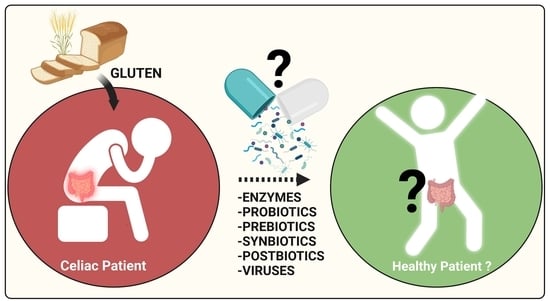
Celiac Disease and Possible Dietary Interventions: From Enzymes and Probiotics to Postbiotics and Viruses
Abstract
1. Introduction
1.1. Celiac Disease: Gluten Proteins and the Triggers of Disease
1.2. Is a Gluten-Free Diet Enough for CeD Treatment?
2. Dietary Interventions for Complementing the GFD and Beyond
2.1. Enzymes as a Nutritional Supplement Therapy for CeD
2.2. Human Microbiota and Dysbiosis in CeD
2.3. Probiotics
2.4. Prebiotics
2.5. Synbiotics
2.6. Postbiotics
2.7. Viruses
3. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Caio, G.; Volta, U.; Sapone, A.; Leffler, D.A.; de Giorgio, R.; Catassi, C.; Fasano, A. Celiac disease: A comprehensive current review. BMC Med. 2019, 17, 142. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Arora, A.; Strad, T.; Leffler, D.A.; Catassi, C.; Green, P.H.; Kelly, C.P.; Ahuja, V.; Makharia, G.K. Global prevalence of celiac disease: Systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 2018, 16, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Volta, U.; Caio, G.; Stanghellini, V.; De Giorgio, R. The changing clinical profile of celiac disease: A 15-year experience (1998–2012) in an Italian referral centre. BMC Gastroenterol. 2014, 14, 194. [Google Scholar] [CrossRef] [PubMed]
- Ciacci, C.; Cirillo, M.; Sollazzo, R.; Savino, G.; Sabbatini, F.; Mazzacca, G. Gender and clinical presentation in adult celiac disease. Scand. J. Gastroenterol. 1995, 30, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Ciccocioppo, R.; Kruzliak, P.; Cangemi, G.; Pohanka, M.; Betti, E.; Lauret, E.; Rodrigo, L. The spectrum of differences between childhood and adulthood celiac disease. Nutrients 2015, 7, 8733–8751. [Google Scholar] [CrossRef] [PubMed]
- Hochegger, R.; Mayer, W.; Prochaska, M. Comparison of R5 and G12 Antibody419 Based ELISA Used for the Determination of the Gluten Content in Official Food 420 Samples. Foods 2015, 4, 654–664. [Google Scholar] [CrossRef]
- Constantin, C.; Huber, W.; Granditsch, G.; Weghofer, M.; Valenta, R. Different profiles of wheat antigens are recognised by patients suffering from coeliac disease and IgE-mediated food allergy. Int. Arch. Allergy Immunol. 2005, 138, 257–266. [Google Scholar] [CrossRef]
- Green, P.; Lebwohl, B.; Greywoode, R. Celiac disease. J. Allergy Clin. Immunol. 2015, 135, 1099–1106. [Google Scholar] [CrossRef]
- Lebwohl, B.; Granath, F.; Ekbom, A.; Smedby, K.E.; Murray, J.A.; Neugut, A.I.; Green, P.H.R.; Ludvigsson, J.F. Mucosal healing and risk for lymphoproliferative malignancy in celiac disease: A population-based cohort study. Ann. Intern. Med. 2013, 159, 169–175. [Google Scholar] [CrossRef]
- Severance, E.; Yolken, R.; Eaton, W. Autoimmune diseases, gastrointestinal disorders and the microbiome in schizophrenia: More than a gut feeling. Schizophr. Res. 2014, 176, 23–35. [Google Scholar] [CrossRef]
- Ventura, A.; Neri, E.; Ughi, C.; Leopaldi, A.; Città, A.; Not, T. Gluten-dependent diabetes-related and thyroid-related autoantibodies in patients with celiac disease. J. Pediatr. 2000, 137, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Haupt-Jorgensen, M.; Holm, L.; Josefsen, K.; Buschard, K. Possible Prevention of Diabetes with a Gluten-Free Diet. Nutrients 2018, 13, 1746. [Google Scholar] [CrossRef] [PubMed]
- Levescot, A.; Malamut, G.; Cerf-Bensussan, N. Immunopathogenesis and environmental triggers in coeliac disease. Gut 2022. [Google Scholar] [CrossRef] [PubMed]
- Wieser, H. The precipitating factor in coeliac disease. Bailliere’s Clin. Gastroenterol. 1995, 9, 191–207. [Google Scholar] [CrossRef]
- Papista, C.; Gerakopoulos, V.; Kourelis, A.; Sounidaki, M.; Kontana, A.; Berthelot, L.; Moura, I.C.; Monteiro, R.C.; Yiangou, M. Gluten induces coeliac-like disease in sensitised mice involving IgA, cd71 and transglutaminase 2 interactions that are prevented by probiotics. Lab. Investig. 2012, 92, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Mamone, G.; Ferranti, P.; Rossi, M.; Roepstorff, P.; Fierro, O.; Malorni, A.; Addeo, F. Identification of a peptide from α-gliadin resistant to digestive enzymes: Implications for celiac disease. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 85, 236–241. [Google Scholar] [CrossRef]
- Gass, J.; Khosla, C. Prolyl endopeptidases. Cell. Mol. Life Sci. 2007, 64, 345–355. [Google Scholar] [CrossRef]
- Helmerhorst, E.; Zamakhchari, M.; Schuppan, D.; Oppenheim, F. Discovery of a novel and rich source of gluten-degrading microbial enzymes in the oral cavity. PLoS ONE 2010, 5, e13264. [Google Scholar] [CrossRef]
- Herrera, M.; Veuthey, T.; Dodero, V. Self-organization of gliadin in aqueous media under physiological digestive pHs. Colloids Surfaces. B Biointerfaces 2016, 141, 565–575. [Google Scholar] [CrossRef]
- Herrera, M.; Vazquez, D.; Sreij, R.; Drechsler, M.; Hertle, Y.; Hellweg, T.; Dodero, V. Insights into gliadin supramolecular organization at digestive pH 3.0. Colloids Surfaces. B Biointerfaces 2018, 165, 363–370. [Google Scholar] [CrossRef]
- Markgren, J.; Rasheed, F.; Hedenqvist, M.; Skepö, M.; Johansson, E. Clustering and cross-linking of the wheat storage protein α-gliadin: A combined experimental and theoretical approach. Int. J. Biol. Macromol. 2022, 211, 592–615. [Google Scholar] [CrossRef]
