Copper Amine Oxidase (CuAO)-Mediated Polyamine Catabolism Plays Potential Roles in Sweet Cherry (Prunus avium L.) Fruit Development and Ripening
Abstract
1. Introduction
2. Results
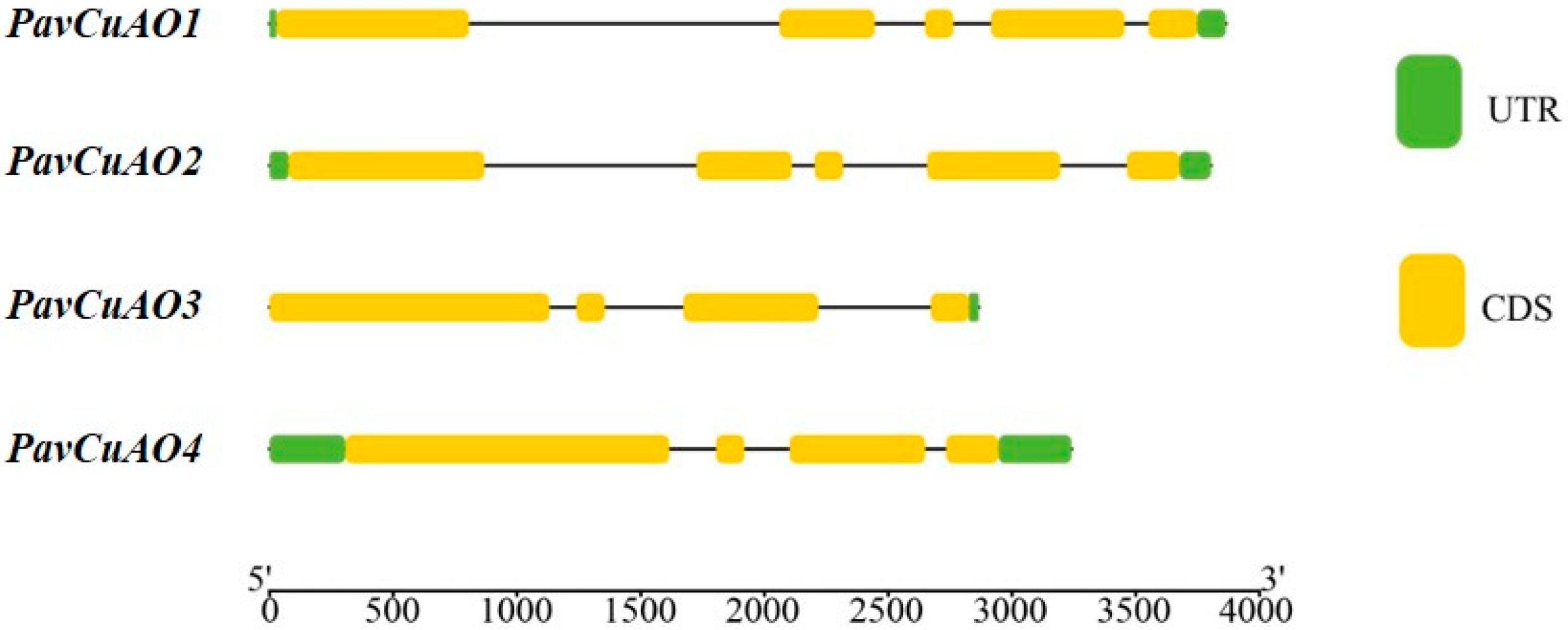
2.1. Identification and Gene Structure Analysis of PavCuAOs
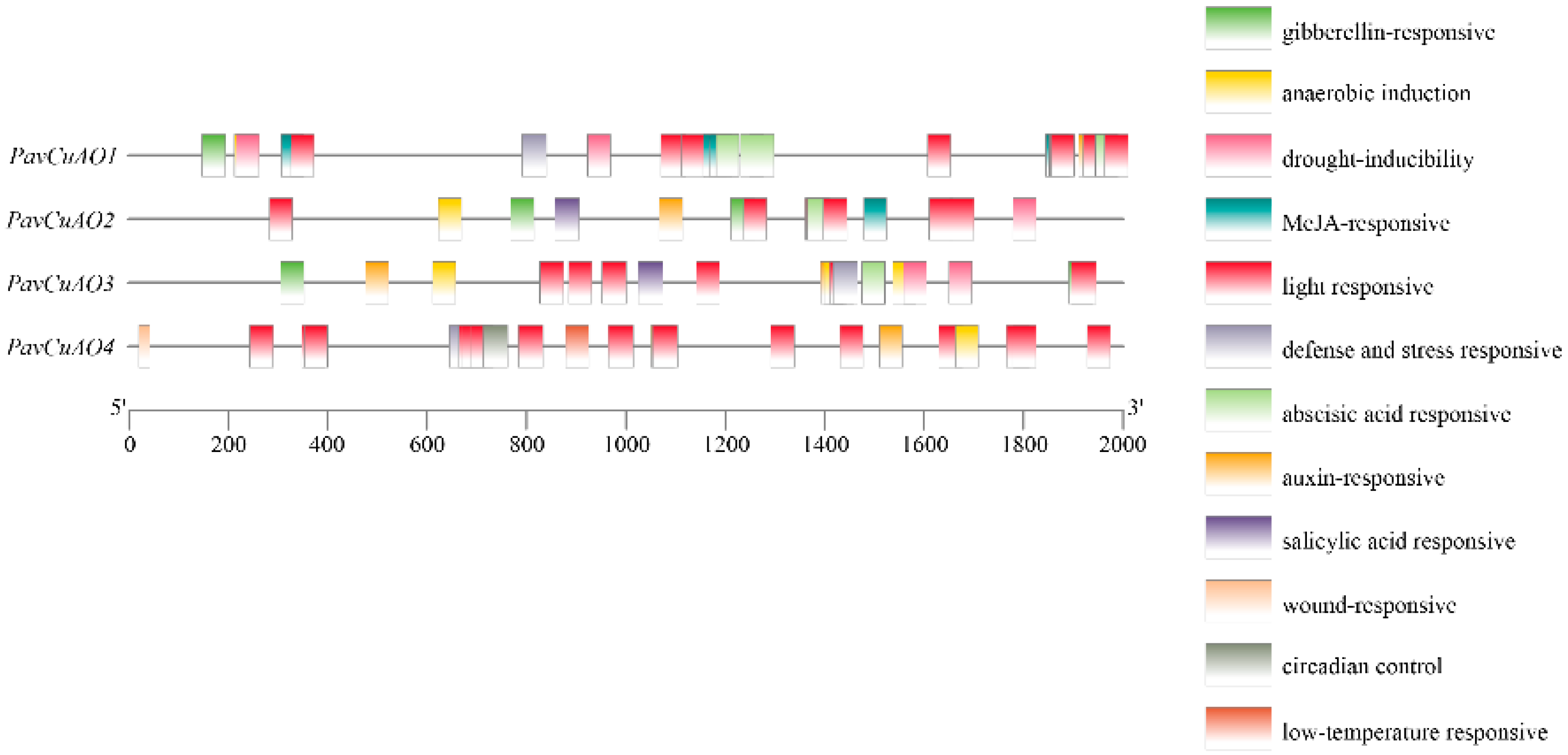
2.2. PavCuAOs Promoter Analysis
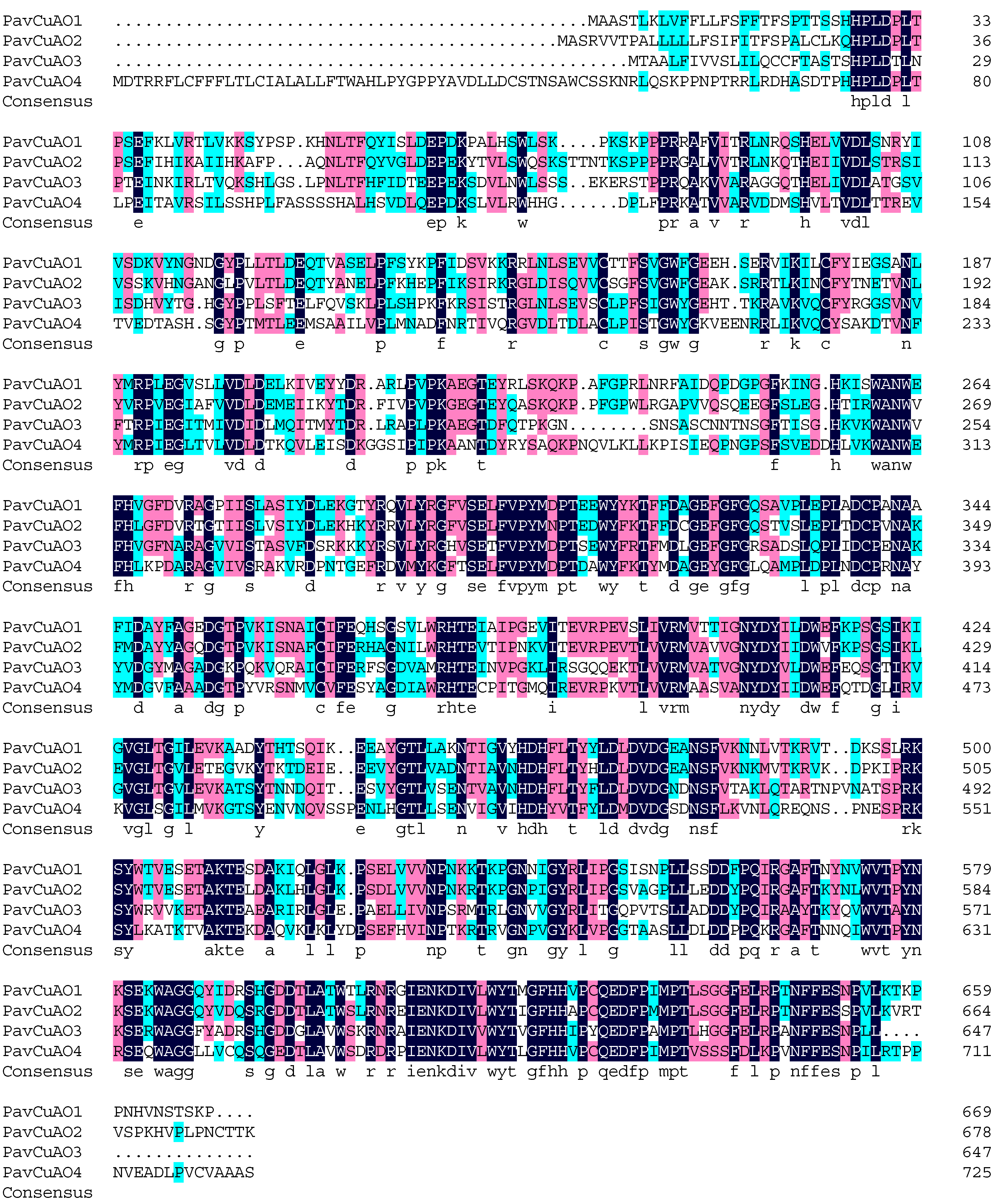
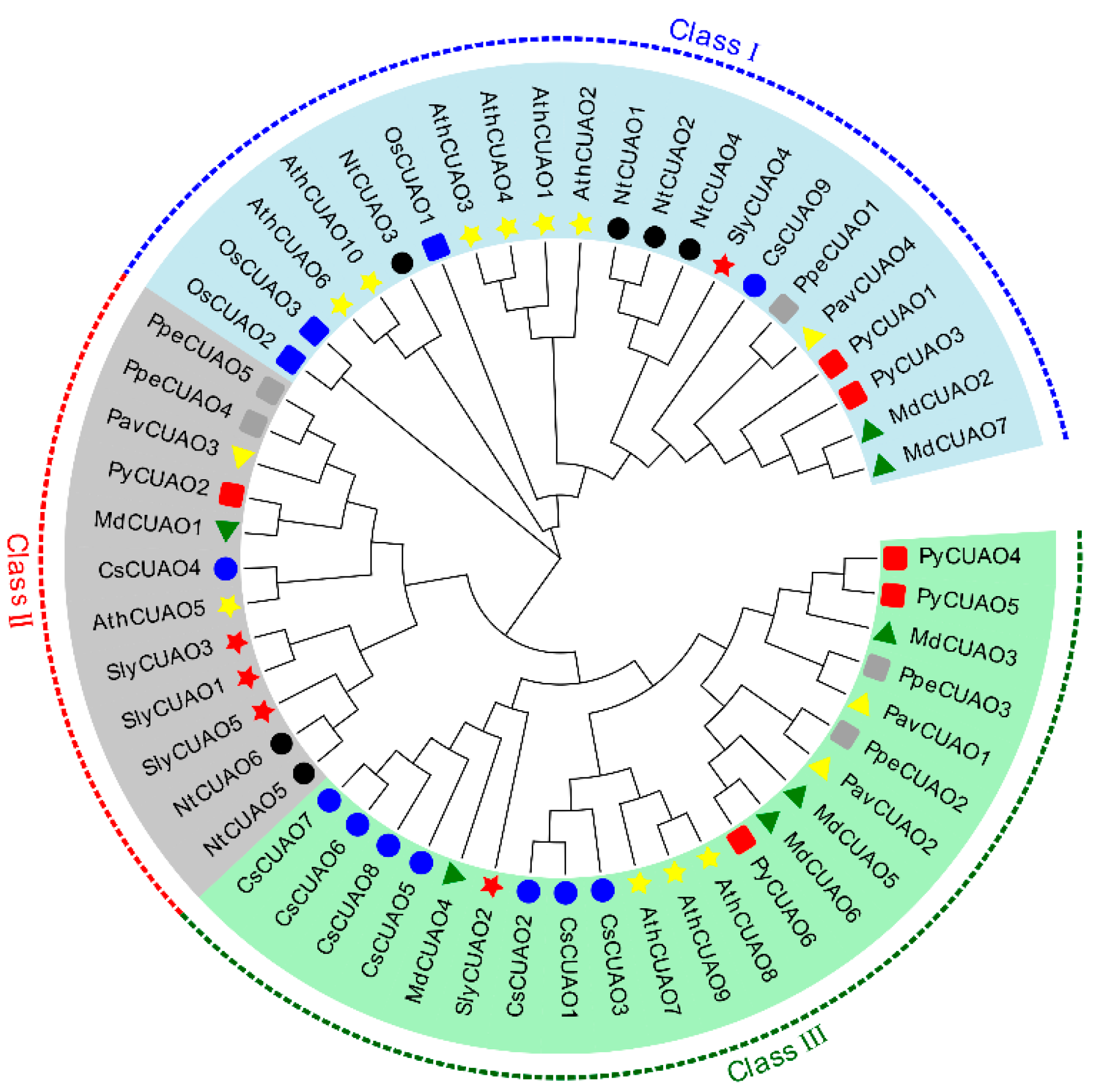
2.3. PavCuAOs Protein Multiplex Sequence Alignment and Phylogenetic Analysis
2.4. Expression Analysis of PavCuAOs in Different Tissues and Fruits at Different Developmental Stages
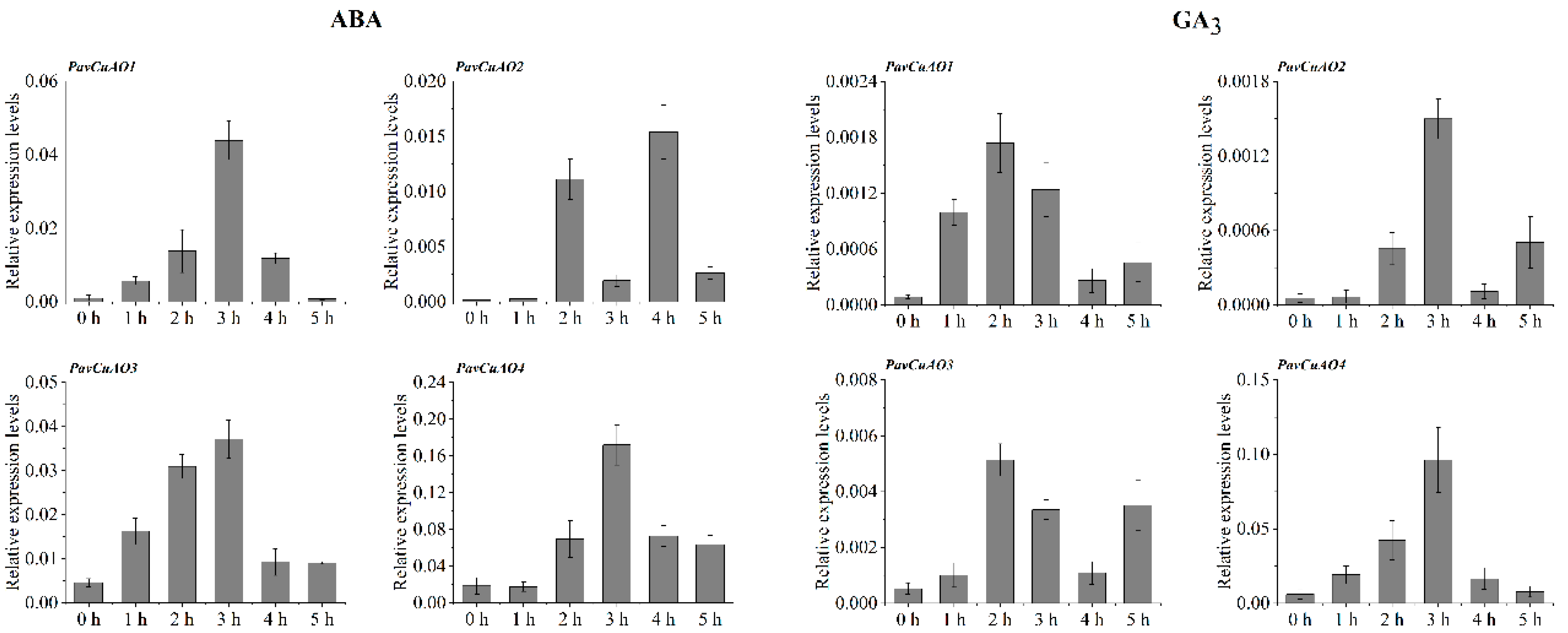
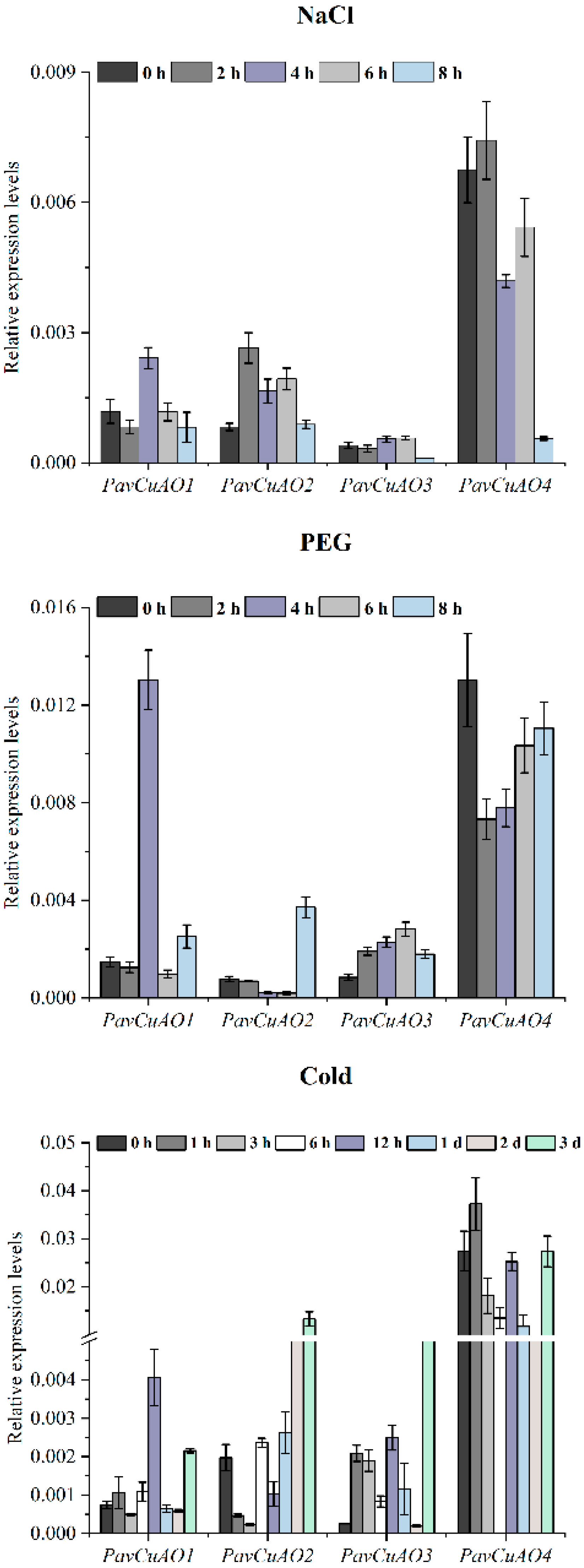
2.5. Expression Patterns of PavCuAO Genes under ABA, GA3, and Abiotic Stress (NaCl, PEG, and Cold) Treatments
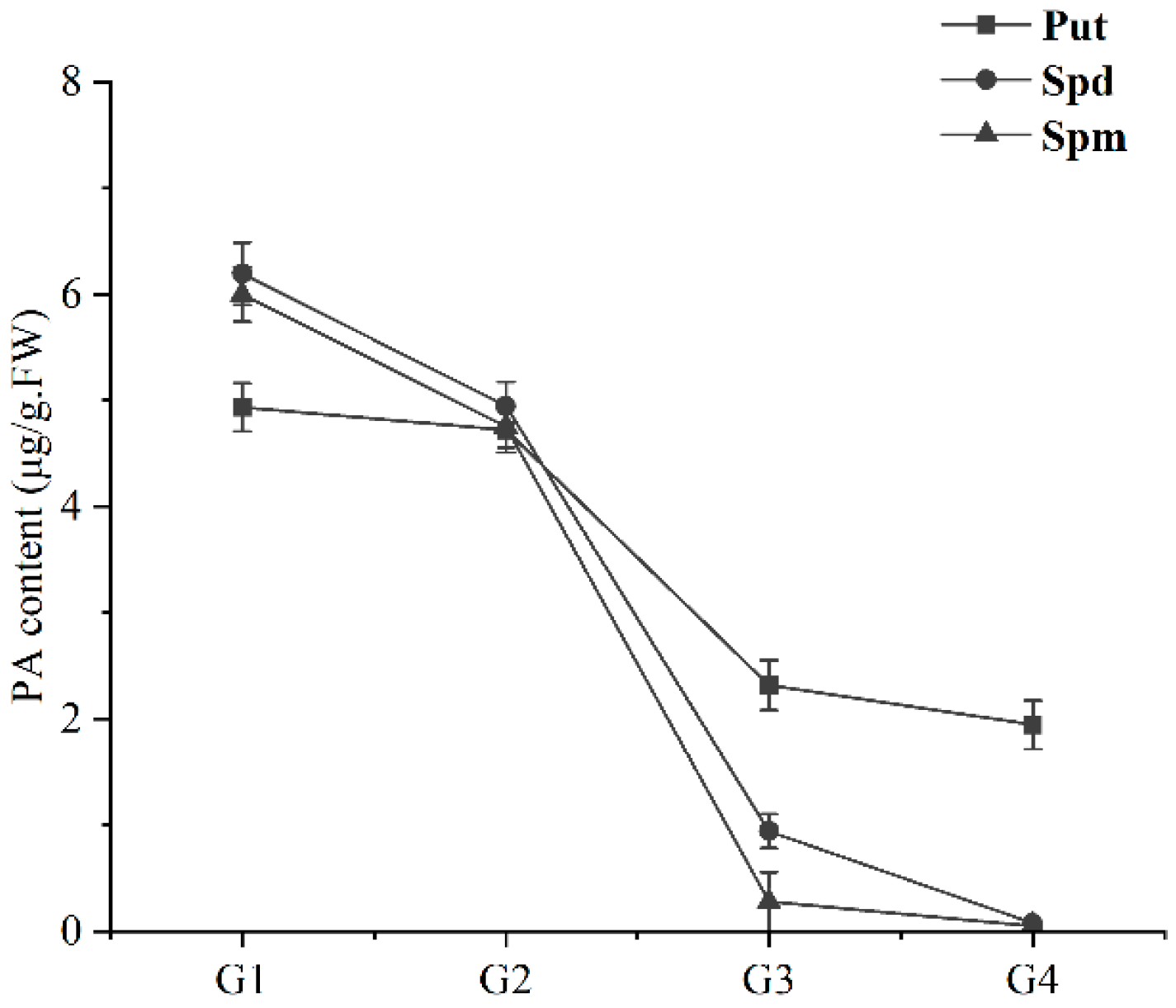
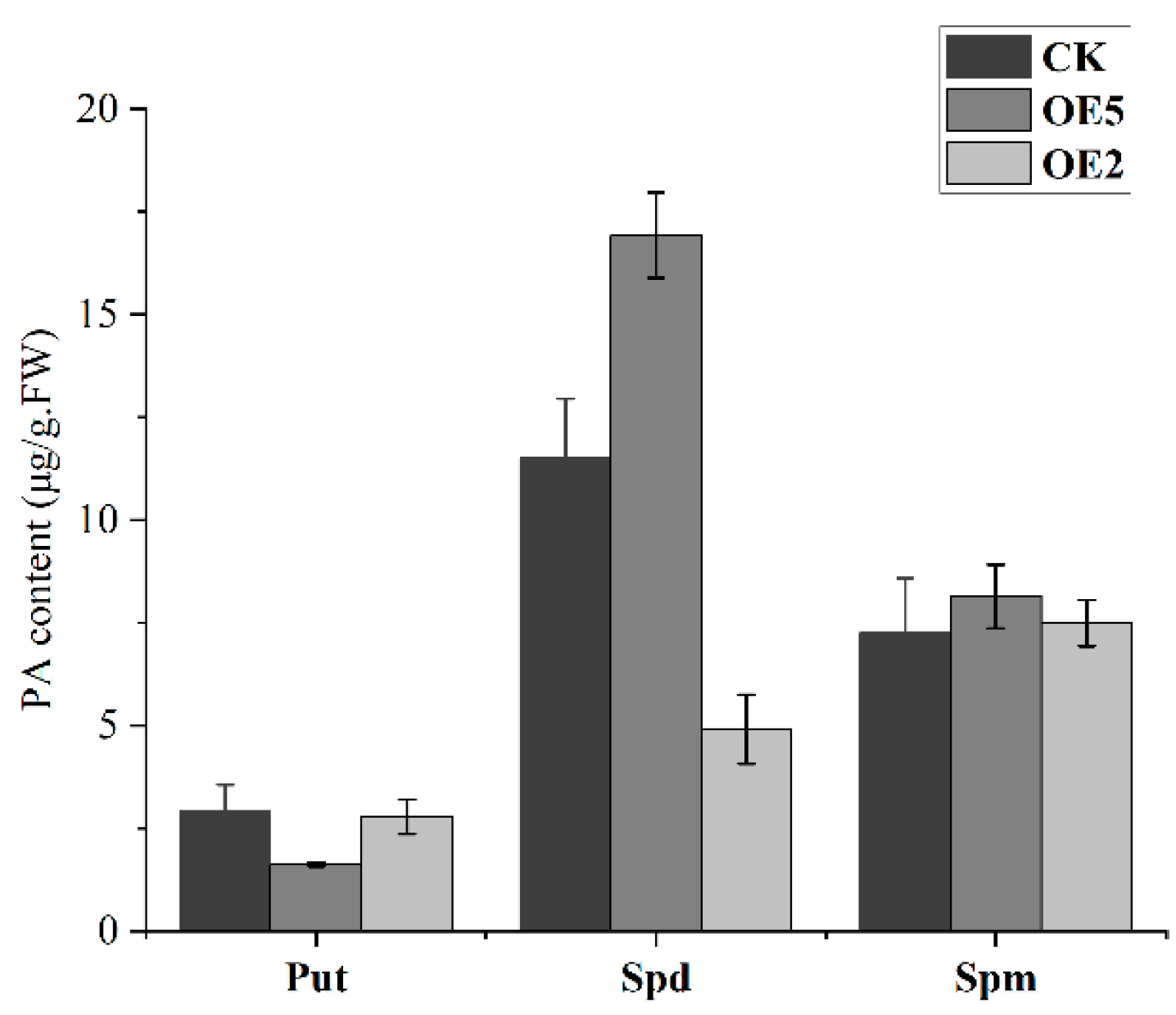
2.6. Detection of PA Content in the Ripening Process of Sweet Cherry Fruit
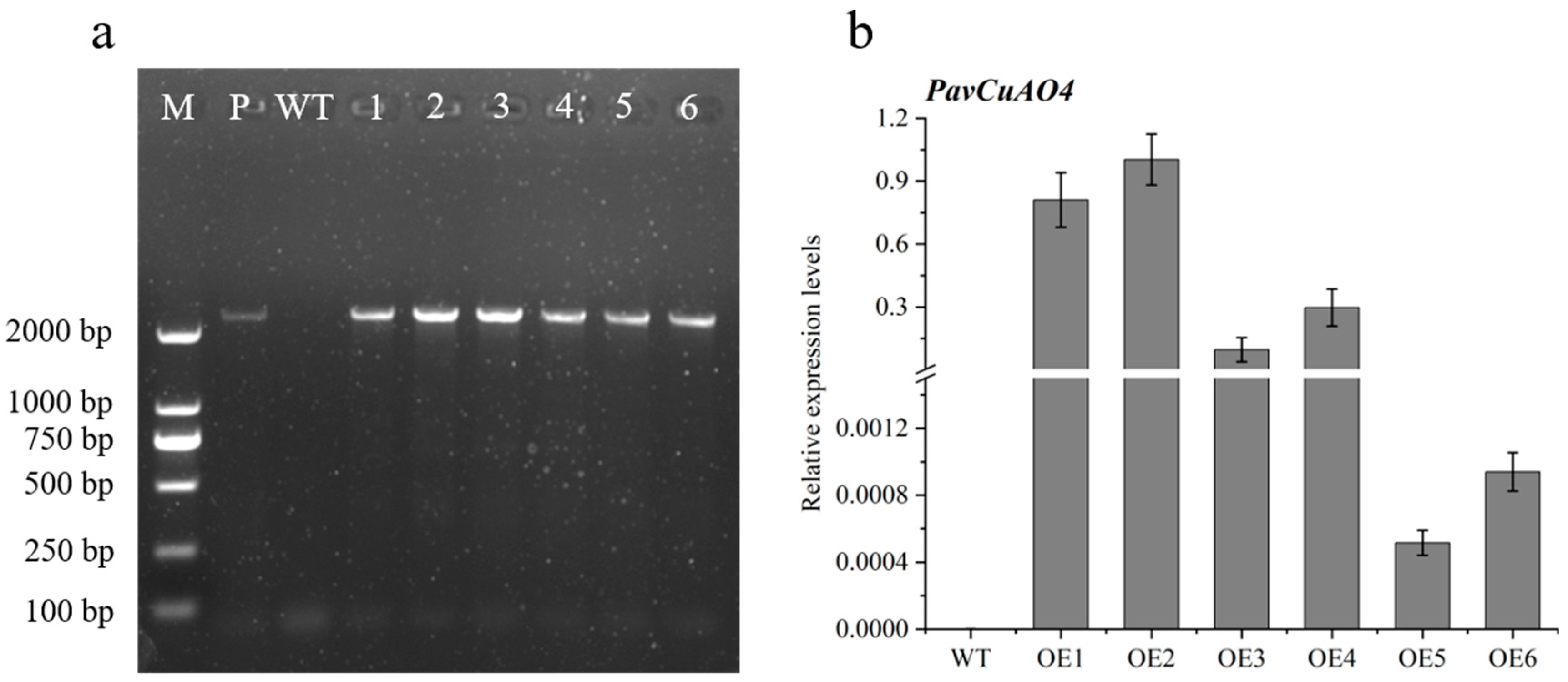
2.7. Genetic Transformation of Arabidopsis thaliana with PavCuAO4 and Detection of Free PAs in Transgenic Plants
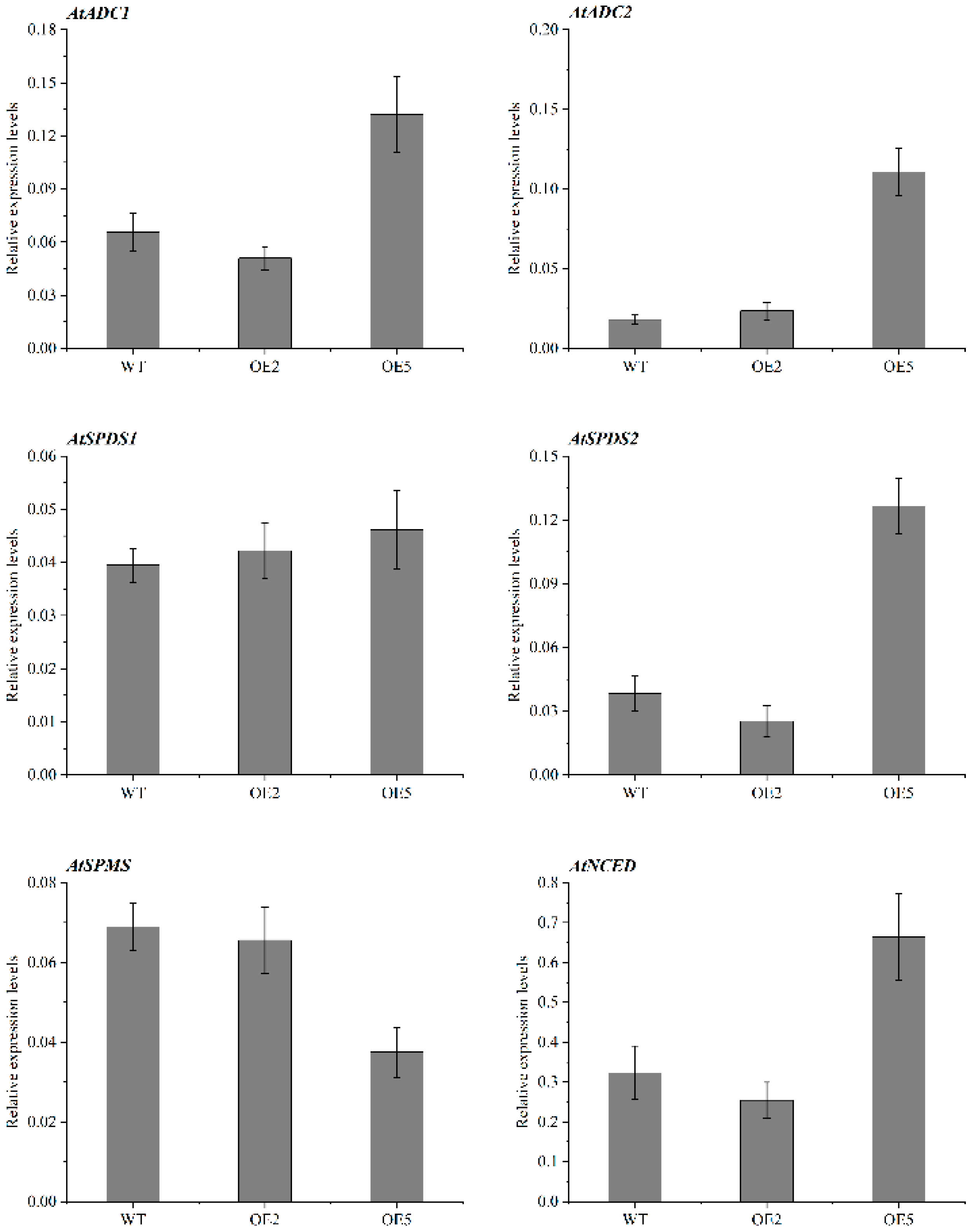
2.8. Expression Patterns of Genes Related to PA and ABA Synthesis in Overexpressed Plants
2.9. H2O2 In Situ Histochemical Staining
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Determination of PA in Sweet Cherry Fruit and Transgenic Plants
4.3. Identification of PavCuAO Gene Family Members
4.4. Gene Expression Analysis
4.5. Genetic Transformation and Transient Expression
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, W.; Wu, H.; Liu, J.-H. Genome-Wide Identification and Expression Profiling of Copper-Containing Amine Oxidase Genes in Sweet Orange (Citrus sinensis). Tree Genet. Genomes 2017, 13, 31. [Google Scholar] [CrossRef]
