Comparative Analysis of Environment-Responsive Alternative Splicing in the Inflorescences of Cultivated and Wild Tomato Species
Abstract
1. Introduction
2. Results
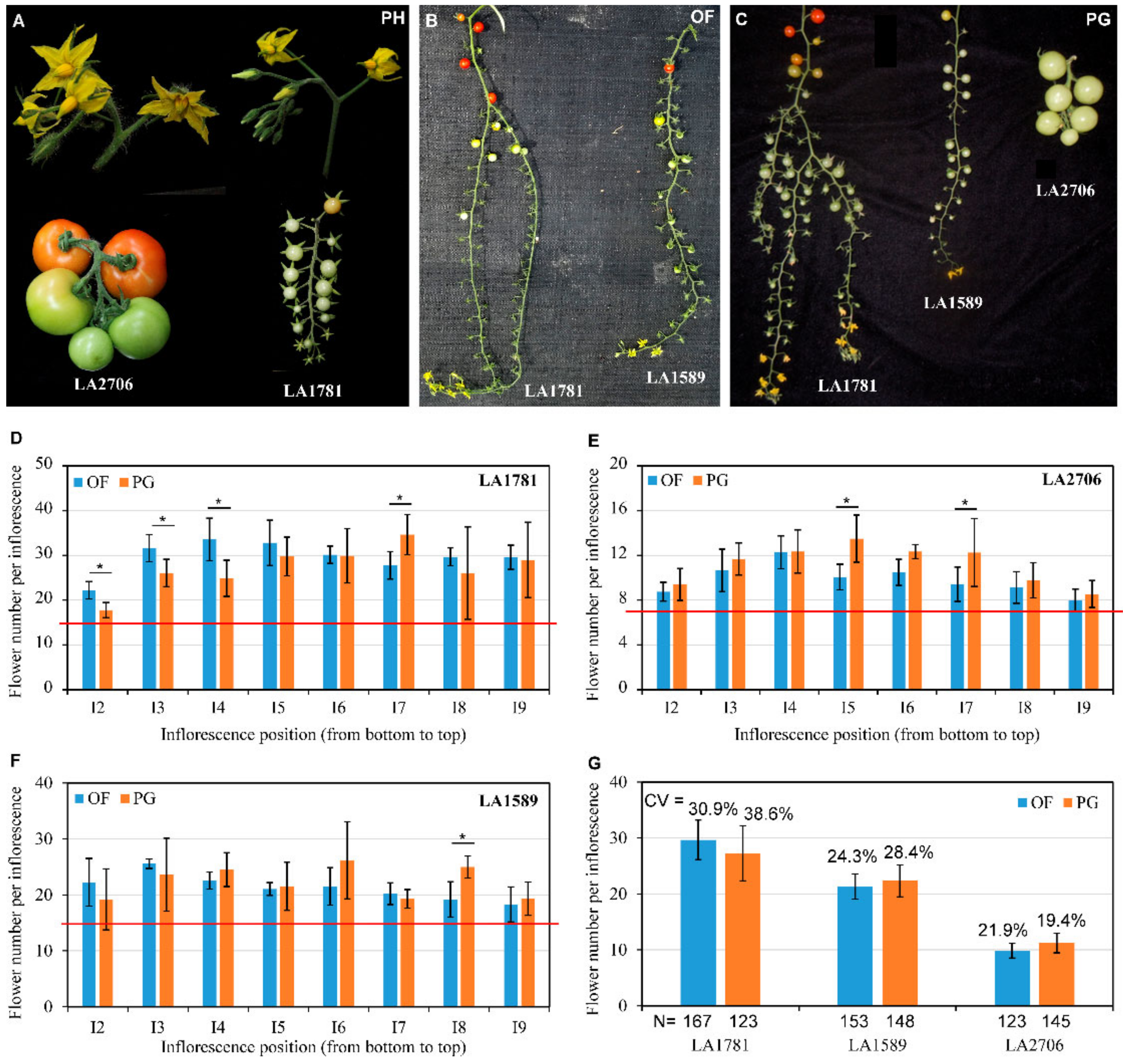
2.1. The Transcriptome Dynamics of Inflorescences in Three Growth Environments
2.2. Alternative Splicing in Response to Growth Conditions
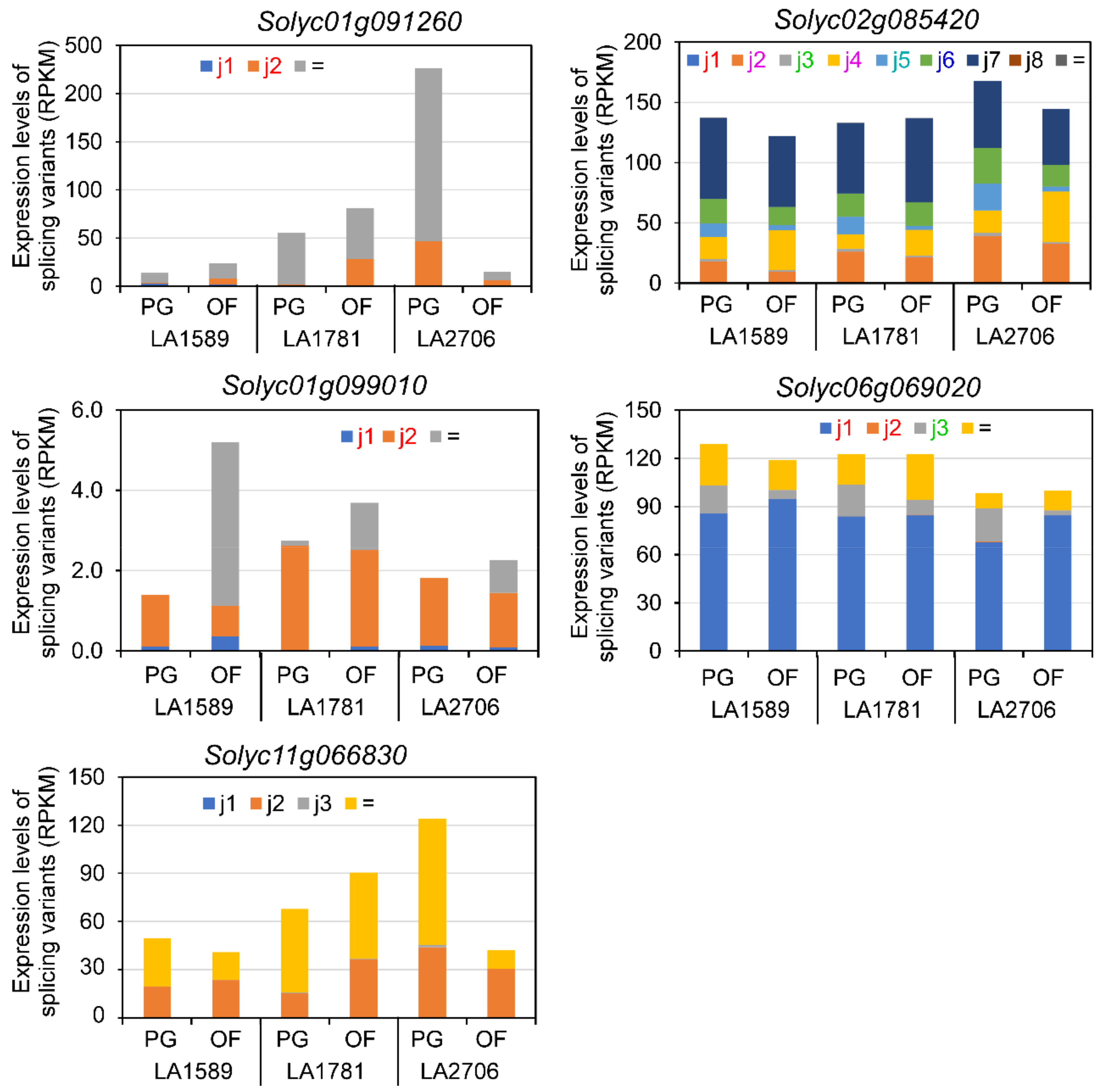
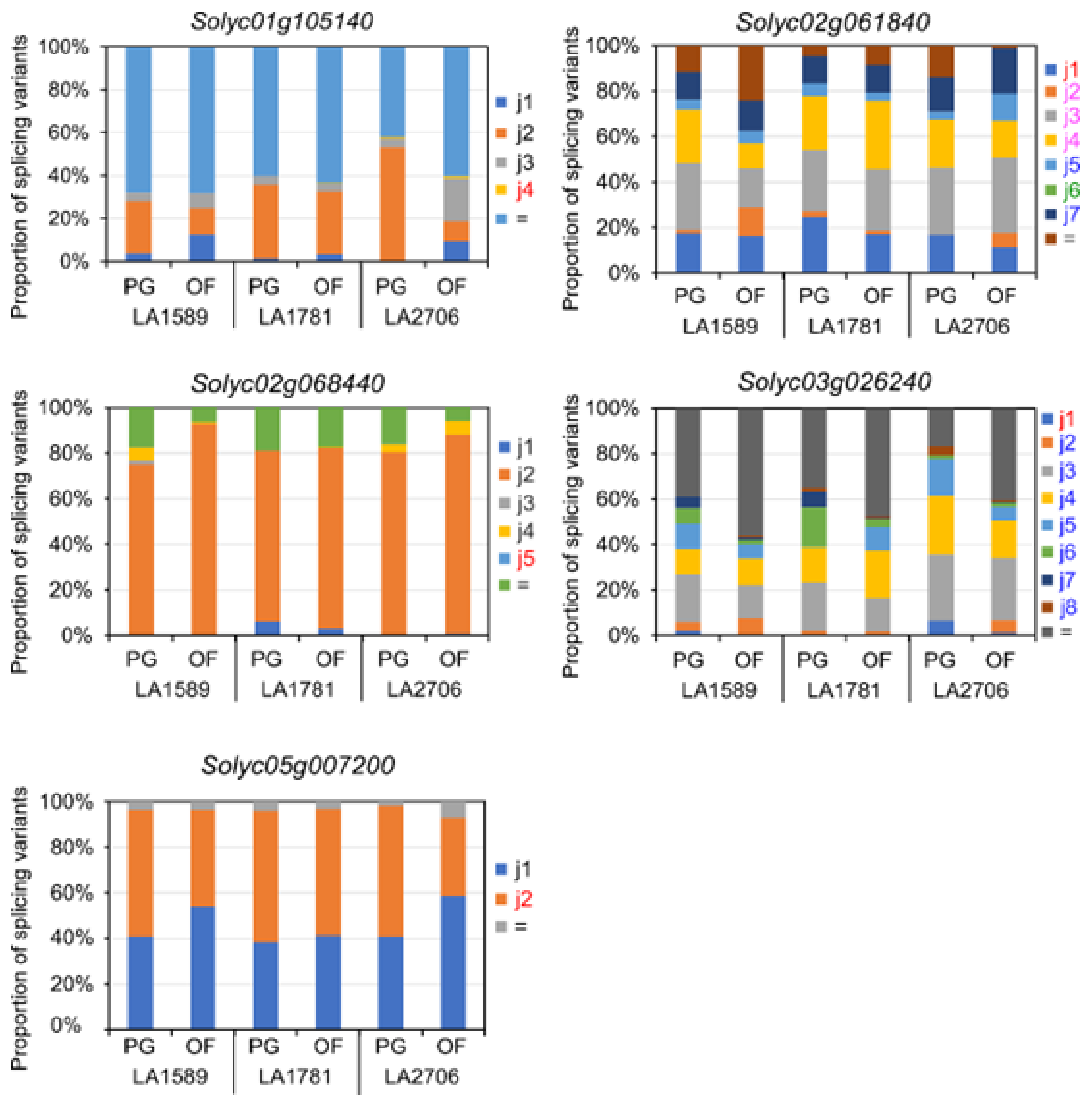
2.3. Expression Features of Splice Variants of DAGs
2.4. Differentially Expressed Genes and AS Variants Associated with the Plasticity of Inflorescence Development
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. RNA Isolation and Library Construction
4.3. Data Processing
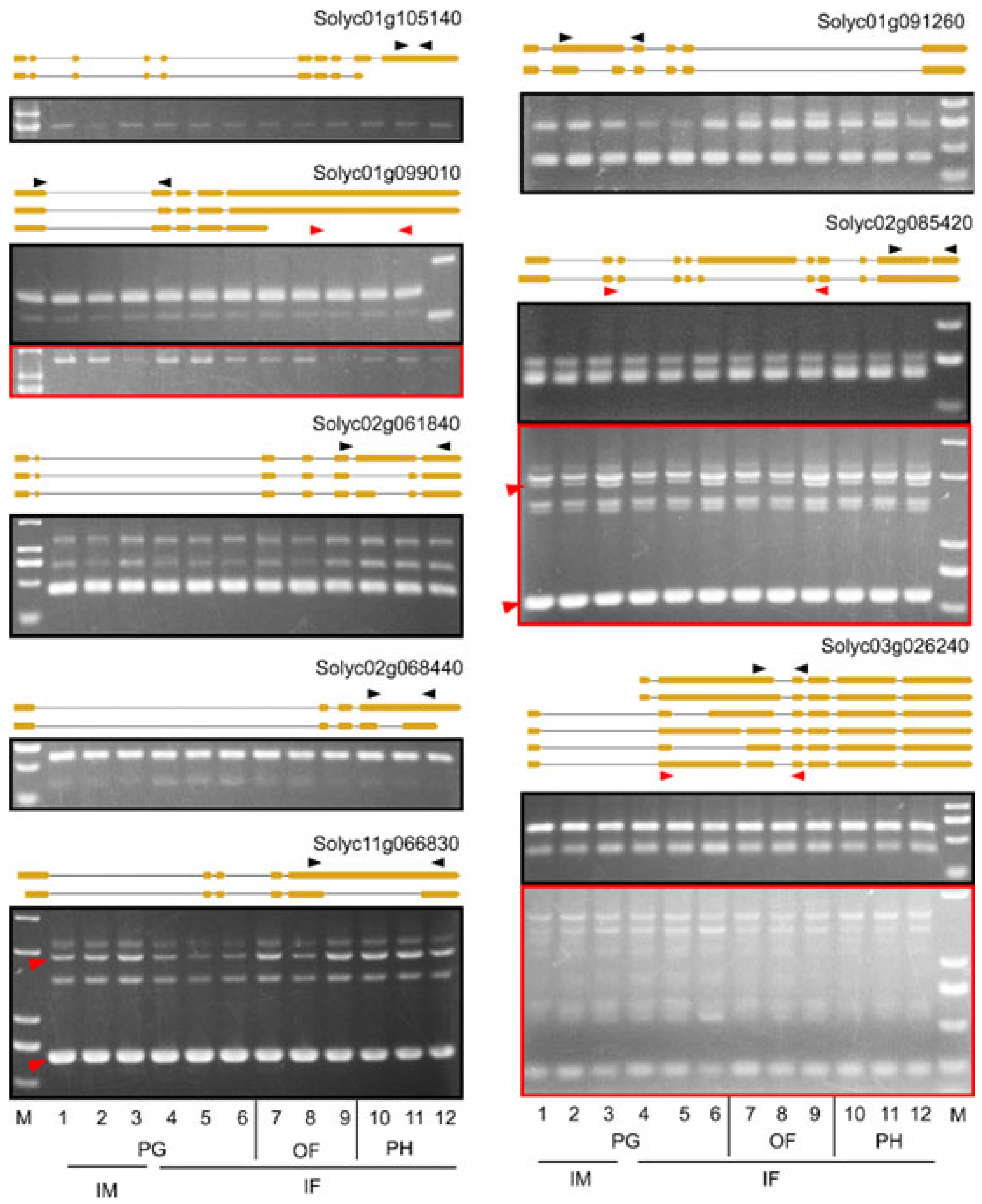
4.4. RT-PCR Verification of AS Events
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, Y.J.; Lee, J.H.; Kim, J.Y.; Park, C.M. Alternative RNA splicing expands the developmental plasticity of flowering transition. Front. Plant Sci. 2019, 10, 606. [Google Scholar] [CrossRef] [PubMed]
- Dal Santo, S.; Tornielli, G.B.; Zenoni, S.; Fasoli, M.; Farina, L.; Anesi, A.; Guzzo, F.; Delledonne, M.; Pezzotti, M. The plasticity of the grapevine berry transcriptome. Genome Biol. 2013, 14, r54. [Google Scholar] [CrossRef]
- Nagano, A.J.; Kawagoe, T.; Sugisaka, J.; Honjo, M.N.; Iwayama, K.; Kudoh, H. Annual transcriptome dynamics in natural environments reveals plant seasonal adaptation. Nat. Plants 2019, 5, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Nagano, A.J.; Sato, Y.; Mihara, M.; Antonio, B.A.; Motoyama, R.; Itoh, H.; Nagamura, Y.; Izawa, T. Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 2012, 151, 1358–1369. [Google Scholar] [CrossRef] [PubMed]
- Plessis, A.; Hafemeister, C.; Wilkins, O.; Gonzaga, Z.J.; Meyer, R.S.; Pires, I.; Muller, C.; Septiningsih, E.M.; Bonneau, R.; Purugganan, M. Multiple abiotic stimuli are integrated in the regulation of rice gene expression under field conditions. eLife 2015, 4, e08411. [Google Scholar] [CrossRef] [PubMed]
- de Klerk, E.; t Hoen, P.A. Alternative mRNA transcription, processing, and translation: Insights from RNA sequencing. Trends Genet. 2015, 31, 128–139. [Google Scholar] [CrossRef]