- Herrera, M.; Nicoletti, F.; Gras, M.; Dörfler, P.; Tonali, N.; Hannappel, Y.; Ennen, I.; Hütten, A.; Hellweg, T.; Lammers, K.; et al. Pepsin Digest of Gliadin Forms Spontaneously Amyloid-Like Nanostructures Influencing the Expression of Selected Pro-Inflammatory, Chemoattractant, and Apoptotic Genes in Caco-2 Cells: Implications for Gluten-Related Disorders. Mol. Nutr. Food Res. 2021, 65, 2100200. [Google Scholar] [CrossRef] [PubMed]
- Barone, M.; Della Valle, N.; Rosania, R.; Facciorusso, A.; Trotta, A.; Cantatore, F.P.; Falco, S.; Pignatiello, S.; Viggiani, M.T.; Amoruso, A.; et al. A comparison of the nutritional status between n adult celiac patients on a long-term, strictly gluten-free diet and healthy subjects. Eur. J. Clin. Nutr. 2016, 70, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Qiao, S.; Arentz-Hansen, H.; Molberg, O.; Gray, G.; Sollid, L.; Khosla, C. Identification and analysis of multivalent proteolytically resistant peptides from gluten: Implications for celiac sprue. Proteome Res. 2005, 4, 1732–1741. [Google Scholar] [CrossRef] [PubMed]
- Vilasi, S.; Sirangelo, I.; Irace, G.; Caputo, I.; Barone, M.; Esposito, C.; Ragone, R. Interaction of ‘toxic’ and ‘immunogenic’ A-gliadin peptides with a membrane-mimetic environment. J. Mol. Recognit. 2010, 23, 322–328. [Google Scholar] [CrossRef]
- Barone, M.; Zimmer, K. Endocytosis and transcytosis of gliadin peptides. Mol. Cell Pediatr. 2016, 3, 1–5. [Google Scholar] [CrossRef]
- Dieterich, W.; Ehnis, T.; Bauer, M.; Donner, P.; Volta, U.; Riecken, E.; Schuppan, D. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 1997, 3, 797–801. [Google Scholar] [CrossRef]
- Lammers, K.; Lu, R.; Brownley, J.; Lu, B.; Gerard, C.; Thomas, K.; Rallabhandi, P.; Shea-Donohue, T.; Tamiz, A.; Alkan, S.; et al. Gliadin induces an increase in intestinal permeability and zonulin release by binding to the chemokine receptor cxcr3. Gastroenterology 2008, 135, 194–204. [Google Scholar] [CrossRef]
- Van Heel, D.; West, J. Recent advances in coeliac disease. Gut 2006, 55, 1037–1046. [Google Scholar] [CrossRef]
- Schalk, K.; Lang, C.; Wieser, H.; Koehler, P.; Scherf, K.A. Quantitation of the immunodominant 33-mer peptide from α-gliadin in wheat flours by liquid chromatography-tandem mass spectrometry. Sci. Rep. 2017, 7, 45092. [Google Scholar] [CrossRef]
- Qiao, S.; Bergseng, E.; Molberg, O.; Xia, J.; Fleckenstein, B.; Khosla, C.; Sollid, L. Antigen Presentation to Celiac Lesion-Derived T Cells of a 33-Mer Gliadin Peptide Naturally Formed by Gastrointestinal Digestion. J. Immunol. 2004, 173, 1757–1762. [Google Scholar] [CrossRef] [PubMed]
- Ozuna, C.; Iehisa, J.; Giménez, M.; Alvarez, J.; Sousa, C.; Barro, F. Diversification of the celiac disease α-gliadin complex in wheat: A 33-mer peptide with six overlapping epitopes, evolved following polyploidization. Plant J. Cell Mol. Biol. 2015, 82, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Molberg, O.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.; Sollid, L.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 27, 2275–2279. [Google Scholar] [CrossRef]
- Hausch, F.; Shan, L.; Santiago, N.; Gray, G.; Khosla, C. Intestinal digestive resistance of immunodominant gliadin peptides. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 283, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.; Benedini, L.; Lonez, C.; Schilardi, P.; Hellweg, T.; Ruysschaert, J.; Dodero, V. Self-assembly of 33-mer gliadin peptide oligomers. Soft Matter. 2015, 11, 8648–8660. [Google Scholar] [CrossRef]
- Herrera, M.; Zamarreno, F.; Costabel, M.; Ritacco, H.; Hutten, A.; Sewald, N.; Dodero, V. Circular Dichroism and Electron Microscopy Studies In Vitro of 33-mer Gliadin Peptide Revealed Secondary Structure Transition and Supramolecular Organization. Biopolymers 2014, 101, 96–106. [Google Scholar] [CrossRef]
- Herrera, M.; Pizzuto, M.; Lonez, C.; Rott, K.; Hütten, A.; Sewald, N.; Ruysschaert, J.-M.; Dodero, V.I. Large supramolecular structures of 33-mer gliadin peptide activate toll-like receptors in macrophages. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1417–1427. [Google Scholar] [CrossRef]
- Liang, L.; Pinier, M.; Leroux, J.; Subirade, M. Interaction of alpha-gliadin with poly (HEMA-co-SS): Structural characterization and biological implication. Biopolymers 2009, 91, 69–178. [Google Scholar] [CrossRef]
- Pinier, M.; Fuhrmann, G.; Galipeau, H.; Rivard, N.; Murray, J.A.; David, C.S.; Drasarova, H.; Tuckova, L.; Leroux, J.; Verdu, E.F. The copolymer P (HEMA-co-SS) binds gluten and reduces immune response in gluten-sensitized mice and human tissues. Gastroenterology 2012, 142, 316–325. [Google Scholar] [CrossRef]
- Gayathri, D.; Rashmi, B. Development of celiac disease; pathogenesis and strategies to control: A molecular approach. J. Nutr. Food Sci. 2014, 4, 310. [Google Scholar] [CrossRef]