- Planas-Portell, J.; Gallart, M.; Tiburcio, A.F.; Altabella, T. Copper-Containing Amine Oxidases Contribute to Terminal Polyamine Oxidation in Peroxisomes and Apoplast of Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Zarei, A.; Trobacher, C.P.; Cooke, A.R.; Meyers, A.J.; Hall, J.C.; Shelp, B.J. Apple Fruit Copper Amine Oxidase Isoforms: Peroxisomal MdAO1 Prefers Diamines as Substrates, Whereas Extracellular MdAO2 Exclusively Utilizes Monoamines. Plant Cell Physiol. 2015, 56, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Cona, A.; Rea, G.; Angelini, R.; Federico, R.; Tavladoraki, P. Functions of Amine Oxidases in Plant Development and Defence. Trends Plant Sci. 2006, 11, 80–88. [Google Scholar] [CrossRef]
- Mirdehghan, S.H.; Rahimi, S. Pre-Harvest Application of Polyamines Enhances Antioxidants and Table Grape (Vitis vinifera L.) Quality during Postharvest Period. Food Chem. 2016, 196, 1040–1047. [Google Scholar] [CrossRef] [PubMed]
- Nambeesan, S.; Datsenka, T.; Ferruzzi, M.G.; Malladi, A.; Mattoo, A.K.; Handa, A.K. Overexpression of Yeast Spermidine Synthase Impacts Ripening, Senescence and Decay Symptoms in Tomato. Plant J. 2010, 63, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Agudelo-Romero, P.; Bortolloti, C.; Pais, M.S.; Tiburcio, A.F.; Fortes, A.M. Plant Physiology and Biochemistry Study of Polyamines during Grape Ripening Indicate an Important Role of Polyamine Catabolism. Plant Physiol. Biochem. 2013, 67, 105–119. [Google Scholar] [CrossRef]
- Guo, J.; Wang, S.; Yu, X.; Dong, R.; Li, Y.; Mei, X.; Shen, Y. Polyamines Regulate Strawberry Fruit Ripening by Abscisic Acid, Auxin, and Ethylene. Plant Physiol. 2018, 177, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, R.; Altabella, T.; Marco, F.; Bortolotti, C.; Reymond, M.; Koncz, C.; Carrasco, P.; Tiburcio, A.F. Polyamines: Molecules with Regulatory Functions in Plant Abiotic Stress Tolerance. Planta 2010, 231, 1237–1249. [Google Scholar] [CrossRef]
