The Integration of Metabolomics and Transcriptomics Provides New Insights for the Identification of Genes Key to Auxin Synthesis at Different Growth Stages of Maize
Abstract
1. Introduction
2. Results
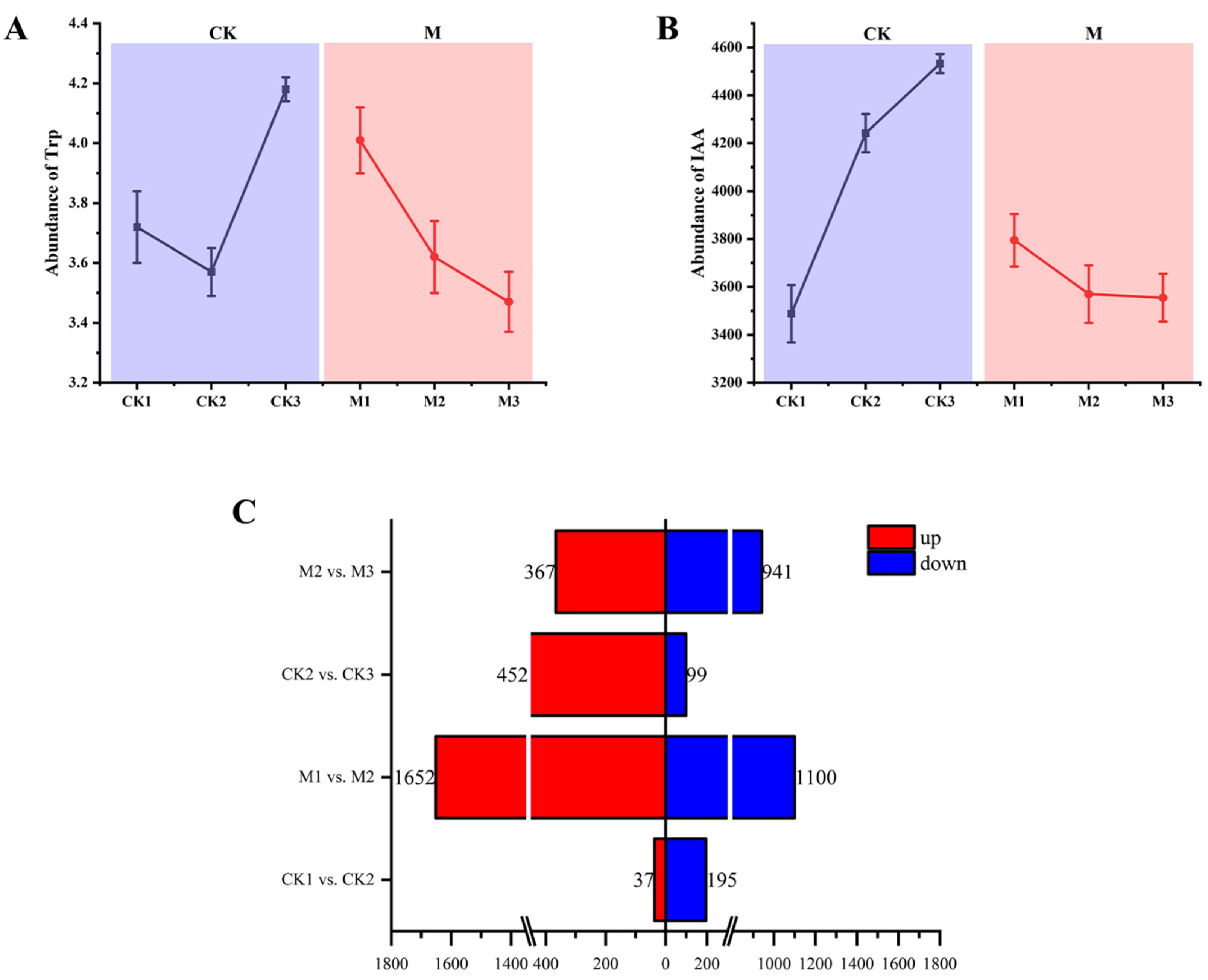
2.1. Determination of Trp and IAA Contents and Identification of DEGs
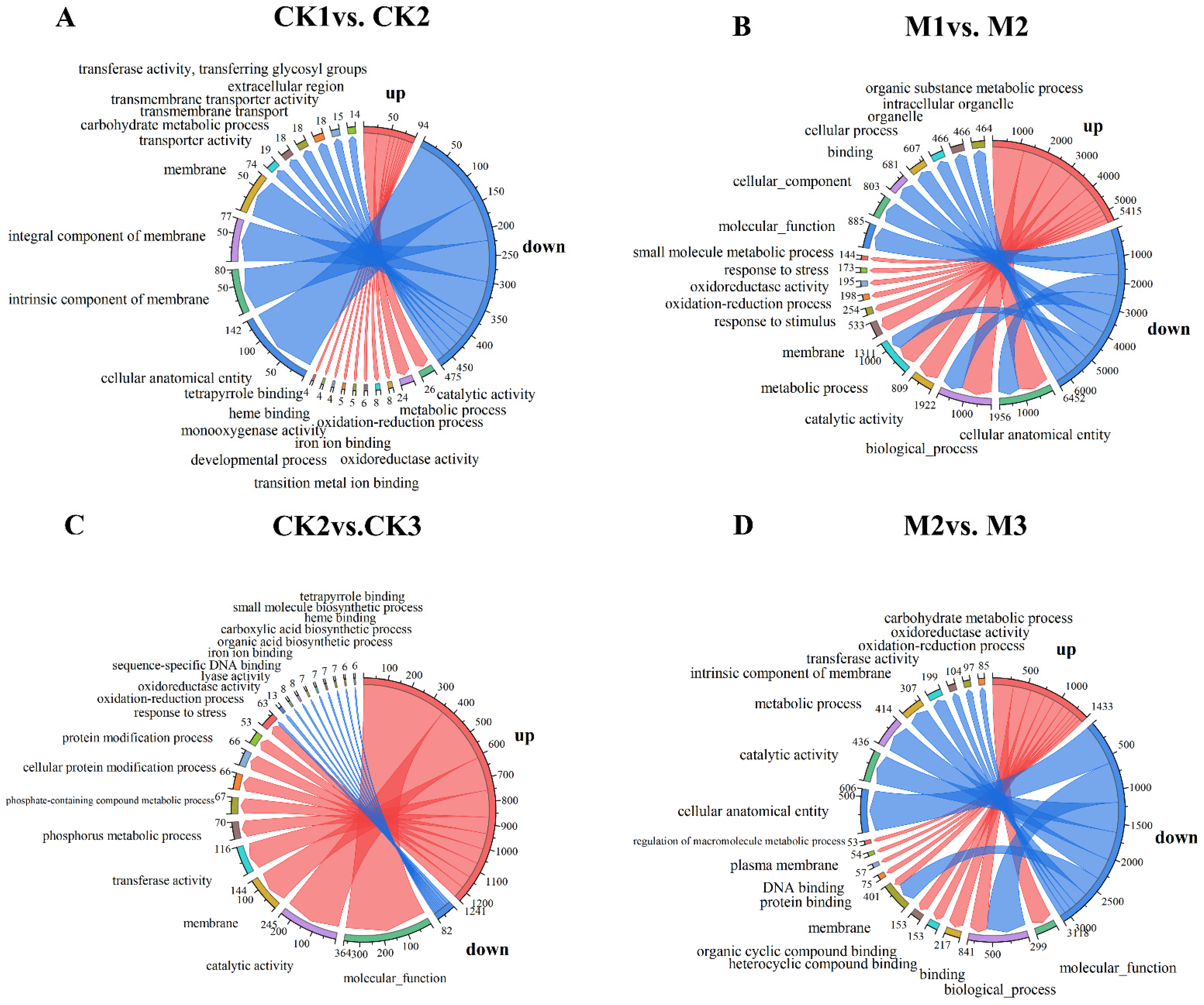
2.2. GO Enrichment Analysis of DEGs
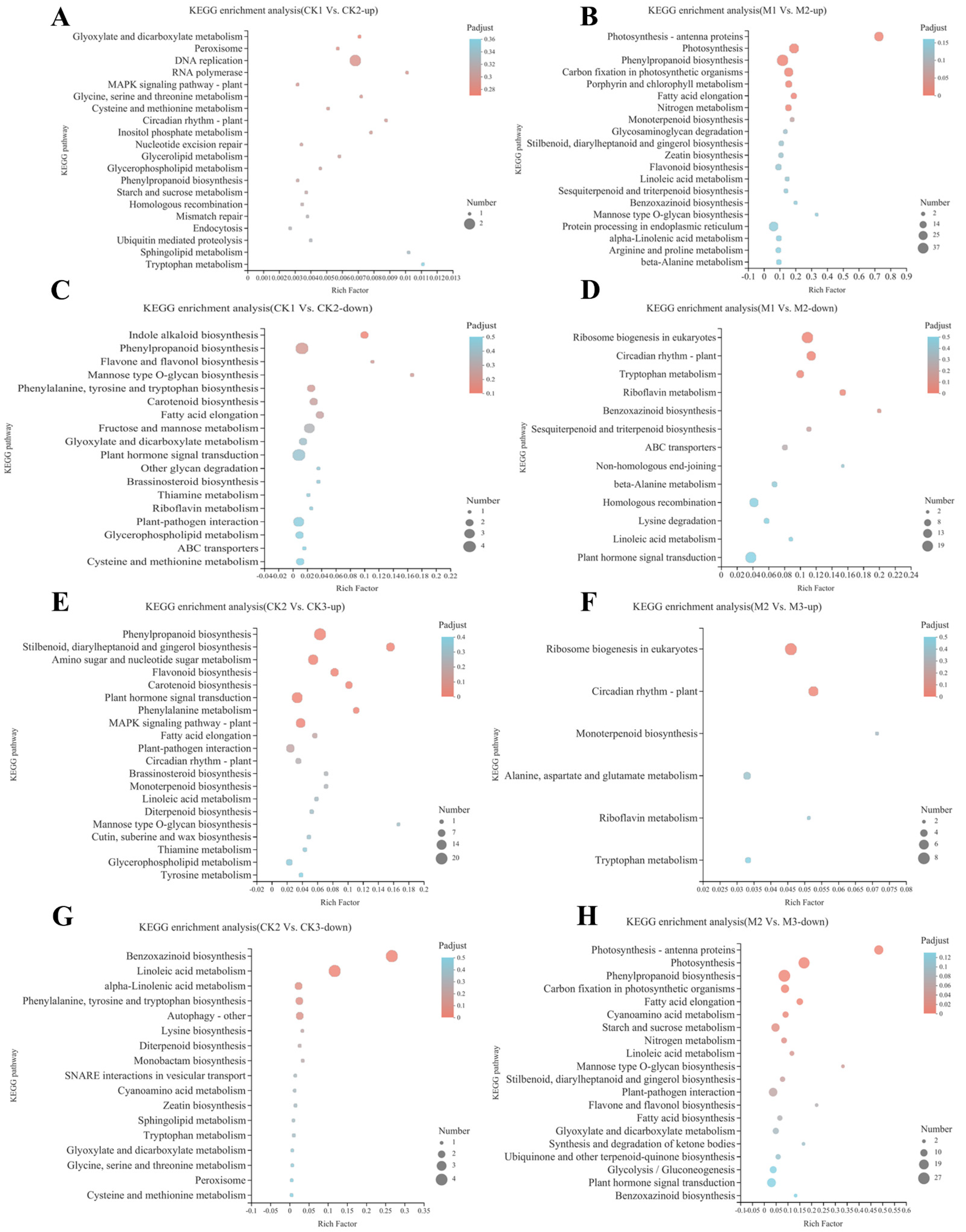
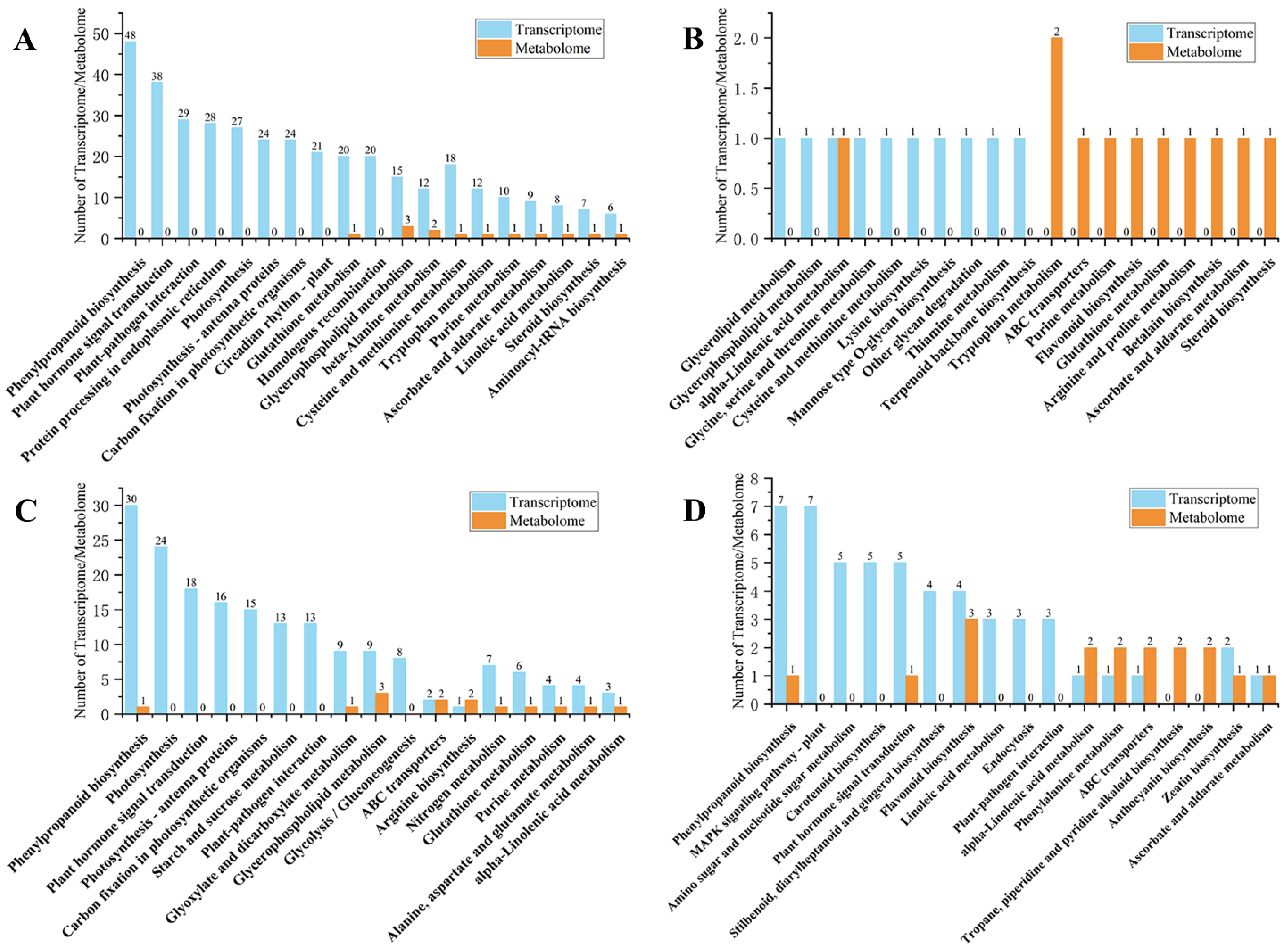
2.3. KEGG Pathway Enrichment Analysis of DEGs
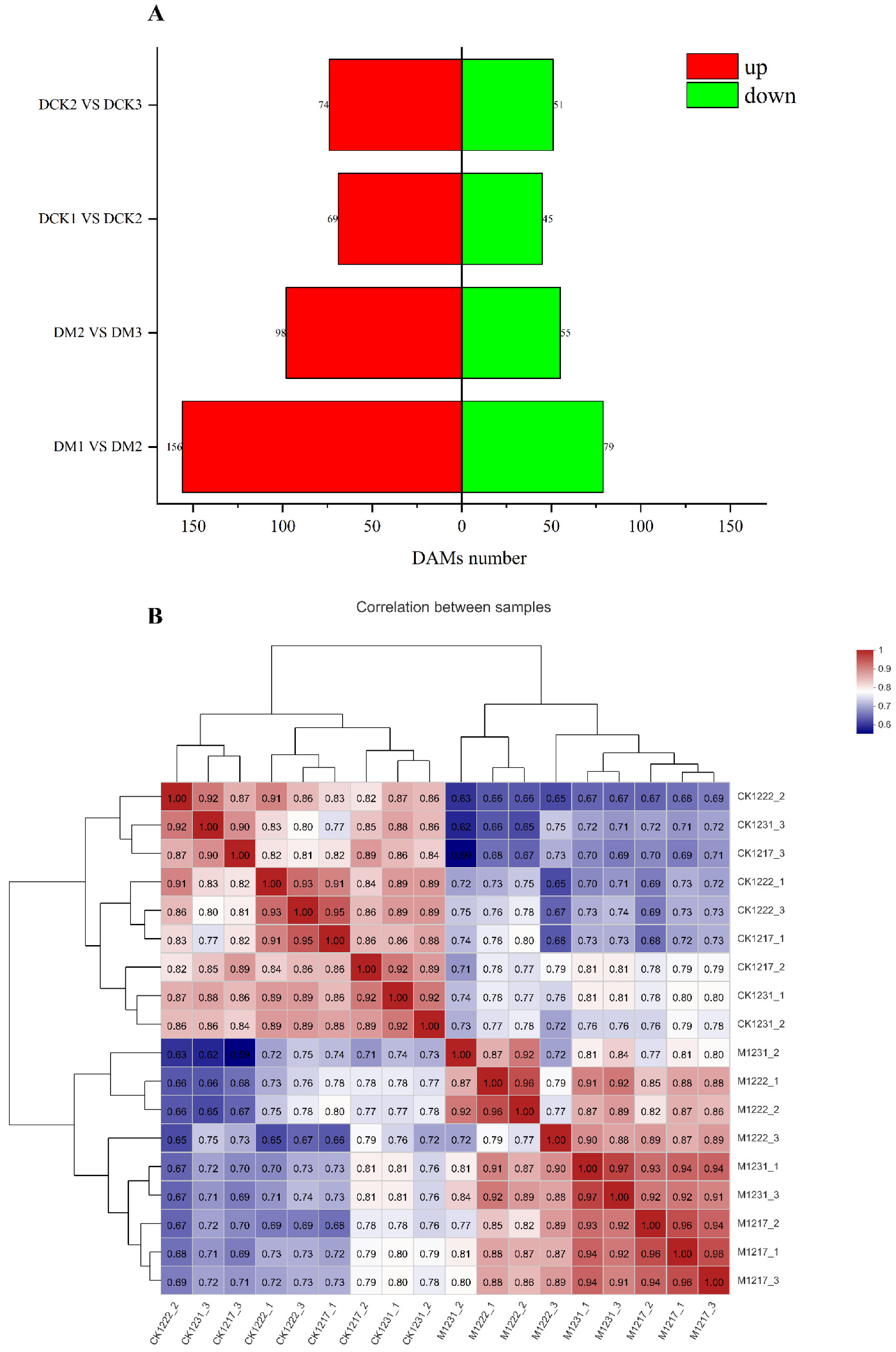
2.4. Widely Targeted Metabolomics Analysis and Overall Metabolite Identification
2.5. Integration of Transcriptomic and Metabonomic Analyses
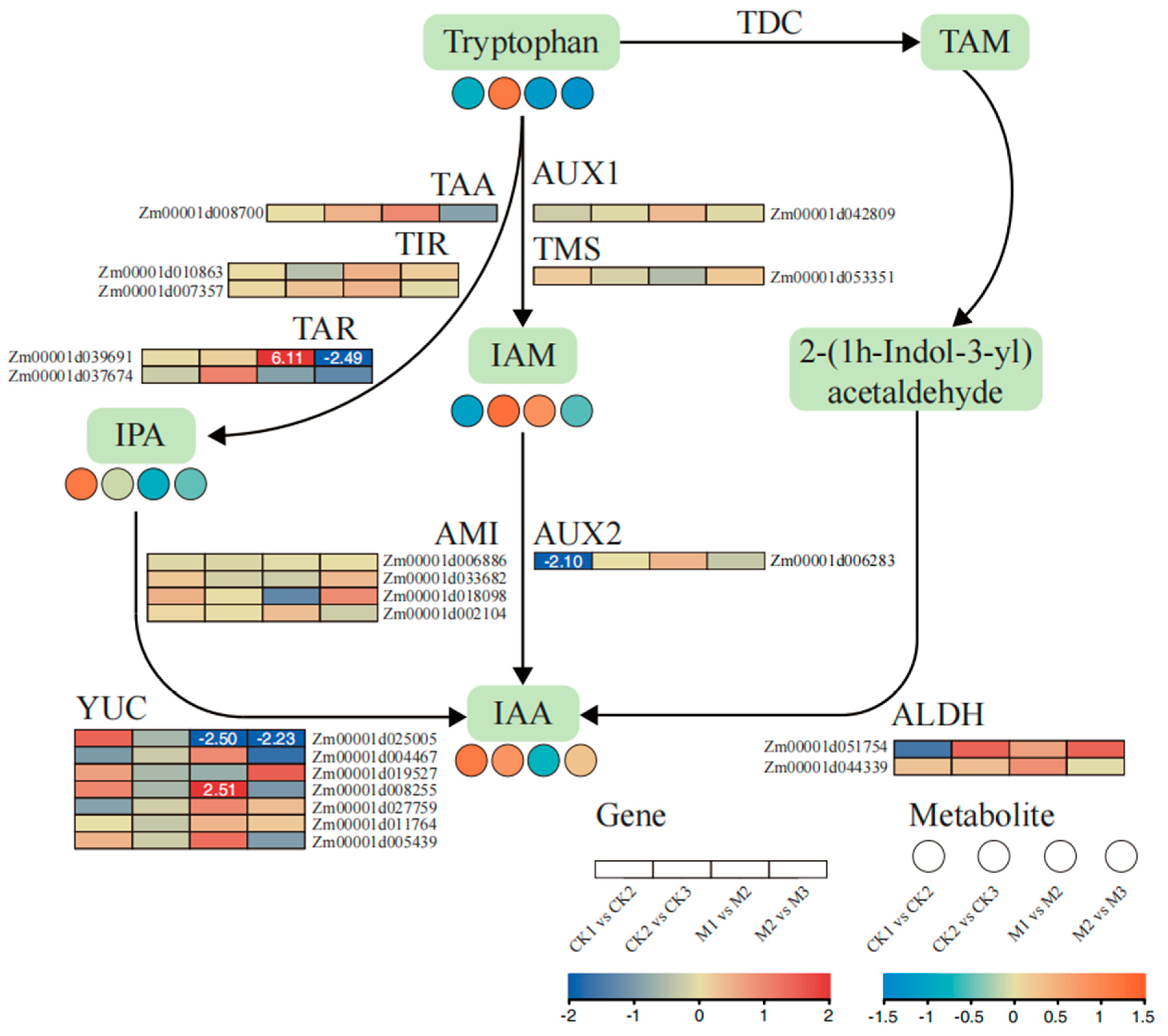
2.6. Analysis of Auxin Biosynthesis-Related Metabolites and Gene Expression
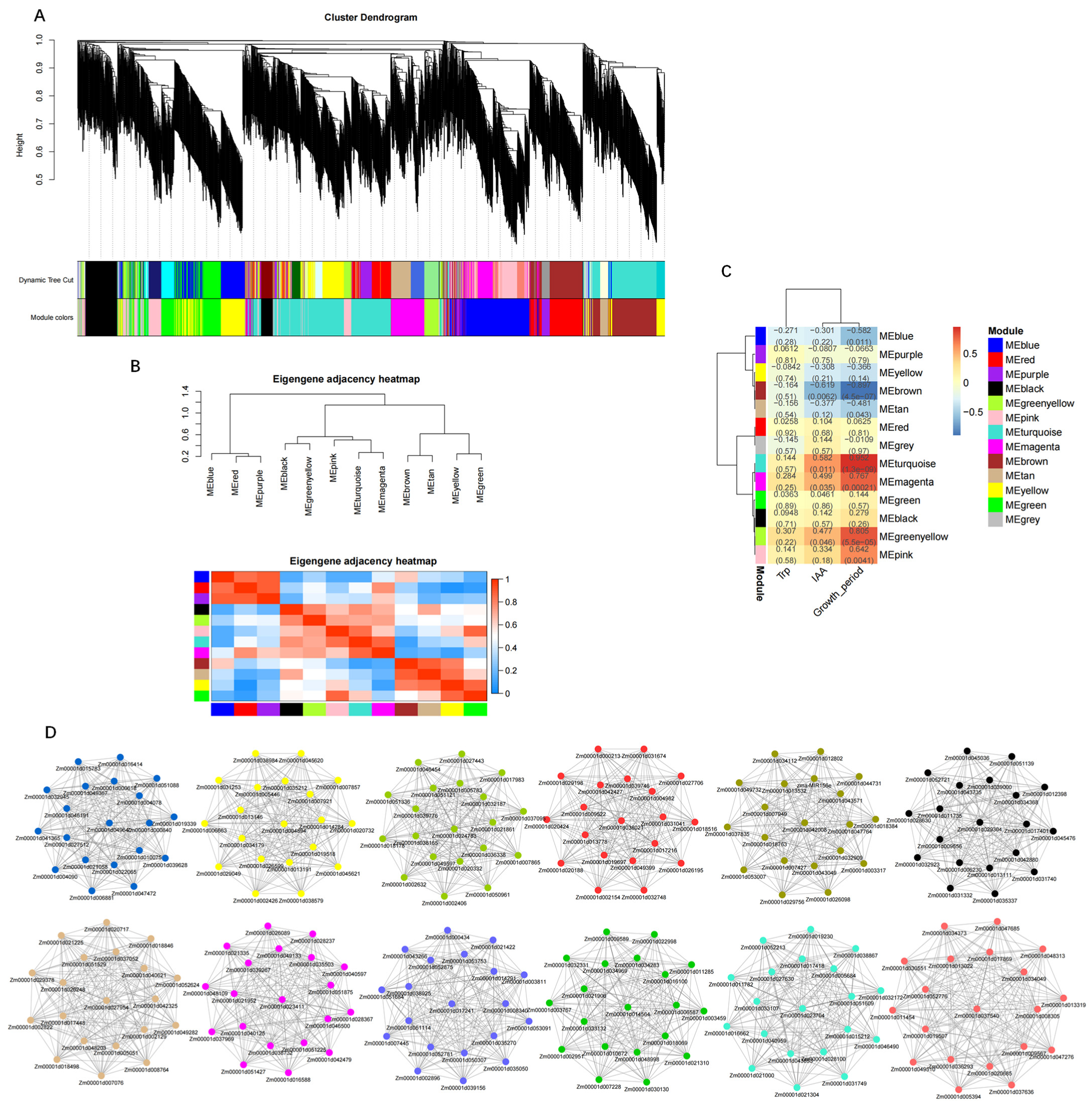
2.7. Weighted Gene Co-Expression Network Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Auxin and Trp Contents
4.3. RNA-Sequencing and Differential Expression Gene Analysis in Maize
4.4. Widely Targeted Metabonomic Analysis
4.5. Transcriptome and Metabolome Analysis
4.6. Construction of Gene Co-Expression Network
4.7. Real-Time Fluorescent Quantitative PCR Verification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fedoroff, N.V. About maize transposable elements and development. Cell 1989, 56, 181–191. [Google Scholar] [CrossRef]
- Blázquez, M.A.; Nelson, D.C.; Weijers, D. Evolution of Plant Hormone Response Pathways. Annu. Rev. Plant Biol. 2020, 71, 327–353. [Google Scholar] [CrossRef] [PubMed]
