Enhancer RNA (eRNA) in Human Diseases
Abstract
1. Introduction
2. Role of eRNAs in Cancers
2.1. The Contribution and Mechanisms of eRNAs in Cancer
2.2. The Potential Clinical Utility of eRNAs in Cancer
3. Role of eRNAs in Neurodegenerative Disorders
3.1. The Contribution and Mechanisms of eRNAs in Neurodegenerative Disorders
3.2. The Potential Clinical Utility of eRNAs in Neurodegenerative Disorders
4. Role of eRNAs in Cardiovascular Diseases
4.1. The Contribution and Mechanisms of eRNAs in Cardiovascular Diseases
4.2. The Potential Clinical Utility of eRNAs in Cardiovascular Diseases
5. Role of eRNAs in Metabolic Diseases
5.1. The Contribution and Mechanisms of eRNAs in Metabolic diseases
5.2. The Potential Clinical Utility of eRNAs in Metabolic diseases
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2001, 2, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Behjati, S.; Tarpey, P.S. What is next generation sequencing? Arch. Dis. Child. Educ. Pract. Ed. 2013, 98, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.D. Non-coding RNA: A new frontier in regulatory biology. Natl. Sci. Rev. 2014, 1, 190–204. [Google Scholar] [CrossRef]
- Sharp, S.J.; Schaack, J.; Cooley, L.; Burke, D.J.; Soil, D. Structure and Transcription of Eukaryotic tRNA Gene. RNA Biol. 1985, 18, 738–746. [Google Scholar] [CrossRef]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef]
- Elliott, B.A.; Ho, H.T.; Ranganathan, S.V.; Vangaveti, S.; Ilkayeva, O.; Abou Assi, H.; Choi, A.K.; Agris, P.F.; Holley, C.L. Modification of messenger RNA by 2’-O-methylation regulates gene expression in vivo. Nat. Commun. 2019, 10, 3401. [Google Scholar] [CrossRef]
- Kiss, T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001, 20, 3617–3622. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A Brief Review on the Mechanisms of miRNA Regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Khashkhashi Moghadam, S.; Bakhshinejad, B.; Khalafizadeh, A.; Mahmud Hussen, B.; Babashah, S. Non-coding RNA-associated competitive endogenous RNA regulatory networks: Novel diagnostic and therapeutic opportunities for hepatocellular carcinoma. J. Cell Mol. Med. 2022, 26, 287–305. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Suzuki, H.; Tsukahara, T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int. J. Mol. Sci. 2014, 15, 9331–9342. [Google Scholar] [CrossRef]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. MicroRNA sponges: Progress and possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Halley, P.; Kadakkuzha, B.M.; Faghihi, M.A.; Magistri, M.; Zeier, Z.; Khorkova, O.; Coito, C.; Hsiao, J.; Lawrence, M.; Wahlestedt, C. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014, 6, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Lu, J.Y.; Liu, L.; Yin, Y.; Chen, C.; Han, X.; Wu, B.; Xu, R.; Liu, W.; Yan, P.; et al. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell 2016, 18, 637–652. [Google Scholar] [CrossRef]
- Niazi, F.; Valadkhan, S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. RNA 2012, 18, 825–843. [Google Scholar] [CrossRef]