- Cherian, S.; Figueroa, C.R.; Nair, H. “Movers and Shakers” in the Regulation of Fruit Ripening: A Cross-Dissection of Climacteric versus Non-Climacteric Fruit. J. Exp. Bot. 2014, 65, 4705–4722. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jia, H.; Chai, Y.; Shen, Y. Abscisic Acid Perception and Signaling Transduction in Strawberry: A Model for Non-Climacteric Fruit Ripening. Plant Signal. Behav. 2011, 6, 1950–1953. [Google Scholar] [CrossRef]
- Liu, M.; Pirrello, J.; Chervin, C.; Roustan, J.-P.; Bouzayen, M. Ethylene Control of Fruit Ripening: Revisiting the Complex Network of Transcriptional Regulation. Plant Physiol. 2015, 169, 2380–2390. [Google Scholar] [CrossRef]
- Wimalasekera, R.; Villar, C.; Begum, T.; Scherer, G.F.E. COPPER AMINE OXIDASE1 (CuAO1) of Arabidopsis thaliana Contributes to Abscisic Acid and Polyamine-Induced Nitric Oxide Biosynthesis and Abscisic Acid Signal Transduction. Mol. Plant 2011, 4, 663–678. [Google Scholar] [CrossRef]
- Wang, W.; Zheng, X.; Liu, S.; Tan, B.; Cheng, J.; Ye, X.; Li, J.; Feng, J. Polyamine Oxidase (PAO)–Mediated Polyamine Catabolism Plays Potential Roles in Peach (Prunus persica L.) Fruit Development and Ripening. Tree Genet. Genomes 2021, 17, 10. [Google Scholar] [CrossRef]
- Wang, X.; Zeng, W.; Ding, Y.; Wang, Y.; Niu, L.; Yao, J.; Pan, L.; Lu, Z.; Cui, G.; Li, G.; et al. Peach Ethylene Response Factor PpeERF2 Represses the Expression of ABA Biosynthesis and Cell Wall Degradation Genes during Fruit Ripening. Plant Sci. 2019, 283, 116–126. [Google Scholar] [CrossRef]