- Swarup, R.; Crespi, M.; Bennett, M.J. One gene, many proteins: Mapping cell-specific alternative splicing in plants. Dev. Cell 2016, 39, 383–385. [Google Scholar] [CrossRef]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W. Alternative splicing in plants—Coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef]
- Yu, H.; Tian, C.; Yu, Y.; Jiao, Y. Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana. Mol. Plant 2016, 9, 749–752. [Google Scholar] [CrossRef]
- Chaudhary, S.; Jabre, I.; Reddy, A.S.N.; Staiger, D.; Syed, N.H. Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci. 2019, 24, 496–506. [Google Scholar] [CrossRef]
- Chaudhary, S.; Khokhar, W.; Jabre, I.; Reddy, A.S.N.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Alternative splicing and protein diversity: Plants versus animals. Front. Plant Sci. 2019, 10, 708. [Google Scholar] [CrossRef]
- Chen, M.X.; Zhang, K.L.; Zhang, M.; Das, D.; Fang, Y.M.; Dai, L.; Zhang, J.; Zhu, F.Y. Alternative splicing and its regulatory role in woody plants. Tree Physiol. 2020, 40, 1475–1486. [Google Scholar] [CrossRef]
- Clark, S.; Yu, F.; Gu, L.; Min, X.J. Expanding alternative splicing identification by integrating multiple sources of transcription data in tomato. Front. Plant Sci. 2019, 10, 689. [Google Scholar] [CrossRef]
- English, A.C.; Patel, K.S.; Loraine, A.E. Prevalence of alternative splicing choices in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 102. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Priest, H.D.; Givan, S.A.; Shen, R.; Bryant, D.W.; Fox, S.E.; Wong, W.K.; Mockler, T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010, 20, 45–58. [Google Scholar] [CrossRef]
- Lu, T.; Lu, G.; Fan, D.; Zhu, C.; Li, W.; Zhao, Q.; Feng, Q.; Zhao, Y.; Guo, Y.; Huang, X.; et al. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010, 20, 1238–1249. [Google Scholar] [CrossRef]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef]
- Martín, G.; Márquez, Y.; Mantica, F.; Duque, P.; Irimia, M. Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol. 2021, 22, 35. [Google Scholar] [CrossRef]