- Gomes, G.; Scortecci, K.C. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef]
- Santner, A.; Calderon-Villalobos, L.I.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Dziewit, K.; Pěnčík, A.; Dobrzyńska, K.; Novák, O.; Szal, B.; Podgórska, A. Spatiotemporal auxin distribution in Arabidopsis tissues is regulated by anabolic and catabolic reactions under long-term ammonium stress. BMC Plant Biol. 2021, 21, 602. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef]
- Kunkel, B.N.; Johnson, J. Auxin Plays Multiple Roles during Plant-Pathogen Interactions. Cold Spring Harb. Perspect. Biol. 2021, 13, a040022. [Google Scholar] [CrossRef]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Yue, K.; Lingling, L.; Xie, J.; Coulter, J.A.; Luo, Z. Synthesis and regulation of auxin and abscisic acid in maize. Plant Signal. Behav. 2021, 16, 1891756. [Google Scholar] [CrossRef]
- Sugawara, S.; Hishiyama, S.; Jikumaru, Y.; Hanada, A.; Nishimura, T.; Koshiba, T.; Zhao, Y.; Kamiya, Y.; Kasahara, H. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5430–5435. [Google Scholar] [CrossRef] [PubMed]
- Quittenden, L.J.; Davies, N.W.; Smith, J.A.; Molesworth, P.P.; Tivendale, N.D.; Ross, J.J. Auxin biosynthesis in pea: Characterization of the tryptamine pathway. Plant Physiol. 2009, 151, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Parra, B.; Pérez-Alonso, M.M.; Ortiz-García, P.; Moya-Cuevas, J.; Hentrich, M.; Pollmann, S. Accumulation of the Auxin Precursor Indole-3-Acetamide Curtails Growth through the Repression of Ribosome-Biogenesis and Development-Related Transcriptional Networks. Int. J. Mol. Sci. 2021, 22, 2040. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, T.; Hoffmann, M.; Hentrich, M.; Pollmann, S. Indole-3-acetamide-dependent auxin biosynthesis: A widely distributed way of indole-3-acetic acid production? Eur. J. Cell Biol. 2010, 89, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alonso, M.M.; Ortiz-García, P.; Moya-Cuevas, J.; Lehmann, T.; Sánchez-Parra, B.; Björk, R.G.; Karim, S.; Amirjani, M.R.; Aronsson, H.; Wilkinson, M.D.; et al. Endogenous indole-3-acetamide levels contribute to the crosstalk between auxin and abscisic acid, and trigger plant stress responses in Arabidopsis. J. Exp. Bot. 2021, 72, 459–475. [Google Scholar] [CrossRef] [PubMed]
- Schütz, A.; Sandalova, T.; Ricagno, S.; Hübner, G.; König, S.; Schneider, G. Crystal structure of thiamindiphosphate-dependent indolepyruvate decarboxylase from Enterobacter cloacae, an enzyme involved in the biosynthesis of the plant hormone indole-3-acetic acid. Eur. J. Biochem. 2003, 270, 2312–2321. [Google Scholar] [CrossRef]
- Cao, X.; Yang, H.; Shang, C.; Ma, S.; Liu, L.; Cheng, J. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef]
- Qin, M.; Wang, J.; Zhang, T.; Hu, X.; Liu, R.; Gao, T.; Zhao, S.; Yuan, Y.; Zheng, J.; Wang, Z.; et al. Genome-Wide Identification and Analysis on YUCCA Gene Family in Isatis indigotica Fort. and IiYUCCA6-1 Functional Exploration. Int. J. Mol. Sci. 2020, 21, 2188. [Google Scholar] [CrossRef]