- Fortes, A.M.; Teixeira, R.T.; Agudelo-Romero, P. Complex Interplay of Hormonal Signals during Grape Berry Ripening. Molecules 2015, 20, 9326–9343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Leng, P.; Zhang, G.; Li, X. Cloning and Functional Analysis of 9-Cis-Epoxycarotenoid Dioxygenase (NCED) Genes Encoding a Key Enzyme during Abscisic Acid Biosynthesis from Peach and Grape Fruits. J. Plant Physiol. 2009, 166, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.D.; Yu, D.J.; Chung, S.W.; Chea, S.; Lee, H.J. Abscisic Acid Stimulates Anthocyanin Accumulation in “Jersey” Highbush Blueberry Fruits during Ripening. Food Chem. 2018, 244, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.-F.; Chai, Y.-M.; Li, C.-L.; Lu, D.; Luo, J.-J.; Qin, L.; Shen, Y.-Y. Abscisic Acid Plays an Important Role in the Regulation of Strawberry Fruit Ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef]
- Sun, L.; Sun, Y.; Zhang, M.; Wang, L.; Ren, J.; Cui, M.; Wang, Y.; Ji, K.; Li, P.; Li, Q.; et al. Suppression of 9-Cis-Epoxycarotenoid Dioxygenase, which Encodes a Key Enzyme in Abscisic Acid Biosynthesis, Alters Fruit Texture in Transgenic Tomato. Plant Physiol. 2012, 158, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Thakur, R.; Pristijono, P.; Bowyer, M.; Singh, S.P.; Scarlett, C.J.; Stathopoulos, C.E.; Vuong, Q.V. A Starch Edible Surface Coating Delays Banana Fruit Ripening. LWT—Food Sci. Technol. 2019, 100, 341–347. [Google Scholar] [CrossRef]
- Parra-Lobato, M.C.; Gomez-Jimenez, M.C. Polyamine-Induced Modulation of Genes Involved in Ethylene Biosynthesis and Signalling Pathways and Nitric Oxide Production during Olive Mature Fruit Abscission. J. Exp. Bot. 2011, 62, 4447–4465. [Google Scholar] [CrossRef]
- Torrigiani, P.; Bressanin, D.; Ruiz, K.B.; Tadiello, A.; Trainotti, L.; Bonghi, C.; Ziosi, V.; Costa, G. Spermidine Application to Young Developing Peach Fruits Leads to a Slowing down of Ripening by Impairing Ripening-Related Ethylene and Auxin Metabolism and Signaling. Physiol. Plant. 2012, 146, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Moschou, P.N.; Wu, J.; Cona, A.; Tavladoraki, P.; Angelini, R.; Roubelakis-Angelakis, K.A. The Polyamines and Their Catabolic Products Are Significant Players in the Turnover of Nitrogenous Molecules in Plants. J. Exp. Bot. 2012, 63, 5003–5015. [Google Scholar] [CrossRef] [PubMed]