- Johnsson, P.; Lipovich, L.; Grander, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Markaki, Y.; Gan Chong, J.; Wang, Y.; Jacobson, E.C.; Luong, C.; Tan, S.Y.X.; Jachowicz, J.W.; Strehle, M.; Maestrini, D.; Banerjee, A.K.; et al. Xist nucleates local protein gradients to propagate silencing across the X chromosome. Cell 2021, 184, 6174–6192.e6132. [Google Scholar] [CrossRef]
- Beltran, M.; Puig, I.; Pena, C.; Garcia, J.M.; Alvarez, A.B.; Pena, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, I.; Munita, R.; Agirre, E.; Dittmer, T.A.; Gysling, K.; Misteli, T.; Luco, R.F. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat. Struct. Mol. Biol. 2015, 22, 370–376. [Google Scholar] [CrossRef]
- Karakas, D.; Ozpolat, B. The Role of LncRNAs in Translation. Noncoding RNA 2021, 7, 16. [Google Scholar] [CrossRef]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- Cui, M.; You, L.; Ren, X.; Zhao, W.; Liao, Q.; Zhao, Y. Long non-coding RNA PVT1 and cancer. Biochem. Biophys. Res. Commun. 2016, 471, 10–14. [Google Scholar] [CrossRef]
- Dong, J.; Su, M.; Chang, W.; Zhang, K.; Wu, S.; Xu, T. Long non-coding RNAs on the stage of cervical cancer (Review). Oncol. Rep. 2017, 38, 1923–1931. [Google Scholar] [CrossRef]
- Han, P.; Chang, C.P. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015, 12, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Li, W.; Lin, C.H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.Y.; Lin, C.J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Kuo, H.C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019, 26, 29. [Google Scholar] [CrossRef]
- Vijesh, N.; Chakrabarti, S.K.; Sreekumar, J. Modeling of gene regulatory networks: A review. J. Biomed. Sci. Eng. 2013, 06, 223–231. [Google Scholar] [CrossRef]
- Emmert-Streib, F.; Dehmer, M.; Haibe-Kains, B. Gene regulatory networks and their applications: Understanding biological and medical problems in terms of networks. Front. Cell Dev. Biol. 2014, 2, 38. [Google Scholar] [CrossRef]
- Banf, M.; Rhee, S.Y. Computational inference of gene regulatory networks: Approaches, limitations and opportunities. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 41–52. [Google Scholar] [CrossRef]
- Monnier, P.; Martinet, C.; Pontis, J.; Stancheva, I.; Ait-Si-Ali, S.; Dandolo, L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc. Natl. Acad. Sci. USA 2013, 110, 20693–20698. [Google Scholar] [CrossRef]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. BioEssays 2010, 32, 473–480. [Google Scholar] [CrossRef]
- Gabory, A.; Ripoche, M.-A.; Le Digarcher, A.; Watrin, F.o.; Ziyyat, A.; Forné, T.; Jammes, H.L.N.; Ainscough, J.F.X.; Surani, M.A.; Journot, L.; et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 2009, 136, 3413–3421. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Huang, Z.; Yang, R.; Chen, Y.; Wang, Q.; Gao, L. Insights into Enhancer RNAs: Biogenesis and Emerging Role in Brain Diseases. Neurosci. A Rev. J. Bringing Neurobiol. Neurol. Psychiatry 2021, 10738584211046889. [Google Scholar] [CrossRef] [PubMed]