- Petersen, J.; Ciacchi, L.; Tran, M.; Loh, K.L.; Kooy-Winkelaar, Y.; Croft, N.P.; Hardy, M.Y.; Chen, Z.; McCluskey, J.; Anderson, R.P.; et al. T cell receptor cross-reactivity between gliadin and bacterial peptides in celiac disease. Nat. Struct. Mol. Biol. 2020, 27, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, D.; Schilbert, H.; Dodero, V. Molecular and Structural Parallels between Gluten Pathogenic Peptides and Bacterial-Derived Proteins by Bioinformatics Analysis. Int. J. Mol. Sci. 2021, 22, 9278. [Google Scholar] [CrossRef]
- Bethune, M.; Khosla, C. Parallels between pathogens and gluten peptides in celiac sprue. PLoS Pathog. 2008, 4, e34. [Google Scholar] [CrossRef] [PubMed]
- Lindfors, K.; Lin, J.; Lee, H.-S.; Hyoty, H.; Nykter, M.; Kurppa, K.; Liu, E.; Koletzko, S.; Rewers, M.; Hagopian, W.; et al. Metagenomics of the faecalvirome indicate a cumulative effect of enterovirus and gluten amount on the risk of coeliac disease autoimmunity in genetically at risk children: The TEDDY study. Gut 2020, 69, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Bouziat, R.; Hinterleitner, R.; Brown, J.; Stencel-Baerenwald, J.E.; Ikizler, M.; Mayassi, T.; Meisel, M.; Kim, S.M.; Discepolo, V.; Pruijssers, A.J.; et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 2017, 356, 44–50. [Google Scholar] [CrossRef]
- Baggus, E.M.R.; Hadjivassiliou, M.; Cross, S.; Penny, H.; Urwin, H.; Watson, S.; Woodward, J.M.; Sanders, D.S. How to manage adult celiac disease: Perspective from the N.H.S. England rare diseases collaborative network for non-responsive and refractory coeliac disease. Frontline Gastroenterol. 2019, 11, 235–242. [Google Scholar] [CrossRef]
- Syage, J.; Kelly, C.; Dickason, M.; Cebolla, A.; Leon, F.; Dominguez, R.; Sealey-Voyksner, J.A. Determination of gluten consumption in celiac disease patients on a gluten-free diet. Am. J. Clin. Nutr. 2018, 107, 201–207. [Google Scholar] [CrossRef]
- Moron, B.; Cebolla, A.; Manyani, H.; Alvarez-Maqueda, M.; Megías, M.; Thomas, M.; Lopez, M.; Sousa, C. Sensitive Detection of Cereal Fractions That Are Toxic to Celiac Disease Patients by Using Monoclonal Antibodies to a Main Immunogenic Wheat Peptide. Am. J. Clin. Nutr. 2008, 87, 405–414. [Google Scholar] [CrossRef]
- Silvester, J.; Therrien, A.; Kelly, C. Celiac Disease: Fallacies and Facts. Am. J. Gastroenterol. 2021, 1, 1148–1155. [Google Scholar] [CrossRef]
- Diosdado, B.; van Bakel, H.; Strengman, E.; Franke, L.; van Oort, E.; Mulder, C.; Wijmenga, C.; Wapenaar, M. Neutrophil recruitment and barrier impairment in celiac disease: A genomic study. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2007, 5, 574–581. [Google Scholar] [CrossRef]
- Freire, R.; Ingano, L.; Serena, G.; Cetinbas, M.; Anselmo, A.; Sapone, A.; Sadreyev, R.; Fasano, A.; Senger, S. Human gut derived-organoids provide model to study gluten response and effects of microbiota-derived molecules in celiac disease. Sci. Rep. 2019, 9, 7029. [Google Scholar] [CrossRef] [PubMed]
- Silvester, J.; Comino, I.; Kelly, C.; Sousa, C.; Duerksen, D.; DOGGIE BAG Study Group. Most Patients With Celiac Disease on Gluten-Free Diets Consume Measurable Amounts of Gluten. Gastroenterology 2020, 158, 1497–1499.e1. [Google Scholar] [CrossRef] [PubMed]
- Hall, N.; Rubin, G.; Charnock, A. Systematic review: Adherence to a gluten-free diet in adult patients with coeliac disease. Aliment. Pharmacol. Ther. 2009, 30, 315–330. [Google Scholar] [CrossRef]
- Weisbrod, V.; Silverster, J.; Raber, C.; Suslovic, W.; Coburn, S.S.; Raber, B.; McMahon, J.; Damast, A.; Kramer, Z.; Kerzner, B. A quantitative assessment of gluten cross-contact in the school environment for children with celiac disease. J. Pediatr.Gastroenterol. Nutr. 2020, 70, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Daveson, A.; Popp, A.; Taavela, J.; Goldstein, K.E.; Isola, J.; Truitt, K.E.; Mäki, M.; Anderson, R.P.; Adams, A.; Andrews, J.; et al. Baseline quantitative histology in therapeutics trials reveals villus atrophy in most patients with coeliac disease who appear well controlled on gluten-free diet. GastroHep 2020, 2, 22–30. [Google Scholar] [CrossRef]
- Freeman, H. Celiac Disease: A Disorder Emerging from Antiquity, Its Evolving Classification and Risk, and Potential New Treatment Paradigms. Gut Liver 2015, 9, 28–37. [Google Scholar] [CrossRef]
- Tennyson, C.; Simpson, S.; Lebwohl, B.; Lewis, S.; Green, P. Interest in medical therapy for celiac disease. Therap. Adv. Gastroenterol. 2013, 6, 358–364. [Google Scholar] [CrossRef]
- Segura, V.; Ruiz-Carnicer, Á.; Sousa, C.; Moreno, M. New Insights into Non-Dietary Treatment in Celiac Disease: Emerging Therapeutic Options. Nutrients 2021, 13, 2146. [Google Scholar] [CrossRef]
- Serena, G.; D’Avino, P.; Fasano, A. Celiac Disease and Non-celiac Wheat Sensitivity: State of Art of Non-dietary Therapies. Front. Nutr. 2020, 7, 152. [Google Scholar] [CrossRef]
- Rey, M.; Yang, M.; Lee, L.; Zhang, Y.; Sheff, J.G.; Sensen, C.W.; Mrazek, H.; Halada, P.; Man, P.; McCarville, J.; et al. Addressing proteolytic efficiency in enzymatic degradation therapy for celiac disease. Sci. Rep. 2016, 6, 30980. [Google Scholar] [CrossRef]