- Agudelo-Romero, P.; Ali, K.; Choi, Y.H.; Sousa, L.; Verpoorte, R.; Tiburcio, A.F.; Fortes, A.M. Perturbation of Polyamine Catabolism Affects Grape Ripening of Vitis vinifera Cv. Trincadeira. Plant Physiol. Biochem. 2014, 74, 141–155. [Google Scholar] [CrossRef]
- Mo, A.; Xu, T.; Bai, Q.; Shen, Y.; Gao, F.; Guo, J. FaPAO5 Regulates Spm/Spd Levels as a Signaling during Strawberry Fruit Ripening. Plant Direct 2020, 4, e00217. [Google Scholar] [CrossRef]
- Giovannoni, J.J. Genetic Regulation of Fruit Development and Ripening. Plant Cell 2004, 16, S170–S180. [Google Scholar] [CrossRef] [PubMed]
- Ghuge, S.A.; Carucci, A.; Rodrigues-Pousada, R.A.; Tisi, A.; Franchi, S.; Tavladoraki, P.; Angelini, R.; Cona, A. The Apoplastic Copper AMINE OXIDASE1 Mediates Jasmonic Acid-Induced Protoxylem Differentiation in Arabidopsis Roots. Plant Physiol. 2015, 168, 690–707. [Google Scholar] [CrossRef] [PubMed]
- Groß, F.; Rudolf, E.-E.; Thiele, B.; Durner, J.; Astier, J. Copper Amine Oxidase 8 Regulates Arginine-Dependent Nitric Oxide Production in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 2149–2162. [Google Scholar] [CrossRef]
- Tsaniklidis, G.; Kotsiras, A.; Tsafouros, A.; Roussos, P.A.; Aivalakis, G.; Katinakis, P.; Delis, C. Spatial and Temporal Distribution of Genes Involved in Polyamine Metabolism during Tomato Fruit Development. Plant Physiol. Biochem. 2016, 100, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cheng, X.; Cui, Z.; Jiang, Y.; Tan, B.; Cheng, J.; Zhang, L.; Feng, J. Function Identification of Copper Amine Oxidase Gene PpCuAO4 During Ripening of Peach (Prunus persica L.) Fruit. J. Nucl. Agric. Sci. 2022, 36, 1738–1745. [Google Scholar] [CrossRef]