- Nimeth, B.A.; Riegler, S.; Kalyna, M. Alternative Splicing and DNA Damage Response in Plants. Front. Plant Sci. 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhou, Z.; Wang, Z.; Li, W.; Fang, C.; Wu, M.; Ma, Y.; Liu, T.; Kong, L.-A.; Peng, D.-L.; et al. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 2014, 26, 996–1008. [Google Scholar] [CrossRef]
- Sun, Y.; Xiao, H. Identification of alternative splicing events by RNA sequencing in early growth tomato fruits. BMC Genom. 2015, 16, 948. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, S.R.; Zhou, W.; Leonard, A.; Wang, B.B.; Beatty, M.; Zastrow-Hayes, G.; Zhao, X.; Baumgarten, A.; Li, B. Genome-wide analysis of alternative splicing in Zea mays: Landscape and genetic regulation. Plant Cell 2014, 26, 3472–3487. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Zhou, X.; Xie, S.; Gu, J.; Wang, Z.Y. Identification of cassava alternative splicing-related genes and functional characterization of MeSCL30 involvement in drought stress. Plant Physiol. Biochem. PPB 2021, 160, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.W.; Simpson, C.G.; Marquez, Y.; Gadd, G.M.; Barta, A.; Kalyna, M. Lost in translation: Pitfalls in deciphering plant alternative splicing transcripts. Plant Cell 2015, 27, 2083–2087. [Google Scholar] [CrossRef]
- Capovilla, G.; Pajoro, A.; Immink, R.G.; Schmid, M. Role of alternative pre-mRNA splicing in temperature signaling. Curr. Opin. Plant Biol. 2015, 27, 97–103. [Google Scholar] [CrossRef]
- John, S.; Olas, J.J.; Mueller-Roeber, B. Regulation of alternative splicing in response to temperature variation in plants. J. Exp. Bot. 2021, 72, 6150–6163. [Google Scholar] [CrossRef]
- Li, S.; Yu, X.; Cheng, Z.; Zeng, C.; Li, W.; Zhang, L.; Peng, M. Large-scale analysis of the cassava transcriptome reveals the impact of cold stress on alternative splicing. J. Exp. Bot. 2020, 71, 422–434. [Google Scholar] [CrossRef]
- Lin, J.; Zhu, Z. Plant responses to high temperature: A view from pre-mRNA alternative splicing. Plant Mol. Biol. 2021, 105, 575–583. [Google Scholar] [CrossRef]