- Cavaletti, L.; Taravella, A.; Carrano, L.; Carenzi, G.; Sigurtà, A.; Solinas, N.; De Caro, S.; Di Stasio, L.; Picascia, S.; Laezza, M.; et al. E40, a novel microbial protease efficiently detoxifies gluten proteins, for the dietary management of gluten intolerance. Sci. Rep. 2019, 9, 13147. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.; Dodero, V. Gliadin proteolytical resistant peptides: The interplay between structure and self-assembly in gluten-related disorders. Biophys. Rev. 2021, 13, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Tian, N.; Valery, A.; Zhong, Y.; Schuppan, D.; Helmerhorst, E. Identification of Pseudolysin (lasB) as an Aciduric Gluten-Degrading Enzyme with High Therapeutic Potential for Celiac Disease. Am. J. Gastroenterol. 2015, 110, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Wagh, S.; Gadge, P.; Padul, M. Significant Hydrolysis of Wheat Gliadin by Bacillus tequilensis (10bT/HQ223107): A Pilot Study. Probiotics Antimicrob. Prot. 2018, 10, 662–667. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Rizzello, C.; Fasano, A.; Clemente, M.; Simone, C.; Silano, M.; De Vincenzi, M.; Losito, I.; Gobbetti, M. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for Celiac Sprue probiotics and gluten intolerance. Biochim. Acta Mol. Basis Dis. 2006, 1762, 80–93. [Google Scholar]
- Duar, R.M.; Clark, K.; Patil, P.B.; Hernández, C.; Brüning, S.; Burkey, T.; Madayiputhiya, N.; Taylor, S.; Walter, J. Identification and characterization of intestinal lactobacilli strains capable of degrading immunotoxic peptides present in gluten. J. Appl. Microbiol. 2014, 118, 515–527. [Google Scholar] [CrossRef]
- Shan, L.; Marti, T.; Sollid, M.; Gary, M.; Khosla, C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: Implications for coeliac sprue. Biochem. J. 2004, 383, 311–318. [Google Scholar] [CrossRef]
- Prabucka, B.; Bielawski, W. Purification and partial characterization of a major gliadin-degrading cysteine endopeptidase from germinating triticale seeds. Acta Physiol. Plant. 2004, 26, 383–392. [Google Scholar] [CrossRef]
- Krishnareddy, S.; Stier, K.; Recanati, M.; Lebwohl, B.; Green, P. Commercially available glutenases: A potential hazard in celiac disease. Ther. Adv. Gastroenterol. 2017, 10, 473–481. [Google Scholar] [CrossRef]
- Cornell, H.; Stelmasiak, T. A unified hypothesis of coeliac disease with implications for management of patients. Amino Acids 2007, 33, 43–49. [Google Scholar] [CrossRef][Green Version]
- Cornell, H.; Doharti, W.; Stelmasiak, T. Papaya latex enzymes capable of detoxification of gliadin. Amino Acids 2010, 38, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Gerez, C.; Dallagnol, A.; Rollán, G.; de Valdez, F. A combination of two lactic acid bacteria improves the hydrolysis of gliadin during wheat dough fermentation. Food Microbiol. 2012, 32, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Tack, J.; van de Water, M.; Bruins, M.; Kooy-Winkelaar, E.M.; van Bergen, J.; Bonnet, P.; Vreugdenhil, A.C.E.; Korponay-Szabo, I.; Edens, L.; von Blomberg, B.M.E.; et al. Consumption of gluten with gluten-degrading enzyme by coeliac patients: A pilot-study. World J. Gastroenterol. 2013, 19, 5837–5847. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, G.; Curiel, A.; Nionelli, L.; Vincentini, O.; Di Cagno, R.; Silano, M.; Gobbetti, M.; Coda, R. Use of fungal proteases and selected sourdough lactic acid bacteria for making wheat bread with an intermediate content of gluten. Food Microbiol. 2014, 37, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, B. Impact of food processing and simulated gastrointestinal digestion on gliadin immunoreactivity in rolls. J. Sci. Food Agric. 2018, 987, 3363–3375. [Google Scholar] [CrossRef]
- Tye-Din, A.; Anderson, P.; Ffrench, A.; Brown, G.J.; Hodsman, P.; Siegel, M.; Botwick, W.; Shreeniwas, R. The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin. Immunol. 2010, 134, 289–295. [Google Scholar] [CrossRef]
- Syage, J.; Green, P.; Khosla, C.; Adelman, D.; Sealey-Voyksner, J.; Murray, A. Latiglutenase Treatment for Celiac Disease: Symptom and Quality of Life Improvement for Seropositive Patients on a GlutenFree Diet. GastroHep 2019, 2, 371. [Google Scholar] [CrossRef]
- Lahdeaho, M.; Kaukinen, K.; Laurila, K.; Vuotikka, P.; Koivurova, O.-P.; Kärjä-Lahdensuu, T.; Marcantonio, A.; Adelman, D.C.; Mäki, M. Glutenase ALV003 Attenuates Gluten-Induced Mucosal Injury in Patients With Celiac Disease. Gastroenterology 2014, 146, 1649–1658. [Google Scholar] [CrossRef]
- Murray, A.; Kelly, P.; Green, P.; Marcantonio, A.; Wu, T.; Mäki, M.; Adelman, C. CeliAction Study Group of Investigators. No Difference Between Latiglutenase and Placebo in Reducing Villous Atrophy or Improving Symptoms in Patients With Symptomatic Celiac Disease. Gastroenterology 2017, 152, 787–798.e2. [Google Scholar] [CrossRef]
- Wolf, C.; Siegel, B.; Tinberg, C.; Camarca, A.; Gianfrani, C.; Paski, S.; Guan, R.; Montelione, G.; Baker, D.; Pultz, S. Engineering of Kuma030: A Gliadin Peptidase That Rapidly Degrades Immunogenic Gliadin Peptides in Gastric Conditions. J. Am. Chem. Soc. 2015, 137, 13106–13113. [Google Scholar] [CrossRef]