- Liu, J.H.; Moriguchi, T. Changes in Free Polyamine Titers and Expression of Polyamine Biosynthetic Genes during Growth of Peach In Vitro Callus. Plant Cell Rep. 2007, 26, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Atanasov, K.E.; Arafaty, N.; Murillo, E.; Tiburcio, A.F.; Zeier, J.; Alcázar, R. Putrescine Elicits ROS-Dependent Activation of the Salicylic Acid Pathway in Arabidopsis thaliana. Plant Cell Environ. 2020, 43, 2755–2768. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.U.; Singh, Z. Endogenous Free Polyamines of Mangos in Relation to Development and Ripening. J. Am. Soc. Hortic. Sci. 2004, 129, 280–286. [Google Scholar] [CrossRef]
- Fraudentali, I.; Rodrigues-Pousada, R.A.; Angelini, R.; Ghuge, S.A.; Cona, A. Plant Copper Amine Oxidases: Key Players in Hormone Signaling Leading to Stress-Induced Phenotypic Plasticity. Int. J. Mol. Sci. 2021, 22, 5136. [Google Scholar] [CrossRef]
- Leng, P.; Zhang, G.; Li, X.; Wang, L.; Zheng, Z. Cloning of 9-Cis-Epoxycarotenoid Dioxygenase (NCED) Gene Encoding a Key Enzyme during Abscisic Acid (ABA) Biosynthesis and ABA-Regulated Ethylene Production in Detached Young Persimmon Calyx. Chin. Sci. Bull. 2009, 54, 2830–2838. [Google Scholar] [CrossRef][Green Version]
- Urano, K.; Yoshiba, Y.; Nanjo, T.; Igarashi, Y.; Seki, M.; Sekiguchi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of Arabidopsis Genes Involved in Biosynthesis of Polyamines in Abiotic Stress Responses and Developmental Stages. Plant Cell Environ. 2003, 26, 1917–1926. [Google Scholar] [CrossRef]
- Cuevas, J.C.; López-Cobollo, R.; Alcázar, R.; Zarza, X.; Koncz, C.; Altabella, T.; Salinas, J.; Tiburcio, A.F.; Ferrando, A. Putrescine Is Involved in Arabidopsis Freezing Tolerance and Cold Acclimation by Regulating Abscisic Acid Levels in Response to Low Temperature. Plant Physiol. 2008, 148, 1094–1105. [Google Scholar] [CrossRef]