- Raisner, R.; Kharbanda, S.; Jin, L.; Jeng, E.; Chan, E.; Merchant, M.; Haverty, P.M.; Bainer, R.; Cheung, T.; Arnott, D.; et al. Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell Rep. 2018, 24, 1722–1729. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.; Gardini, A.; Zhang, A.; Shiekhattar, R. Integrator mediates the biogenesis of enhancer RNAs. Nature 2015, 525, 399–403. [Google Scholar] [CrossRef]
- Lee, J.H.; Xiong, F.; Li, W. Enhancer RNAs in cancer: Regulation, mechanisms and therapeutic potential. RNA Biol. 2020, 17, 1550–1559. [Google Scholar] [CrossRef]
- Han, Z.; Li, W. Enhancer RNA: What we know and what we can achieve. Cell Prolif. 2022, 55, e13202. [Google Scholar] [CrossRef]
- Mattick, J.S.; De Santa, F.; Barozzi, I.; Mietton, F.; Ghisletti, S.; Polletti, S.; Tusi, B.K.; Muller, H.; Ragoussis, J.; Wei, C.-L.; et al. A Large Fraction of Extragenic RNA Pol II Transcription Sites Overlap Enhancers. PLoS Biol. 2010, 8, e1000384. [Google Scholar] [CrossRef]
- Shi, L.; Li, S.; Maurer, K.; Zhang, Z.; Petri, M.; Sullivan, K.E. Enhancer RNA and NFκB-dependent P300 regulation of ADAMDEC1. Mol. Immunol. 2018, 103, 312–321. [Google Scholar] [CrossRef]
- de Lara, J.C.-F.; Arzate-Mejía, R.G.; Recillas-Targa, F. Enhancer RNAs: Insights Into Their Biological Role. Epigenetics Insights 2019, 12, 2516865719846093. [Google Scholar] [CrossRef]
- Theodorou, V.; Stark, R.; Menon, S.; Carroll, J.S. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2013, 23, 12–22. [Google Scholar] [CrossRef]
- Bernardo, G.M.; Bebek, G.; Ginther, C.L.; Sizemore, S.T.; Lozada, K.L.; Miedler, J.D.; Anderson, L.A.; Godwin, A.K.; Abdul-Karim, F.W.; Slamon, D.J.; et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene 2012, 32, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Stees, J.; Varn, F.; Huang, S.; Strouboulis, J.; Bungert, J. Recruitment of Transcription Complexes to Enhancers and the Role of Enhancer Transcription. Biology 2012, 1, 778–793. [Google Scholar] [CrossRef]
- Sartorelli, V.; Lauberth, S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020, 27, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Furlong, E.E.M. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Kreibich, E.; Krebs, A.R. Cofactors: A new layer of specificity to enhancer regulation. Trends Biochem. Sci. 2022. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Tamura, I.; Fujimura, T.; Doi-Tanaka, Y.; Shirafuta, Y.; Mihara, Y.; Maekawa, R.; Taketani, T.; Sato, S.; Tamura, H.; et al. Transcriptional coactivator PGC-1α contributes to decidualization by forming a histone-modifying complex with C/EBPβ and p300. J. Biol. Chem. 2022, 298, 101874. [Google Scholar] [CrossRef]
- Wang, W.; Qiao, S.; Li, G.; Cheng, J.; Yang, C.; Zhong, C.; Stovall, D.B.; Shi, J.; Teng, C.; Li, D.; et al. A histidine cluster determines YY1-compartmentalized coactivators and chromatin elements in phase-separated enhancer clusters. Nucleic Acids Res. 2022, 50, 4917–4937. [Google Scholar] [CrossRef]