- Ling, Y.; Alshareef, S.; Butt, H.; Lozano-Juste, J.; Li, L.; Galal, A.A.; Moustafa, A.; Momin, A.A.; Tashkandi, M.; Richardson, D.N.; et al. Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. Cell Mol. Biol. 2017, 89, 291–309. [Google Scholar] [CrossRef]
- Ling, Y.; Mahfouz, M.M.; Zhou, S. Pre-mRNA alternative splicing as a modulator for heat stress response in plants. Trends Plant Sci. 2021, 26, 1153–1170. [Google Scholar] [CrossRef]
- Ling, Y.; Serrano, N.; Gao, G.; Atia, M.; Mokhtar, M.; Woo, Y.H.; Bazin, J.; Veluchamy, A.; Benhamed, M.; Crespi, M.; et al. Thermopriming triggers splicing memory in Arabidopsis. J. Exp. Bot. 2018, 69, 2659–2675. [Google Scholar] [CrossRef]
- Punzo, P.; Grillo, S.; Batelli, G. Alternative splicing in plant abiotic stress responses. Biochem. Soc. Trans. 2020, 48, 2117–2126. [Google Scholar] [CrossRef]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef]
- Hartmann, L.; Drewe-Boss, P.; Wiessner, T.; Wagner, G.; Geue, S.; Lee, H.C.; Obermuller, D.M.; Kahles, A.; Behr, J.; Sinz, F.H.; et al. Alternative splicing substantially diversifies the transcriptome during early photomorphogenesis and correlates with the energy availability in Arabidopsis. Plant Cell 2016, 28, 2715–2734. [Google Scholar] [CrossRef]
- Palusa, S.G.; Reddy, A.S. Differential recruitment of splice variants from SR pre-mRNAs to polysomes during development and in response to stresses. Plant Cell Physiol. 2015, 56, 421–427. [Google Scholar] [CrossRef]
- Wang, G.; Weng, L.; Li, M.; Xiao, H. Response of gene expression and alternative splicing to distinct growth environments in tomato. Int. J. Mol. Sci. 2017, 18, 475. [Google Scholar] [CrossRef]
- Xiao, H.; Radovich, C.; Welty, N.; Hsu, J.; Li, D.; Meulia, T.; van der Knaap, E. Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biol. 2009, 9, 49. [Google Scholar] [CrossRef]
- Jiang, X.; Lubini, G.; Hernandes-Lopes, J.; Rijnsburger, K.; Veltkamp, V.; de Maagd, R.A.; Angenent, G.C.; Bemer, M. FRUITFULL-like genes regulate flowering time and inflorescence architecture in tomato. Plant Cell 2022, 34, 1002–1019. [Google Scholar] [CrossRef]
- Lippman, Z.B.; Cohen, O.; Alvarez, J.P.; Abu-Abied, M.; Pekker, I.; Paran, I.; Eshed, Y.; Zamir, D. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol. 2008, 6, e288. [Google Scholar] [CrossRef]