- An, Z.; Jing, W.; Liu, Y.; Zhang, W. Hydrogen Peroxide Generated by Copper Amine Oxidase Is Involved in Abscisic Acid-Induced Stomatal Closure in Vicia faba. J. Exp. Bot. 2008, 59, 815–825. [Google Scholar] [CrossRef]
- Courtois, C.; Besson, A.; Dahan, J.; Bourque, S.; Dobrowolska, G.; Pugin, A.; Wendehenne, D. Nitric Oxide Signalling in Plants: Interplays with Ca2+ and Protein Kinases. J. Exp. Bot. 2008, 59, 155–163. [Google Scholar] [CrossRef]
- Yue, P.; Lu, Q.; Liu, Z.; Lv, T.; Li, X.; Bu, H.; Liu, W.; Xu, Y.; Yuan, H.; Wang, A. Auxin-Activated MdARF5 Induces the Expression of Ethylene Biosynthetic Genes to Initiate Apple Fruit Ripening. New Phytol. 2020, 226, 1781–1795. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, F.; Rao, J. Effect of Putrescine Treatment on Chilling Injury, Fatty Acid Composition and Antioxidant System in Kiwifruit. PLoS ONE 2016, 11, e0162159. [Google Scholar] [CrossRef]
- Ziosi, V.; Bregoli, A.M.; Fregola, F.; Costa, G.; Torrigani, P. Jasmonate-Induced Ripening Delay Is Associated with up-Regulation of Polyamine Levels in Peach Fruit. J. Plant Physiol. 2009, 166, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, G.; Wei, R. CE-ECL Analysis of Endo-Spermine During the Development of Blueberry Fruits and Its Relationship with the Preservation Quality. Mol. Plant Breed. 2019, 17, 2356–2362. [Google Scholar] [CrossRef]
- Abdullah; Faraji, S.; Heidari, P.; Poczai, P. The BAHD Gene Family in Cacao (Theobroma cacao, Malvaceae): Genome-Wide Identification and Expression Analysis. Front. Ecol. Evol. 2021, 9, 707708. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z. Selection and Validation of Reference Genes in Sweet Cherry Flower Bud at Different Development Stages. Seed 2020, 39, 37–43. (In Chinese) [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Daudi, A.; O’Brien, J.A. Detection of Hydrogen Peroxide by DAB Staining in Arabidopsis Leaves. Bio-protocol 2012, 2, e263. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | GRAVY | CDS Length (bp) | PI | MW (kDa) | Chromosome LOCATION |