- Kim, T.-K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A Class of Long Noncoding RNAs Synthesized at Enhancers: Figure 1. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Herz, H.M.; Hu, D.; Shilatifard, A. Enhancer malfunction in cancer. Mol. Cell 2014, 53, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, K.; Fukaya, T. Molecular architecture of enhancer-promoter interaction. Curr. Opin. Cell Biol. 2022, 74, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, M.M.; Ahmadiyeh, N.; Jia, L.; Herman, P.; Verzi, M.P.; Doddapaneni, H.; Beckwith, C.A.; Chan, J.A.; Hills, A.; Davis, M.; et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009, 41, 882–884. [Google Scholar] [CrossRef] [PubMed]
- Grisanzio, C.; Freedman, M.L. Chromosome 8q24-Associated Cancers and MYC. Genes Cancer 2010, 1, 555–559. [Google Scholar] [CrossRef]
- Gu, Y.; Lin, X.; Kapoor, A.; Chow, M.J.; Jiang, Y.; Zhao, K.; Tang, D. The Oncogenic Potential of the Centromeric Border Protein FAM84B of the 8q24.21 Gene Desert. Genes 2020, 11, 312. [Google Scholar] [CrossRef]
- Walavalkar, K.; Saravanan, B.; Singh, A.K.; Jayani, R.S.; Nair, A.; Farooq, U.; Islam, Z.; Soota, D.; Mann, R.; Shivaprasad, P.V.; et al. A rare variant of African ancestry activates 8q24 lncRNA hub by modulating cancer associated enhancer. Nat. Commun. 2020, 11, 3598. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef]
- Heintzman, N.D.; Hon, G.C.; Hawkins, R.D.; Kheradpour, P.; Stark, A.; Harp, L.F.; Ye, Z.; Lee, L.K.; Stuart, R.K.; Ching, C.W.; et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009, 459, 108–112. [Google Scholar] [CrossRef]
- Jia, L.; Landan, G.; Pomerantz, M.; Jaschek, R.; Herman, P.; Reich, D.; Yan, C.; Khalid, O.; Kantoff, P.; Oh, W.; et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009, 5, e1000597. [Google Scholar] [CrossRef]
- Sole, X.; Hernandez, P.; de Heredia, M.L.; Armengol, L.; Rodriguez-Santiago, B.; Gomez, L.; Maxwell, C.A.; Aguilo, F.; Condom, E.; Abril, J.; et al. Genetic and genomic analysis modeling of germline c-MYC overexpression and cancer susceptibility. BMC Genom. 2008, 9, 12. [Google Scholar] [CrossRef] [PubMed]
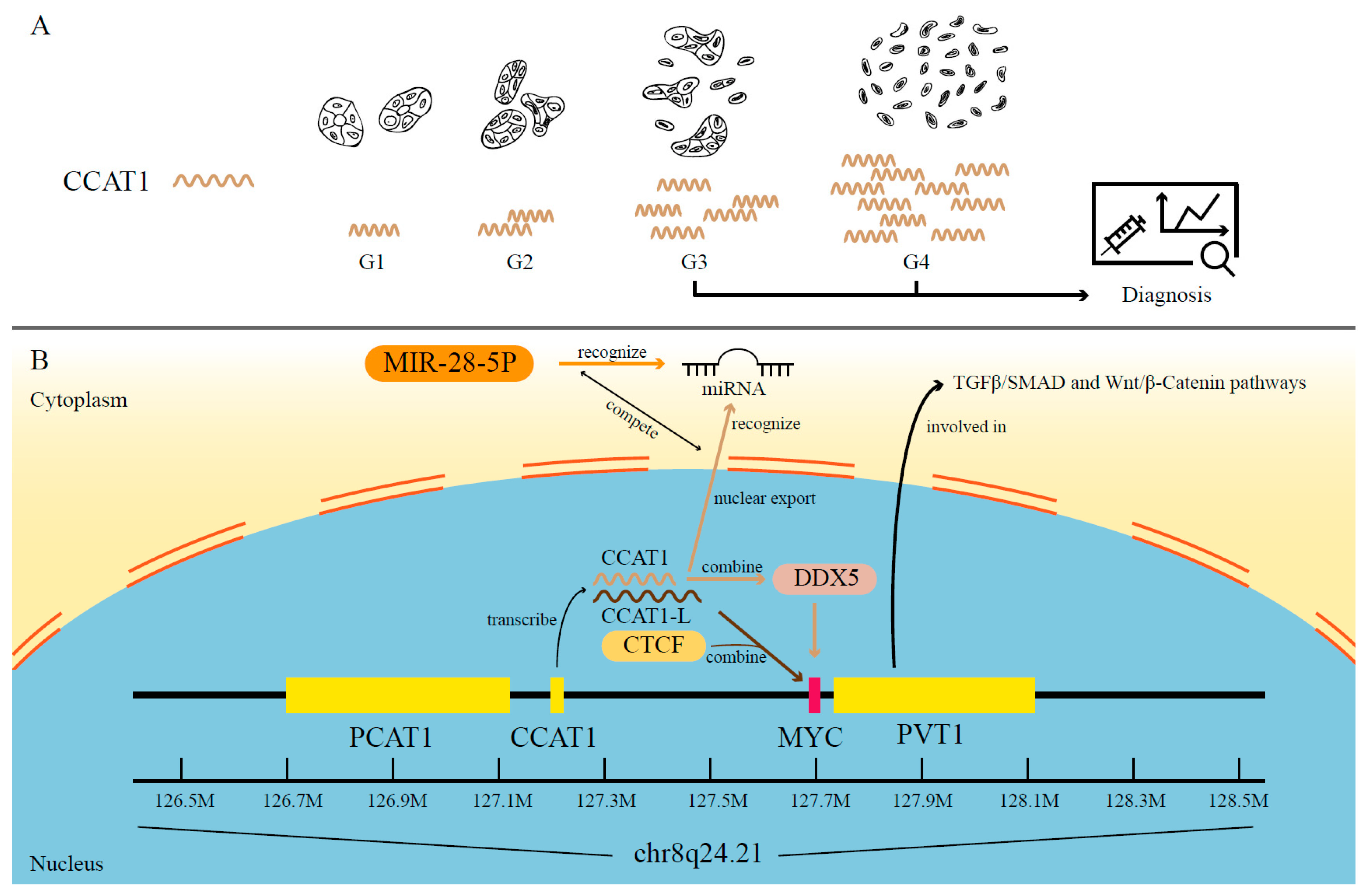
- You, Z.; Liu, C.; Wang, C.; Ling, Z.; Wang, Y.; Wang, Y.; Zhang, M.; Chen, S.; Xu, B.; Guan, H.; et al. LncRNA CCAT1 Promotes Prostate Cancer Cell Proliferation by Interacting with DDX5 and MIR-28-5P. Mol. Cancer Ther. 2019, 18, 2469–2479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Liu, T.; Li, Y.; Li, S. Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 9440–9445. [Google Scholar]
- Hu, M.; Zhang, Q.; Tian, X.H.; Wang, J.L.; Niu, Y.X.; Li, G. lncRNA CCAT1 is a biomarker for the proliferation and drug resistance of esophageal cancer via the miR-143/PLK1/BUBR1 axis. Mol. Carcinog. 2019, 58, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Clark, E.L.; Hadjimichael, C.; Temperley, R.; Barnard, A.; Fuller-Pace, F.V.; Robson, C.N. p68/DdX5 supports beta-catenin & RNAP II during androgen receptor mediated transcription in prostate cancer. PLoS ONE 2013, 8, e54150. [Google Scholar] [CrossRef]
- Clark, E.L.; Coulson, A.; Dalgliesh, C.; Rajan, P.; Nicol, S.M.; Fleming, S.; Heer, R.; Gaughan, L.; Leung, H.Y.; Elliott, D.J.; et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008, 68, 7938–7946. [Google Scholar] [CrossRef]
- Wu, G.; Xing, Z.; Tran, E.J.; Yang, D. DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. USA 2019, 116, 20453–20461. [Google Scholar] [CrossRef]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef]