- Pultz, S.; Hill, M.; Vitanza, M.; Wolf, C.; Saaby, L.; Liu, T.; Winkle, P.; Leffler, A. Gluten Degradation, Pharmacokinetics, Safety, and Tolerability of TAK-062, an Engineered Enzyme to Treat Celiac Disease. Gastroenterology 2021, 161, 81–93.e3. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Fabiani, E.; Iacono, G.; D’Agate, C.; Francavilla, R.; Biagi, F.; Volta, U.; Accomando, S.; Picarelli, A.; De Vitis, I.; et al. A prospective, double-blind, placebo-controlled trial to establish a safe gluten threshold for patients with celiac disease. Am. J. Clin. Nutr. 2007, 85, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Leonard, M.M.; Silvester, J.A.; Leffler, D.; Fasano, A.; Kelly, C.P.; Lewis, S.K.; Goldsmith, J.D.; Greenblatt, E.; Kwok, W.W.; McAuliffe, W.J.; et al. Evaluating Responses to Gluten Challenge: A Randomized, Double-Blind, 2-Dose Gluten Challenge Trial. Gastroenterology 2021, 160, 720–733.e8. [Google Scholar] [CrossRef] [PubMed]
- Kunz, C.; Kuntz, S.; Rudloff, S. Intestinal flora. Adv. Exp. Med. Biol. 2009, 639, 67–79. [Google Scholar]
- Morelli, L. Postnatal development of intestinal microflora as influenced by infant nutrition. J. Nutr. 2008, 138, 1791–1795. [Google Scholar] [CrossRef]
- Neish, S. Microbes in gastrointestinal health and disease. Gastroenterology 2009, 136, 65–80. [Google Scholar] [CrossRef]
- Sekirov, I.; Shannon, L.; Russell, L.; Caetano, M.; Antunes, B.; Finlay, B. Gut Microbiota in Health and Disease. Physiol. Rev. 2009, 90, 859–904. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-P.; , Yu-Jie. Impacts of Gut Bacteria on Human Health and Diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef]
- Mtasher, S.; Abdulhussein, J.; Mutlag, H. Probiotics and Prebiotics. Int. J. Curr. Res. 2018, 10, 75341–77535. [Google Scholar]
- Chiller, K.; Selkin, A.; Murakawa, J. Skin microflora and bacterial infections of the skin. J. Investig. Dermatol. Symp. Proc. 2001, 6, 170–174. [Google Scholar] [CrossRef]
- Lewis, B.; Buffie, G.; Carter, R.; Leiner, I.; Toussaint, N.C.; Miller, L.C.; Gobourne, A.; Ling, L.; Pamer, E.G. Loss of microbiota-mediated colonization resistance to clostridium difficile infection is greater following oral vancomycin as compared with metronidazole. J. Infect. Dis. 2015, 212, 1656–1665. [Google Scholar] [CrossRef] [PubMed]
- Perez-Cobas, E.; Moya, A.; Gosalbes, J.; Latorre, A. Colonization resistance of the gut microbiota against clostridium difficile. Antibiotics 2015, 4, 337–357. [Google Scholar] [CrossRef] [PubMed]
- Pecora, F.; Persico, F.; Gismondi, P.; Fornaroli, F.; Iuliano, S.; de’Angelis, G.; Esposito, S. Gut Microbiota in Celiac Disease: Is There Any Role for Probiotics? Front. Immunol. 2020, 11, 957. [Google Scholar] [CrossRef] [PubMed]
- Laterza, L.; Rizzatti, G.; Gaetani, E.; Chiusolo, P.; Gasbarrini, A. The Gut Microbiota and Immune System Relationship in Human Graft-versus-Host Disease. Mediterr. J. Hematol. Infect. Dis. 2016, 8, e2016025. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.; Gasbarrini, A.; Mele, M. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Verdu, F.; Galipeau, J.; Jabri, B. Novel players in coeliac disease pathogenesis: Role of the gut microbiota. Nat. Reviews. Gastroenterol. Hepatol. 2015, 12, 497–506. [Google Scholar] [CrossRef]
- Wu, X.; Qian, L.; Liu, K.; Wu, J.; Shan, Z. Gastrointestinal microbiome and gluten in celiac disease. Ann Med. 2021, 53, 1797–1805. [Google Scholar] [CrossRef]
- Elsouri, K.; Arboleda, V.; Heise, S.; Kesselman, M.; Demory-Beckler, M. Microbiome in Rheumatoid Arthritis and Celiac Disease: A Friend or Foe. Cureus 2021, 9, e15543. [Google Scholar] [CrossRef]
- Caminero, A.; Meisel, M.; Jabri, B.; Verdu, E. Mechanisms by which gut microorganisms influence food sensitivities. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 7–18. [Google Scholar] [CrossRef]
- Caminero, A.; Galipeau, H.; McCarville, J.; Johnston, C.W.; Bernier, S.P.; Russell, A.K.; Jury, J.; Herran, A.R.; Casqueiro, J.; Tye-Din, J.A.; et al. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology 2016, 151, 670–683. [Google Scholar] [CrossRef]
- Hiippala, K.; Jouhten, J.; Ronkein, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef] [PubMed]
- Huibregtse, I.; Marrieta, E.; Rashtak, S.; Koning, F.; Rottiers, P.; David, C.S.; van Deventer, S.J.H.; Murray, J.A. Induction of antigen-specific tolerance by oral administration of Lactococcus lactis delivered immunodominant DQ8-restricted gliadin peptide in sensitized nonobese diabetic Abo Dq8 transgenic mice. J. Immunol. 2009, 183, 2390–2396. [Google Scholar] [CrossRef] [PubMed]
- Helmerhorst, E.; Wei, G. Experimental Strategy to Discover Microbes with Gluten-degrading Enzyme Activities. Conference paper at international society of optical engineering. In Proceedings of the SPIE 9112, Sensing Technologies for Global Health, Military Medicine, and Environmental Monitoring IV, Baltimore, MD, USA, 5 June 2014. [Google Scholar] [CrossRef]