- Uc-Chuc, M.A.; Pérez-Hernández, C.; Galaz-Ávalos, R.M.; Brito-Argaez, L.; Aguilar-Hernández, V.; Loyola-Vargas, V.M. YUCCA-Mediated Biosynthesis of the Auxin IAA Is Required during the Somatic Embryogenic Induction Process in Coffea canephora. Int. J. Mol. Sci. 2020, 21, 4751. [Google Scholar] [CrossRef]
- Leontovyčová, H.; Trdá, L.; Dobrev, P.I.; Šašek, V.; Gay, E.; Balesdent, M.H.; Burketová, L. Auxin biosynthesis in the phytopathogenic fungus Leptosphaeria maculans is associated with enhanced transcription of indole-3-pyruvate decarboxylase LmIPDC2 and tryptophan aminotransferase LmTAM1. Res. Microbiol. 2020, 171, 174–184. [Google Scholar] [CrossRef]
- Lavy, M.; Estelle, M. Mechanisms of auxin signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef] [PubMed]
- Sadok, I.; Gamian, A.; Staniszewska, M.M. Chromatographic analysis of tryptophan metabolites. J. Sep. Sci. 2017, 40, 3020–3045. [Google Scholar] [CrossRef] [PubMed]
- Gruß, H.; Sewald, N. Late-Stage Diversification of Tryptophan-Derived Biomolecules. Chem. A Eur. J. 2020, 26, 5328–5340. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chen, L.; Zeng, A.P. CRISPR/Cas9-facilitated engineering with growth-coupled and sensor-guided in vivo screening of enzyme variants for a more efficient chorismate pathway in E. coli. Metab. Eng. Commun. 2019, 9, e00094. [Google Scholar] [CrossRef] [PubMed]
- Xiu, Z.L.; Chang, Z.Y.; Zeng, A.P. Nonlinear dynamics of regulation of bacterial trp operon: Model analysis of integrated effects of repression, feedback inhibition, and attenuation. Biotechnol. Prog. 2002, 18, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Shao, X.; Li, J. Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2000, 24, 327–333. [Google Scholar] [CrossRef]