- Shigeyasu, K.; Toden, S.; Ozawa, T.; Matsuyama, T.; Nagasaka, T.; Ishikawa, T.; Sahoo, D.; Ghosh, P.; Uetake, H.; Fujiwara, T.; et al. The PVT1 lncRNA is a novel epigenetic enhancer of MYC, and a promising risk-stratification biomarker in colorectal cancer. Mol. Cancer 2020, 19, 155. [Google Scholar] [CrossRef]
- Zhao, M.; Mishra, L.; Deng, C.X. The role of TGF-beta/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018, 14, 111–123. [Google Scholar] [CrossRef]
- Fodde, R.; Brabletz, T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr. Opin. Cell Biol. 2007, 19, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Lancho, O.; Herranz, D. The MYC Enhancer-ome: Long-Range Transcriptional Regulation of MYC in Cancer. Trends Cancer 2018, 4, 810–822. [Google Scholar] [CrossRef] [PubMed]
- Morgado-Pascual, J.L.; Rayego-Mateos, S.; Tejedor, L.; Suarez-Alvarez, B.; Ruiz-Ortega, M. Bromodomain and Extraterminal Proteins as Novel Epigenetic Targets for Renal Diseases. Front. Pharmacol. 2019, 10, 1315. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Chitsaz, F.; Abbasi, A.; Misteli, T.; Ozato, K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 2003, 100, 8758–8763. [Google Scholar] [CrossRef] [PubMed]
- Loven, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef]
- Kanno, T.; Kanno, Y.; LeRoy, G.; Campos, E.; Sun, H.W.; Brooks, S.R.; Vahedi, G.; Heightman, T.D.; Garcia, B.A.; Reinberg, D.; et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat. Struct. Mol. Biol. 2014, 21, 1047–1057. [Google Scholar] [CrossRef]
- Zou, Z.; Huang, B.; Wu, X.; Zhang, H.; Qi, J.; Bradner, J.; Nair, S.; Chen, L.F. Brd4 maintains constitutively active NF-kappaB in cancer cells by binding to acetylated RelA. Oncogene 2014, 33, 2395–2404. [Google Scholar] [CrossRef]
- Hajmirza, A.; Emadali, A.; Gauthier, A.; Casasnovas, O.; Gressin, R.; Callanan, M.B. BET Family Protein BRD4: An Emerging Actor in NFkappaB Signaling in Inflammation and Cancer. Biomedicines 2018, 6, 16. [Google Scholar] [CrossRef]
- Rahnamoun, H.; Lee, J.; Sun, Z.; Lu, H.; Ramsey, K.M.; Komives, E.A.; Lauberth, S.M. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat. Struct. Mol. Biol. 2018, 25, 687–697. [Google Scholar] [CrossRef]
- Wu, X.; Xiao, Y.; Zhou, Y.; Zhou, Z.; Yan, W. LncRNA FOXP4-AS1 is activated by PAX5 and promotes the growth of prostate cancer by sequestering miR-3184-5p to upregulate FOXP4. Cell Death Dis. 2019, 10, 472. [Google Scholar] [CrossRef]
- Ye, J.; Fu, Y.; Wang, Z.; Yu, J. Long non-coding RNA FOXP4-AS1 facilitates the biological functions of hepatocellular carcinoma cells via downregulating ZC3H12D by mediating H3K27me3 through recruitment of EZH2. Cell Biol. Toxicol. 2021, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Wang, A.; Ma, Y.; Liu, H. Long non-coding RNA FOXP4-AS1 is a prognostic biomarker and associated with immune infiltrates in ovarian serous cystadenocarcinoma. Medicine 2021, 100, e27473. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lian, Y.; Yan, C.; Cai, Z.; Ding, J.; Ma, Z.; Peng, P.; Wang, K. Long non-coding RNA FOXP4-AS1 is an unfavourable prognostic factor and regulates proliferation and apoptosis in colorectal cancer. Cell Prolif. 