- Girbovan, A.; Sur, G.; Samasca, G.; Lupan, I. Dysbiosis a risk factor for celiac disease. Med Microbiol Immunol. 2017, 206, 83–91. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Casaburi, G.; Precone, V.; Pagliuca, C.; Colicchio, R.; Sarnataro, D.; Discepolo, V.; Kim, S.M.; Russo, I.; Blanco, G.D.V.; et al. Metagenomics reveals dysbiosis and a potentially pathogenic N. flavescens strain in duodenum of adult celiac patients. Am. J. Gastroenterol. 2016, 111, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Iaffaldano, L.; Granata, I.; Pagliuca, C.; Esposito, M.V.; Casaburi, G.; Salerno, G.; Colicchio, R.; Piccirillo, M.; Ciacci, C.; Blanco, G.D.V.; et al. Oropharyngeal microbiome evaluation highlights Neisseria abundance in active celiac patients. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Di Biase, A.; Marasco, G.; Ravaioli, F.; Dajti, E.; Colecchia, L.; Righi, B.; D’Amico, V.; Festi, D.; Iughetti, L.; Colecchia, A. Gut microbiota signatures and clinical manifestations in celiac disease children at onset: A pilot study. J. Gastroenterol. Hepatol. 2021, 36, 446–454. [Google Scholar] [CrossRef]
- Palmieri, O.; Castellana, S.; Bevilacqua, A.; Latiano, A.; Latiano, T.; Panza, A.; Fontana, R.; Ippolito, A.M.; Biscaglia, G.; Gentile, A.; et al. Adherence to Gluten-Free Diet Restores Alpha Diversity in Celiac People but the Microbiome Composition Is Different to Healthy People. Nutrients 2022, 14, 2452. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Gagliardi, A.; Totino, V.; Cacciotti, F.; Iebba, V.; Neroni, B.; Bonfiglio, G.; Trancassini, M.; Passariello, C.; Pantanella, F.; Schippa, S. Rebuilding the Gut Microbiota Ecosystem. Int. J. Environ. Res. Public Health 2018, 15, 1679. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, P.; Zhang, X. Probiotics Regulate Gut Microbiota: An Effective Method to Improve Immunity. Molecules 2021, 26, 6076. [Google Scholar] [CrossRef]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterology. 2013, 6, 39–51. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Siragusa, S.; Vacca, M.; Di Cagno, R.; Cristofori, F.; Schwarm, M.; Pelzer, S.; Flügel, M.; Speckmann, B.; Francavilla, R.; et al. Selection of Gut-Resistant Bacteria and Construction of Microbial Consortia for Improving Gluten Digestion under Simulated Gastrointestinal Conditions. Nutrients 2021, 13, 992. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Guevarra, B.; Kim, T.; Kwon, J.; Kim, H.; Cho, H.; Kim, M.; Lee, H. Role of probiotics in the human gut microbiome-associated diseases. J. Microbiol. Biotechnol. 2019, 29, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Rubio, R.; Jofre, A.; Martin, B.; Aymerich, T.; Garriga, M. Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages. Food Microbiol. 2014, 38, 303–331. [Google Scholar] [CrossRef] [PubMed]
- Khochamit, N.; Siripornadulsil, S.; Sukon, P.; Siripornadulsi, W. Antibacterial activity and genotypic-phenotypic characteristics of bacteriocin-producing Bacillus subtilis KKU213: Potential as a probiotic strain. Microbiol. Res. 2015, 170, 36–50. [Google Scholar] [CrossRef]
- Swain, R.; Anandharaj, M.; Ray, C.; Rani, P. Fermented fruits and vegetables of Asia: A potential source of probiotics. Biotechnol. Res. Int. 2014, 2014, 1–19. [Google Scholar] [CrossRef]
- Francavilla, R.; Cristofori, F.; Tripaldi, E.; Indro, F. Intervention for disbiosis in children born by C- section. Ann. Nutr. Metab. 2018, 73, 33–39. [Google Scholar] [CrossRef]
- Scott, P.; Antoine, M.; Midtvedt, T.; Hemert, V. Manipulating the gut microbiota to maintain health and treat disease. Microb. Ecol. Health Dis. 2015, 26, 25877. [Google Scholar] [CrossRef]
- Kristensen, B.; Bryrup, T.; Allin, H.; Nielsen, T.; Hansen, T.H.; Pedersen, O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: A systematic review of randomized control trials. Genome Med. 2016, 8, 1–11. [Google Scholar] [CrossRef]
- Kobyliak, N.; Conte, C.; Cammarota, G.; Haley, A.P.; Styriak, I.; Gaspar, L.; Fusek, J.; Rodrigo, L.; Kruzliak, P. Probiotics in prevention and treatment of obesity: A critical view. Nutr. Metab. 2016, 13, 1e13. [Google Scholar] [CrossRef]
- Besselink, M.G.; Van Santvoort, H.C.; Buskens, E.; Boermeester, M.A.; Van Goor, H.; Timmerman, H.M.; Nieuwenhuijs, V.B.; Bollen, T.L.; van Ramshorst, B.; Witterman, B.J.; et al. Dutch Acute Pancreatitis Study Grp (2008). Probiotic prophylaxis in predicted severe acute pancreatitis: A randomised, double-blind, placebo-controlled trial. Lancet 2008, 371, 651–659. [Google Scholar] [CrossRef]
- Plaza- Diaz, J.; Rulz- Ozeda, F.; Gil-Campos, M.; Gil, A. Mechanisms of action of Probiotics. Am. Soc. Nutr. 2019, 10 (Suppl. 1), S49–S66. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.; Russo, P.; Duenas, M.; Lopez, P.; Spano, G. Lactic acid bacteria producing B-group vitamins: A great potential for functional cereals products. Appl. Microbiol. Biotechnol. 2012, 96, 1383–1394. [Google Scholar] [CrossRef] [PubMed]
- Terpou, A.; Papadaki, A.; Lappa, I.; Kachrimanidou, V.; Bosnea, L.; Kopsahelis, N. Probiotics in food systems: Significance and emerging strategies towards improved viability and delivery of enhanced beneficial value. Nutrients 2019, 11, 1591. [Google Scholar] [CrossRef]
- Cristofori, F.; Francavilla, R.; Capobianco, D.; Dargenio, N.; Filardo, S.; Mastromarino, P. Bacterial-Based Strategies to Hydrolyze Gluten Peptides and Protect Intestinal Mucosa. Front. Immunol. 2020, 11, 567801. [Google Scholar] [CrossRef]
- De Almeida, N.E.C.; Esteves, F.G.; Dos Santos-Pinto, J.R.A.; De Paula, C.P.; Da Cunha, A.F.; Malavazi, I.; Palma, M.S.; Rodrigues-Filho, E. Digestion of intact gluten proteins by Bifidobacterium species: Reduction of cytotoxicity and pro-inflammatory responses. J. Agric. Food Chem. 2020, 68, 4485–4492. [Google Scholar] [CrossRef]