- MacAlister, C.A.; Park, S.J.; Jiang, K.; Marcel, F.; Bendahmane, A.; Izkovich, Y.; Eshed, Y.; Lippman, Z.B. Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat. Genet. 2012, 44, 1393–1398. [Google Scholar] [CrossRef]
- Soyk, S.; Lemmon, Z.H.; Oved, M.; Fisher, J.; Liberatore, K.L.; Park, S.J.; Goren, A.; Jiang, K.; Ramos, A.; van der Knaap, E.; et al. Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 2017, 169, 1142–1155.e1112. [Google Scholar] [CrossRef]
- Soyk, S.; Lemmon, Z.H.; Sedlazeck, F.J.; Jiménez-Gómez, J.M.; Alonge, M.; Hutton, S.F.; Van Eck, J.; Schatz, M.C.; Lippman, Z.B. Duplication of a domestication locus neutralized a cryptic variant that caused a breeding barrier in tomato. Nat. Plants 2019, 5, 471–479. [Google Scholar] [CrossRef]
- Thouet, J.; Quinet, M.; Lutts, S.; Kinet, J.M.; Périlleux, C. Repression of floral meristem fate is crucial in shaping tomato inflorescence. PLoS ONE 2012, 7, e31096. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Z.; Sun, S.; Wu, J.; Li, R.; Wang, H.; Cui, X. SISTER OF TM3 activates FRUITFULL1 to regulate inflorescence branching in tomato. Hortic. Res. 2021, 8, 251. [Google Scholar] [CrossRef]
- Zheng, H.; Kawabata, S. Identification and validation of new alleles of FALSIFLORA and COMPOUND INFLORESCENCE genes controlling the number of branches in tomato inflorescence. Int. J. Mol. Sci. 2017, 18, 1572. [Google Scholar] [CrossRef]
- Ganie, S.A.; Reddy, A.S.N. Stress-induced changes in alternative splicing landscape in rice: Functional significance of splice isoforms in stress tolerance. Biology 2021, 10, 309. [Google Scholar] [CrossRef]
- Maillot, P.; Velt, A.; Rustenholz, C.; Butterlin, G.; Merdinoglu, D.; Duchêne, E. Alternative splicing regulation appears to play a crucial role in grape berry development and is also potentially involved in adaptation responses to the environment. BMC Plant Biol. 2021, 21, 487. [Google Scholar] [CrossRef] [PubMed]
- Paajanen, P.; Lane de Barros Dantas, L.; Dodd, A.N. Layers of crosstalk between circadian regulation and environmental signalling in plants. Curr. Biol. CB 2021, 31, R399–R413. [Google Scholar] [CrossRef]