2017, 50, e12312. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sheppard, D.; Dong, X.; Hu, X.; Chen, M.; Chen, R.; Chakrabarti, J.; Zavros, Y.; Peek, R.M.; Chen, L.F.H. pylori infection confers resistance to apoptosis via Brd4-dependent BIRC3 eRNA synthesis. Cell Death Dis. 2020, 11, 667. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Tai, J.; Zhou, N.; Huang, T.; Xue, Y.; Quan, Z. A prospective study revealing the role of an immune-related eRNA, WAKMAR2, in breast cancer. Sci. Rep. 2021, 11, 15328. [Google Scholar] [CrossRef]
- Qu, Q.H.; Jiang, S.Z.; Li, X.Y. LncRNA TBX5-AS1 Regulates the Tumor Progression Through the PI3K/AKT Pathway in Non-Small Cell Lung Cancer. Onco Targets Ther. 2020, 13, 7949–7961. [Google Scholar] [CrossRef]
- Cheng, L.; Han, T.; Chen, B.; Nie, K.; Peng, W. TBX5-AS1, an enhancer RNA, is a potential novel prognostic biomarker for lung adenocarcinoma. BMC Cancer 2021, 21, 794. [Google Scholar] [CrossRef]
- Hsieh, C.L.; Fei, T.; Chen, Y.; Li, T.; Gao, Y.; Wang, X.; Sun, T.; Sweeney, C.J.; Lee, G.S.; Chen, S.; et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. USA 2014, 111, 7319–7324. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Chen, C.; Zhang, R.; Wang, K. Pan-cancer analysis of long non-coding RNA NEAT1 in various cancers. Genes Dis. 2018, 5, 27–35. [Google Scholar] [CrossRef]
- Zhang, Z.; Lee, J.H.; Ruan, H.; Ye, Y.; Krakowiak, J.; Hu, Q.; Xiang, Y.; Gong, J.; Zhou, B.; Wang, L.; et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun. 2019, 10, 4562. [Google Scholar] [CrossRef]
- McCleland, M.L.; Mesh, K.; Lorenzana, E.; Chopra, V.S.; Segal, E.; Watanabe, C.; Haley, B.; Mayba, O.; Yaylaoglu, M.; Gnad, F.; et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J. Clin. Investig. 2016, 126, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Thean, L.F.; Blocker, C.; Li, H.H.; Lo, M.; Wong, M.; Tang, C.L.; Tan, E.K.W.; Rozen, S.G.; Cheah, P.Y. Enhancer-derived long non-coding RNAs CCAT1 and CCAT2 at rs6983267 has limited predictability for early stage colorectal carcinoma metastasis. Sci. Rep. 2021, 11, 404. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Wang, S.; Qie, J.B.; Sun, P.L. SPRY4-AS1, A Novel Enhancer RNA, Is a Potential Novel Prognostic Biomarker and Therapeutic Target for Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 765484. [Google Scholar] [CrossRef]
- Sun, Y.; Han, J.; Wang, Z.; Li, X.; Sun, Y.; Hu, Z. Safety and Efficacy of Bromodomain and Extra-Terminal Inhibitors for the Treatment of Hematological Malignancies and Solid Tumors: A Systematic Study of Clinical Trials. Front. Pharmacol. 2020, 11, 621093. [Google Scholar] [CrossRef]
- Léveillé, N.; Melo, C.A.; Agami, R. Enhancer-associated RNAs as therapeutic targets. Expert Opin. Biol. Ther. 2015, 15, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Dugger, B.N.; Dickson, D.W. Pathology of Neurodegenerative Diseases. Cold Spring Harb. Perspect. Biol. 2017, 9, a028035. [Google Scholar] [CrossRef] [PubMed]