|---|---|---|---|---|---|---|
| PavCuAO1 | FUN_000165-T1 | −0.306 | 2010 | 6.41 | 75.71 | 1 |
| PavCuAO2 | FUN_000166-T1 | −0.239 | 2040 | 6.82 | 76.86 | 1 |
| PavCuAO3 | FUN_000167-T1 | −0.276 | 1944 | 7.66 | 72.38 | 1 |
| PavCuAO4 | FUN_024869-T1 | −0.268 | 2181 | 5.7 | 81.43 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, X.; Wen, Z.; Shang, C.; Cai, X.; Hou, Q.; Qiao, G. Copper Amine Oxidase (CuAO)-Mediated Polyamine Catabolism Plays Potential Roles in Sweet Cherry (Prunus avium L.) Fruit Development and Ripening. Int. J. Mol. Sci. 2022, 23, 12112. https://doi.org/10.3390/ijms232012112
Cao X, Wen Z, Shang C, Cai X, Hou Q, Qiao G. Copper Amine Oxidase (CuAO)-Mediated Polyamine Catabolism Plays Potential Roles in Sweet Cherry (Prunus avium L.) Fruit Development and Ripening. International Journal of Molecular Sciences. 2022; 23(20):12112. https://doi.org/10.3390/ijms232012112
Chicago/Turabian StyleCao, Xuejiao, Zhuang Wen, Chunqiong Shang, Xiaowei Cai, Qiandong Hou, and Guang Qiao. 2022. "Copper Amine Oxidase (CuAO)-Mediated Polyamine Catabolism Plays Potential Roles in Sweet Cherry (Prunus avium L.) Fruit Development and Ripening" International Journal of Molecular Sciences 23, no. 20: 12112. https://doi.org/10.3390/ijms232012112
APA StyleCao, X., Wen, Z., Shang, C., Cai, X., Hou, Q., & Qiao, G. (2022). Copper Amine Oxidase (CuAO)-Mediated Polyamine Catabolism Plays Potential Roles in Sweet Cherry (Prunus avium L.) Fruit Development and Ripening. International Journal of Molecular Sciences, 23(20), 12112. https://doi.org/10.3390/ijms232012112