- Jijón-Moreno, S.; Marcos-Jiménez, C.; Pedraza, R.O.; Ramírez-Mata, A.; de Salamone, I.G.; Fernández-Scavino, A.; Vásquez-Hernández, C.A.; Soto-Urzúa, L.; Baca, B.E. The ipdC, hisC1 and hisC2 genes involved in indole-3-acetic production used as alternative phylogenetic markers in Azospirillum brasilense. Antonie Van Leeuwenhoek 2015, 107, 1501–1517. [Google Scholar] [CrossRef]
- Skirycz, A.; Reichelt, M.; Burow, M.; Birkemeyer, C.; Rolcik, J.; Kopka, J.; Zanor, M.I.; Gershenzon, J.; Strnad, M.; Szopa, J.; et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. Cell Mol. Biol. 2006, 47, 10–24. [Google Scholar] [CrossRef]
- Ferrer, L.; Mindt, M.; Suarez-Diez, M.; Jilg, T.; Zagorščak, M.; Lee, J.H.; Gruden, K.; Wendisch, V.F.; Cankar, K. Fermentative Indole Production via Bacterial Tryptophan Synthase Alpha Subunit and Plant Indole-3-Glycerol Phosphate Lyase Enzymes. J. Agric. Food Chem. 2022, 70, 5634–5645. [Google Scholar] [CrossRef]
- Li, R.; Jiang, J.; Jia, S.; Zhu, X.; Su, H.; Li, J. Overexpressing broccoli tryptophan biosynthetic genes BoTSB1 and BoTSB2 promotes biosynthesis of IAA and indole glucosinolates. Physiol. Plant. 2020, 168, 174–187. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, K.F.; D’Amico, R.N.; Sahu, D.; Boehr, D.D. Distinct conformational dynamics and allosteric networks in alpha tryptophan synthase during active catalysis. Protein Sci. A Publ. Protein Soc. 2021, 30, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Abu-Zaitoon, Y.M.; Abu-Zaiton, A.; Tawaha, A.; Fandi, K.G.; Alnaimat, S.M.; Pati, S.; Almomani, F.A. Evidence from Co-expression Analysis for the Involvement of Amidase and INS in the Tryptophan-Independent Pathway of IAA Synthesis in Arabidopsis. Appl. Biochem. Biotechnol. 2022, 194, 4673–4682. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.D.; Sampson, M.B.; Neuffer, M.G.; Michalczuk, L.; Slovin, J.P.; Cohen, J.D. Indole-3-Acetic Acid Biosynthesis in the Mutant Maize orange pericarp, a Tryptophan Auxotroph. Science 1991, 254, 998–1000. [Google Scholar] [CrossRef]
- Xu, F.; He, S.; Zhang, J.; Mao, Z.; Wang, W.; Li, T.; Yang, H.Q. Photoactivated CRY1 and phyB Interact Directly with AUX/IAA Proteins to Inhibit Auxin Signaling in Arabidopsis. Mol. Plant 2018, 11, 523–541. [Google Scholar] [CrossRef]
- Jing, Y.; Lin, R. Transcriptional regulatory network of the light signaling pathways. New Phytol. 2020, 227, 683–697. [Google Scholar] [CrossRef]
- Pollmann, S.; Neu, D.; Lehmann, T.; Berkowitz, O.; Schäfer, T.; Weiler, E.W. Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta 2006, 224, 1241–1253. [Google Scholar] [CrossRef]
- Nemoto, K.; Hara, M.; Suzuki, M.; Seki, H.; Muranaka, T.; Mano, Y. The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Lett. 2009, 583, 487–492. [Google Scholar] [CrossRef]
- Pollmann, S.; Düchting, P.; Weiler, E.W. Tryptophan-dependent indole-3-acetic acid biosynthesis by ‘IAA-synthase’ proceeds via indole-3-acetamide. Phytochemistry 2009, 70, 523–531. [Google Scholar] [CrossRef]