- Giorgi, A.; Cerrone, R.; Capobianco, D.; Filardo, S.; Mancini, P.; Fanelli, S.; Mastromarino, P.; Mosca, L. A probiotic preparation hydrolyzes gliadin and protects intestinal cells from the toxicity of pro-inflammatory peptides. Nutrients 2020, 12, 495. [Google Scholar] [CrossRef]
- Heeney, D.; Gareau, G.; Marco, L. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opin. Biotechnol. 2018, 49, 140–147. [Google Scholar] [CrossRef]
- D’Arienzo, R.; Maurano, F.; Luongo, D.; Mazzarella, G.; Stefanile, R.; Troncone, R.; Auricchio, S.; Ricca, E.; David, C.; Rossi, M. Adjuvant effect of Lactobacillus casei in a mouse model of gluten sensitivity. Immunol. Lett. 2008, 119, 78–83. [Google Scholar] [CrossRef]
- D’Arienzo, R.; Stefanile, R.; Maurano, F.; Mazzarella, G.; Ricca, E.; Troncone, R.; Auricchio, S.; Rossi, M. Immunomodulatory effects of Lactobacillus casei administration in a mouse model of gliadin-sensitive enteropathy. Scand. J. Immunol. 2011, 74, 335–341. [Google Scholar] [CrossRef]
- Olivares, M.; Laparra, M.; Sanz, Y. Oral administration of Bifidobacterium longum CECT 7347 modulates jejunal proteome in an in vivo gliadin-induced enteropathy animal model. J. Proteom. 2012, 77, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Laparra, M.; Olivares, M.; Gallina, O.; Sanz, Y. Bifidobacterium longum CECT 7347 modulates immune responses in a gliadin-induced enteropathy animal model. PLoS ONE 2012, 7, e30744. [Google Scholar] [CrossRef]
- Akobeng, A.; Singh, P.; Kumar, M.; Al Khodor, S. Role of the gut microbiota in the pathogenesis of coeliac disease and potential therapeutic implications. Eur. J. Nutr. 2020, 59, 3369–3390. [Google Scholar] [CrossRef]
- Marasco, G.; Cirota, G.; Rossini, B.; Lungaro, L.; Di Biase, A.R.; Colecchia, A.; Volta, U.; de Giorgio, R.; Festi, D.; Caio, G. Probiotics, Prebiotics and Other Dietary Supplements for Gut Microbiota Modulation in Celiac Disease Patients. Nutrients 2020, 12, 2674. [Google Scholar] [CrossRef] [PubMed]
- Gibson, R.; Roberfroid, B. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Drabinska, N.; Jarocka-Cyrta, E.; Markiewicz, H.; Krupa-Kozak, U. The effect of oligofructose-enriched inulin on faecal bacterial counts and microbiota associated characteristics in celiac disease children following a gluten-free diet: Results of a randomized, placebo-controlled trial. Nutrients 2018, 10, 201. [Google Scholar] [CrossRef]
- Magge, S.; Lembo, A. Low-FODMAP diet for treatment of irritable bowel syndrome. Gastroenterol. Hepatol. 2012, 8, 39–45. [Google Scholar]
- Aziz, I.; Sanders, S. The irritable bowel syndrome-celiac disease connection. Gastrointest. Endosc. Clin. North Am. 2012, 22, 623–637. [Google Scholar] [CrossRef]
- Testa, A.; Imperatore, N.; Rispo, A.; Rea, M.; Tortora, R.; Nardone, O.M.; Lucci, L.; Accarino, G.; Caporaso, N.; Castiglione, F. Beyond irritable bowel syndrome: The efficacy of the low fodmap diet for improving symptoms in inflammatory bowel diseases and Celiac disease. Dig Dis. 2018, 36, 271–280. [Google Scholar] [CrossRef]
- Chen, Q.; Ren, Y.; Lu, J.; Bartlett, M.; Chen, L.; Zhang, Y.; Guo, X.; Liu, C. A Novel prebiotic blend product prevents irritable bowel syndrome in mice by improving gut microbiota and modulating immune response. Nutrients 2017, 9, 1341. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, A.; Milani, E.; Madadlou, A.; Mortazavi, S.A.; Mokarram, R.R.; Salarbashi, D. Synbiotic yogurt-ice cream produced via incorporation of microencapsulated lactobacillus acidophilus (la-5) and fructooligosaccharide. J. Food Sci. Technol. 2014, 51, 1568–1574. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients 2017, 15, 1021. [Google Scholar] [CrossRef] [PubMed]
- Wilms, E.; Gerritsen, J.; Smidt, H.; Besseling-van der Vaart, I.; Rijkers, G.T.; Fuentes, A.R.G.; Masclee, A.A.M.; Troost, F.J. Effects of Supplementation of the Synbiotic Ecologic® 825/FOS P6 on Intestinal Barrier Function in Healthy Humans: A Randomized Controlled Trial. PLoS ONE 2016, 11, e0167775. [Google Scholar] [CrossRef]
- Demiroren, K. Can a Synbiotic Supplementation Contribute to Decreasing Anti-Tissue Transglutaminase Levels in Children with Potential Celiac Disease? Pediatr. Gastroenterol. Hepatol. Nutr. 2020, 23, 397–404. [Google Scholar] [CrossRef]
- Ugural, A.; Akyol, A. Can pseudocereals modulate microbiota by functioning as probiotics or prebiotics? Crit. Rev. Food Sci. Nutr. 2022, 62, 1725–1739. [Google Scholar] [CrossRef] [PubMed]
- Gullón, B.; Gullón, P.; Tavaria, F.; Yáñez, R. Assessment of the prebiotic effect of quinoa and amaranth in the human intestinal ecosystem. Food Funct. 2016, 7, 3782–3788. [Google Scholar] [CrossRef]
- Rad, A.; Aghebati-Maleki, L.; Kafil, H.; Gilani, N.; Abbasi, A.; Khani, N. Postbiotics, as dynamic biomolecules, and their promising role in promoting food safety. Biointerface Res. Appl. Chem. 2021, 11, 14529–14544. [Google Scholar]
- Salminen, S.; Collado, M.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.; Sanders, M.; Shamir, R.; Swann, J.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef]