- Yu, H.; Du, Q.; Campbell, M.; Yu, B.; Walia, H.; Zhang, C. Genome-wide discovery of natural variation in pre-mRNA splicing and prioritising causal alternative splicing to salt stress response in rice. New Phytol. 2021, 230, 1273–1287. [Google Scholar] [CrossRef]
- Mesihovic, A.; Ullrich, S.; Rosenkranz, R.R.E.; Gebhardt, P.; Bublak, D.; Eich, H.; Weber, D.; Berberich, T.; Scharf, K.D.; Schleiff, E.; et al. HsfA7 coordinates the transition from mild to strong heat stress response by controlling the activity of the master regulator HsfA1a in tomato. Cell Rep. 2022, 38, 110224. [Google Scholar] [CrossRef]
- Hu, Y.; Mesihovic, A.; Jiménez-Gómez, J.M.; Röth, S.; Gebhardt, P.; Bublak, D.; Bovy, A.; Scharf, K.D.; Schleiff, E.; Fragkostefanakis, S. Natural variation in HsfA2 pre-mRNA splicing is associated with changes in thermotolerance during tomato domestication. New Phytol. 2020, 225, 1297–1310. [Google Scholar] [CrossRef]
- Tognacca, R.S.; Kubaczka, M.G.; Servi, L.; Rodríguez, F.S.; Godoy Herz, M.A.; Petrillo, E. Light in the transcription landscape: Chromatin, RNA polymerase II and splicing throughout Arabidopsis thaliana’s life cycle. Transcription 2020, 11, 117–133. [Google Scholar] [CrossRef] [PubMed]
- Savvides, A.; van Ieperen, W.; Dieleman, J.A.; Marcelis, L.F. Meristem temperature substantially deviates from air temperature even in moderate environments: Is the magnitude of this deviation species-specific? Plant Cell Environ. 2013, 36, 1950–1960. [Google Scholar] [CrossRef] [PubMed]
- Craufurd, P.Q.; Wheeler, T.R. Climate change and the flowering time of annual crops. J. Exp. Bot. 2009, 60, 2529–2539. [Google Scholar] [CrossRef]
- Olas, J.J.; Apelt, F.; Annunziata, M.G.; John, S.; Richard, S.I.; Gupta, S.; Kragler, F.; Balazadeh, S.; Mueller-Roeber, B. Primary carbohydrate metabolism genes participate in heat-stress memory at the shoot apical meristem of Arabidopsis thaliana. Mol. Plant 2021, 14, 1508–1524. [Google Scholar] [CrossRef]
- Ding, F.; Cui, P.; Wang, Z.; Zhang, S.; Ali, S.; Xiong, L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 2014, 15, 431. [Google Scholar] [CrossRef]