- Suzuki, M.; Yamazaki, C.; Mitsui, M.; Kakei, Y.; Mitani, Y.; Nakamura, A.; Ishii, T.; Soeno, K.; Shimada, Y. Transcriptional feedback regulation of YUCCA genes in response to auxin levels in Arabidopsis. Plant Cell Rep. 2015, 34, 1343–1352. [Google Scholar] [CrossRef]
- Bagley, M.C.; Stepanova, A.N.; Ekelöf, M.; Alonso, J.M.; Muddiman, D.C. Development of a relative quantification method for infrared matrix-assisted laser desorption electrospray ionization mass spectrometry imaging of Arabidopsis seedlings. Rapid Commun. Mass Spectrom. 2020, 34, e8616. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G. Auxin signal transduction. Essays Biochem. 2015, 58, 1–12. [Google Scholar] [PubMed]
- Kriechbaumer, V.; Park, W.J.; Gierl, A.; Glawischnig, E. Auxin biosynthesis in maize. Plant Biol. 2006, 8, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Perez, V.C.; Dai, R.; Bai, B.; Tomiczek, B.; Askey, B.C.; Zhang, Y.; Rubin, G.M.; Ding, Y.; Grenning, A.; Block, A.K.; et al. Aldoximes are precursors of auxins in Arabidopsis and maize. New Phytol. 2021, 231, 1449–1461. [Google Scholar] [CrossRef]
- He, W.; Brumos, J.; Li, H.; Ji, Y.; Ke, M.; Gong, X.; Zeng, Q.; Li, W.; Zhang, X.; An, F.; et al. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 2011, 23, 3944–3960. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Yang, H.; Ji, S.; Tian, C.; Chen, N.; Gong, H.; Li, J. Metabolome and Transcriptome Analyses of Anthocyanin Accumulation Mechanisms Reveal Metabolite Variations and Key Candidate Genes Involved in the Pigmentation of Prunus tomentosa Thunb. Cherry Fruit. Front. Plant Sci. 2022, 13, 938908. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Z.; Zhang, H.; Jiao, P.; Wei, X.; Liu, S.; Guan, S.; Ma, Y. The Integration of Metabolomics and Transcriptomics Provides New Insights for the Identification of Genes Key to Auxin Synthesis at Different Growth Stages of Maize. Int. J. Mol. Sci. 2022, 23, 13195. https://doi.org/10.3390/ijms232113195
Jiang Z, Zhang H, Jiao P, Wei X, Liu S, Guan S, Ma Y. The Integration of Metabolomics and Transcriptomics Provides New Insights for the Identification of Genes Key to Auxin Synthesis at Different Growth Stages of Maize. International Journal of Molecular Sciences. 2022; 23(21):13195. https://doi.org/10.3390/ijms232113195
Chicago/Turabian StyleJiang, Zhenzhong, Honglin Zhang, Peng Jiao, Xiaotong Wei, Siyan Liu, Shuyan Guan, and Yiyong Ma. 2022. "The Integration of Metabolomics and Transcriptomics Provides New Insights for the Identification of Genes Key to Auxin Synthesis at Different Growth Stages of Maize" International Journal of Molecular Sciences 23, no. 21: 13195. https://doi.org/10.3390/ijms232113195
APA StyleJiang, Z., Zhang, H., Jiao, P., Wei, X., Liu, S., Guan, S., & Ma, Y. (2022). The Integration of Metabolomics and Transcriptomics Provides New Insights for the Identification of Genes Key to Auxin Synthesis at Different Growth Stages of Maize. International Journal of Molecular Sciences, 23(21), 13195. https://doi.org/10.3390/ijms232113195