- Conte, M.; Porpora, M.; Nigro, F.; Nigro, R.; Budelli, L.; Barone, V.; Nanayakkara, M. Pro-Pre and Postbiotic in Celiac Disease. Appl. Sci. 2021, 11, 8185. [Google Scholar] [CrossRef]
- Sarno, M.; Lania, G.; Cuomo, M.; Nigro, F.; Passannanti, F.; Budelli, A.; Fasano, F.; Troncone, R.; Auricchio, S.; Barone, M.V.; et al. Lactobacillus paracasei CBA L74 interferes with gliadin peptides entrance in Caco-2 cells. Int. J. Food Sci. Nutr. 2014, 65, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.; Nigro, F.; Porpora, M.; Bellomo, C.; Furone, F.; Budelli, A.L.; Nigro, R.; Barone, M.V.; Nanayakkara, M. Gliadin Peptide P31–43 Induces mTOR/NFkβ Activation and Reduces Autophagy: The Role of Lactobacillus paracasei CBA L74 Postbiotc. Int. J. Mol. Sci. 2022, 23, 3655. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Sugimura, N.; Burgermeister, E.; Ebert, M.; Zuo, T.; Lan, P. The gut virome: A new microbiome component in health and disease. EBioMedicine 2022, 81, 104113. [Google Scholar] [CrossRef] [PubMed]
- Mills, S.; Shanahan, F.; Stanton, C.; Hill, C.; Coffey, A.; Ross, R. Movers and shakers: Influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes 2013, 4, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Vitetta, L.; Vitetta, G.; Hall, S. Immunological Tolerance and Function: Associations Between Intestinal Bacteria, Probiotics, Prebiotics, and Phages. Front. Immunol. 2018, 9, 2240. [Google Scholar] [CrossRef]
- El Mouzan, M.; Assiri, A.; Al Sarkhy, A.; Alasmi, M.; Saeed, A.; Al-Hussaini, A.; AlSaleem, B.; Al Mofarreh, M. Viral dysbiosis in children with new-onset celiac disease. PLoS ONE 2022, 17, e0262108. [Google Scholar] [CrossRef]
- Garmaeva, S.; Gulyaeva, A.; Sinha, T.; Shkoporov, A.N.; Clooney, A.G.; Stockdale, S.R.; Spreckels, J.E.; Sutton, T.D.; Draper, L.A.; Dutilh, B.E.; et al. Stability of the human gut virome and effect of gluten-free diet. Cell Rep. 2021, 35, 109132. [Google Scholar] [CrossRef]
- Febvre, H.; Rao, S.; Gindin, M.; Goodwin, N.; Finer, E.; Vivanco, J.; Lu, S.; Manter, D.; Wallace, T.; Weir, T. PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults. Nutrients 2019, 11, 666. [Google Scholar] [CrossRef]
- Lerner, A.; Ramesh, A.; Matthias, T. The Revival of the Battle between David and Goliath in the Enteric Viruses and Microbiota Struggle: Potential Implication for Celiac Disease. Microorganisms 2019, 7, 173. [Google Scholar] [CrossRef]
- SBIR Phase I: Bacteriophage-Based Microbial Gene Therapy Platform for In Situ Engineering of Microbiomes. Available online: https://www.sbir.gov/sbirsearch/detail/1705577 (accessed on 20 September 2022).
- Heder, M. From NASA to EU: The evolution of the TRL scale in Public Sector Innovation. Innov. J. 2017, 22, 1. [Google Scholar]
Gluten avoidance
|
Balanced diet
|
Social restrictions
|
| Probiotic Cultures | Animal Models | Mode of Sensitization | Major Findings | References |
|---|---|---|---|---|
| Lactobacillus casei | Transgenic mice expressing the human DQ8 heterodimer | Chymotryptic digest of gliadin and cholera toxin | Enhanced the gliadin-specific response mediated by CD4 T cells. | [130] |
| Lactobacillus casei | Transgenic mice expressing the HLA-DQ8 molecule without endogenous mouse class II genes, non-transgenic for human CD4. | Wheat gliadin | L. casei can be effective in rescuing the normal mucosal architecture. | [131] |
| Bifidobacterium longum CECT 7347 | Female weanling Wistar rats | Gliadin | Ameliorate the inflammation caused by gliadin. | [132] |
| Bifidobacterium longum CECT 7347 | Female, weaning Wistar rats | IFN-g and fed gliadin | Bifidobacterium longum attenuates the production of inflammatory cytokines, and the CD4 C T-cell mediated immune response. | [133] |
| Saccharomyces boulardii KK1strain | BALB/c mice | Gluten-containing commercial food pellets | Improved enteropathy development In association with a decrease of epithelial cell CD71 expression and local cytokine production. | [15] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagh, S.K.; Lammers, K.M.; Padul, M.V.; Rodriguez-Herrera, A.; Dodero, V.I. Celiac Disease and Possible Dietary Interventions: From Enzymes and Probiotics to Postbiotics and Viruses. Int. J. Mol. Sci. 2022, 23, 11748. https://doi.org/10.3390/ijms231911748
Wagh SK, Lammers KM, Padul MV, Rodriguez-Herrera A, Dodero VI. Celiac Disease and Possible Dietary Interventions: From Enzymes and Probiotics to Postbiotics and Viruses. International Journal of Molecular Sciences. 2022; 23(19):11748. https://doi.org/10.3390/ijms231911748
Chicago/Turabian StyleWagh, Sandip K., Karen M. Lammers, Manohar V. Padul, Alfonso Rodriguez-Herrera, and Veronica I. Dodero. 2022. "Celiac Disease and Possible Dietary Interventions: From Enzymes and Probiotics to Postbiotics and Viruses" International Journal of Molecular Sciences 23, no. 19: 11748. https://doi.org/10.3390/ijms231911748
APA StyleWagh, S. K., Lammers, K. M., Padul, M. V., Rodriguez-Herrera, A., & Dodero, V. I. (2022). Celiac Disease and Possible Dietary Interventions: From Enzymes and Probiotics to Postbiotics and Viruses. International Journal of Molecular Sciences, 23(19), 11748. https://doi.org/10.3390/ijms231911748