- Li, Z.; Tang, J.; Bassham, D.C.; Howell, S.H. Daily temperature cycles promote alternative splicing of RNAs encoding SR45a, a splicing regulator in maize. Plant Physiol. 2021, 186, 1318–1335. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Morris, B.A.; Sutherland, P.; Veit, B.; Yao, J.L. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 2002, 130, 605–617. [Google Scholar] [CrossRef]
- Castelán-Muñoz, N.; Herrera, J.; Cajero-Sánchez, W.; Arrizubieta, M.; Trejo, C.; García-Ponce, B.; Sánchez, M.P.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. MADS-Box genes are key components of genetic regulatory networks involved in abiotic stress and plastic developmental responses in plants. Front. Plant Sci. 2019, 10, 853. [Google Scholar] [CrossRef]
- Lozano, R.; Angosto, T.; Gomez, P.; Payan, C.; Capel, J.; Huijser, P.; Salinas, J.; Martinez-Zapater, J.M. Tomato flower abnormalities induced by low temperatures are associated with changes of expression of MADS-Box genes. Plant Physiol. 1998, 117, 91–100. [Google Scholar] [CrossRef]
- Xiao, H.; Jiang, N.; Schaffner, E.; Stockinger, E.J.; van der Knaap, E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 2008, 319, 1527–1530. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]


| Gene Expression Levels | FPKM ≥ 0.1 | FPKM ≥ 0.5 | FPKM ≥ 1.0 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of expressed multiexon genes | Number of genes with AS | AS Frequency | Number of expressed multiexon genes | Number of genes with AS | AS Frequency | Number of expressed multiexon genes | Number of genes with AS | AS Frequency | |
| LA1589_IF_PG | 15415 | 8005 | 51.93% | 14240 | 7984 | 56.07% | 13629 | 7919 | 58.10% |
| LA1589_IF_OF | 15397 | 7955 | 51.67% | 14238 | 7931 | 55.70% | 13625 | 7860 | 57.69% |
| LA1589_IF_PH | 15309 | 6552 | 42.80% | 14286 | 6545 | 45.81% | 13726 | 6501 | 47.36% |
| LA1589_IM_PG | 15331 | 7946 | 51.83% | 14179 | 7938 | 55.98% | 13565 | 7895 | 58.20% |
| LA1781_IF_PG | 15356 | 8167 | 53.18% | 14285 | 8146 | 57.02% | 13691 | 8066 | 58.91% |
| LA1781_IF_OF | 15439 | 8361 | 54.16% | 14320 | 8289 | 57.88% | 13703 | 8143 | 59.42% |
