR | VIT_02s0025g04660.1 | chr2:4225612..4232509+ | 228 |
| VvSGRL | VIT_18s0001g01210.1 | chr18:1811441..1814300− | 252 |
| VvRCCR | VIT_07s0031g00680.1 | chr7:16847972..1685168+ | 323 |
| VvHCAR | VIT_05s0051g00070.2 | chr5:10300193..10315728− | 458 |
| VvPAO1 | VIT_04s0008g07020.1 | chr4:7106897..7110246− | 545 |
| VvPAO2 | VIT_06s0004g00610.1 | chr6:769345..772779− | 524 |
| VvPAO3 | VIT_06s0004g00620.1 | chr6:780506..783167− | 464 |
| VvPAO4 | VIT_06s0061g00790.1 | chr6:18321351..18327161− | 540 |
| VvPPH1 | VIT_04s0023g02010.1 | chr4:18547350..18550496+ | 368 |
| VvPPH2 | VIT_13s0158g00180.2 | chr13:21072015..21076223− | 525 |
| VvPPH3 | VIT_16s0022g01340.2 | chr6:780506..783167− | 464 |
| VvFtsH6-1 | VIT_12s0028g01600.1 | chr12:2304815..2308584+ | 695 |
| VvFtsH6-2 | VIT_14s0108g00590.1 | chr14:29333100..29335715+ | 677 |
| Element | Function | Cis-Element on SAGs Promoters | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NYC1 | NOL1 | NOL2 | SGRL | SGR | RCCR | HCAR | PAO1 | PAO2 | PAO3 | PAO4 | PPH1 | PPH2 | PPH3 | FtsH6-1 | FtsH6-2 | ||
| ABRE | Abscisic acid responsiveness | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| ACE | Light responsiveness | + | + | ||||||||||||||
| AE-box | Light response | + | + | + | + | + | + | ||||||||||
| ARE | Anaerobic induction | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| CAAT-box | Common cis-acting element in promoter and enhancer regions | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| AuxRR-core | Auxin responsiveness | + | + | ||||||||||||||
| CGTCA-motif | MeJA-responsiveness | + | + | + | + | + | |||||||||||
| G-box | Light responsiveness | + | + | + | + | + | + | + | + | + | + | ||||||
| TATC-box | Gibberellin-responsiveness | + | + | + | + | ||||||||||||
| TC-rich repeats | Defense and stress responsiveness | + | + | + | |||||||||||||
| TCA-element | Salicylic acid responsiveness | + | + | + | + | + | + | + | + | + | + | ||||||
| TCT-motif | Part of a light-responsive element | + | + | + | + | + | + | + | + | + | |||||||
| TGACG-motif | MeJA-responsiveness | + | + | + | + | + | + | ||||||||||
| Box4 | Light responsiveness | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| GARE-motif | Gibberellin-responsive element | + | + | + | + | + | + | + | |||||||||
| P-box | Gibberellin-responsive element | + | + | + | + | + | + | ||||||||||
| GATA-motif | Part of a light-responsive element | + | + | + | |||||||||||||
| MRE | MYB binding site involved in light responsiveness | + | + | + | + | ||||||||||||
| AT1-motif | Part of a light-responsive module | + | + | + | |||||||||||||
| ATCT-motif | Light responsiveness | + | + | ||||||||||||||
| I-box | Light-responsive element | + | + | ||||||||||||||
| chs-CMA2a | Light-responsive element | + | + | + | |||||||||||||
| ABRE3a | + | + | + | + | + | + | + | + | |||||||||
| ABRE4 | + | + | + | + | + | + | + | + | |||||||||
| TCCC-motif | Light-responsive element | + | + | + | + | + | |||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.-M.; Sun, M.-H.; Tang, X.-S.; Wang, C.-P.; Xie, Z.-S. Genome-Wide Identification and Expression Analysis of Senescence-Associated Genes in Grapevine (Vitis vinifera L.) Reveal Their Potential Functions in Leaf Senescence Order. Int. J. Mol. Sci. 2022, 23, 12731. https://doi.org/10.3390/ijms232112731
Li Y-M, Sun M-H, Tang X-S, Wang C-P, Xie Z-S. Genome-Wide Identification and Expression Analysis of Senescence-Associated Genes in Grapevine (Vitis vinifera L.) Reveal Their Potential Functions in Leaf Senescence Order. International Journal of Molecular Sciences. 2022; 23(21):12731. https://doi.org/10.3390/ijms232112731
Chicago/Turabian StyleLi, You-Mei, Meng-Hao Sun, Xuan-Si Tang, Chao-Ping Wang, and Zhao-Sen Xie. 2022. "Genome-Wide Identification and Expression Analysis of Senescence-Associated Genes in Grapevine (Vitis vinifera L.) Reveal Their Potential Functions in Leaf Senescence Order" International Journal of Molecular Sciences 23, no. 21: 12731. https://doi.org/10.3390/ijms232112731
APA StyleLi, Y.-M., Sun, M.-H., Tang, X.-S., Wang, C.-P., & Xie, Z.-S. (2022). Genome-Wide Identification and Expression Analysis of Senescence-Associated Genes in Grapevine (Vitis vinifera L.) Reveal Their Potential Functions in Leaf Senescence Order. International Journal of Molecular Sciences, 23(21), 12731. https://doi.org/10.3390/ijms232112731