| LA1781_IF_PH | 15513 | 6674 | 43.02% | 14391 | 6657 | 46.26% | 13813 | 6613 | 47.88% |
| LA1781_IM_PG | 15260 | 7866 | 51.55% | 14158 | 7853 | 55.47% | 13567 | 7800 | 57.49% |
| LA2706_IF_PG | 15369 | 8097 | 52.68% | 14201 | 8081 | 56.90% | 13616 | 8019 | 58.89% |
| LA2706_IF_OF | 15412 | 8299 | 53.85% | 14265 | 8241 | 57.77% | 13682 | 8101 | 59.21% |
| LA2706_IF_PH | 15499 | 6913 | 44.60% | 14415 | 6895 | 47.83% | 13834 | 6836 | 49.41% |
| LA2706_IM_PG | 15419 | 7746 | 50.24% | 14297 | 7734 | 54.10% | 13693 | 7670 | 56.01% |
| Total | 17100 | 12811 | 74.92% | 15570 | 12720 | 81.70% | 14923 | 12551 | 84.11% |
| PG, plastic greenhouse; OF, open field; PH, phytotron | |||||||||
| LA1589IF | LA1781IF | LA2706IF | |
|---|---|---|---|
| Number of expressed genes (FPKM ≥ 1) | 17947 | 17946 | 17984 |
| Number of DEGs | 871 | 413 | 616 |
| Number of DEIs | 1106 | 599 | 1119 |
| Number of DEVs | 350 | 225 | 529 |
| Number of DEAs | 681 | 342 | 505 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, E.; Wang, G.; Weng, L.; Li, M.; Xiao, H. Comparative Analysis of Environment-Responsive Alternative Splicing in the Inflorescences of Cultivated and Wild Tomato Species. Int. J. Mol. Sci. 2022, 23, 11585. https://doi.org/10.3390/ijms231911585
Zhou E, Wang G, Weng L, Li M, Xiao H. Comparative Analysis of Environment-Responsive Alternative Splicing in the Inflorescences of Cultivated and Wild Tomato Species. International Journal of Molecular Sciences. 2022; 23(19):11585. https://doi.org/10.3390/ijms231911585
Chicago/Turabian StyleZhou, Enbai, Guixiang Wang, Lin Weng, Meng Li, and Han Xiao. 2022. "Comparative Analysis of Environment-Responsive Alternative Splicing in the Inflorescences of Cultivated and Wild Tomato Species" International Journal of Molecular Sciences 23, no. 19: 11585. https://doi.org/10.3390/ijms231911585
APA StyleZhou, E., Wang, G., Weng, L., Li, M., & Xiao, H. (2022). Comparative Analysis of Environment-Responsive Alternative Splicing in the Inflorescences of Cultivated and Wild Tomato Species. International Journal of Molecular Sciences, 23(19), 11585. https://doi.org/10.3390/ijms231911585