- Hill, G.W., 3rd; Purcell, E.K.; Liu, L.; Velkey, J.M.; Altschuler, R.A.; Duncan, R.K. Netrin-1-mediated axon guidance in mouse embryonic stem cells overexpressing neurogenin-1. Stem Cells Dev. 2012, 21, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Alcala-Vida, R.; Awada, A.; Boutillier, A.L.; Merienne, K. Epigenetic mechanisms underlying enhancer modulation of neuronal identity, neuronal activity and neurodegeneration. Neurobiol. Dis. 2021, 147, 105155. [Google Scholar] [CrossRef]
- Achour, M.; Le Gras, S.; Keime, C.; Parmentier, F.; Lejeune, F.X.; Boutillier, A.L.; Neri, C.; Davidson, I.; Merienne, K. Neuronal identity genes regulated by super-enhancers are preferentially down-regulated in the striatum of Huntington’s disease mice. Hum. Mol. Genet. 2015, 24, 3481–3496. [Google Scholar] [CrossRef]
- Salta, E.; De Strooper, B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012, 11, 189–200. [Google Scholar] [CrossRef]
- Tanila, H. The role of BDNF in Alzheimer’s disease. Neurobiol Dis 2017, 97, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Brookes, E.; Alan Au, H.Y.; Varsally, W.; Barrington, C.; Hadjur, S.; Riccio, A. A novel enhancer that regulates Bdnf expression in developing neurons. bioRxiv 2021. [Google Scholar] [CrossRef]
- Yu, C.; Li, C.H.; Chen, S.; Yoo, H.; Qin, X.; Park, H. Decreased BDNF Release in Cortical Neurons of a Knock-in Mouse Model of Huntington’s Disease. Sci. Rep. 2018, 8, 16976. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.A.; Grunseich, C.; Rodriguez, Y.; Liu, Y.; Li, D.; Burdick, J.T.; Bruzel, A.; Crouch, R.J.; Mahley, R.W.; Wilson, S.H.; et al. A transcriptional mechanism involving R-loop, m6A modification and RNA abasic sites regulates an enhancer RNA of APOE. bioRxiv 2022. [Google Scholar] [CrossRef]
- Li, P.; Marshall, L.; Oh, G.; Jakubowski, J.L.; Groot, D.; He, Y.; Wang, T.; Petronis, A.; Labrie, V. Epigenetic dysregulation of enhancers in neurons is associated with Alzheimer’s disease pathology and cognitive symptoms. Nat. Commun. 2019, 10, 2246. [Google Scholar] [CrossRef]
- Dong, X.; Liao, Z.; Gritsch, D.; Hadzhiev, Y.; Bai, Y.; Locascio, J.J.; Guennewig, B.; Liu, G.; Blauwendraat, C.; Wang, T.; et al. Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat. Neurosci. 2018, 21, 1482–1492. [Google Scholar] [CrossRef]
- Li, T.; Lu, D.; Yao, C.; Li, T.; Dong, H.; Li, Z.; Xu, G.; Chen, J.; Zhang, H.; Yi, X.; et al. Kansl1 haploinsufficiency impairs autophagosome-lysosome fusion and links autophagic dysfunction with Koolen-de Vries syndrome in mice. Nat. Commun. 2022, 13, 931, Erratum in Nat. Commun. 2022, 13, 1575. [Google Scholar] [CrossRef]
- Le Gras, S.; Keime, C.; Anthony, A.; Lotz, C.; De Longprez, L.; Brouillet, E.; Cassel, J.C.; Boutillier, A.L.; Merienne, K. Altered enhancer transcription underlies Huntington’s disease striatal transcriptional signature. Sci. Rep. 2017, 7, 42875. [Google Scholar] [CrossRef]
- Bond, A.M.; Vangompel, M.J.; Sametsky, E.A.; Clark, M.F.; Savage, J.C.; Disterhoft, J.F.; Kohtz, J.D. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 2009, 12, 1020–1027. [Google Scholar] [CrossRef]
- Cajigas, I.; Chakraborty, A.; Swyter, K.R.; Luo, H.; Bastidas, M.; Nigro, M.; Morris, E.R.; Chen, S.; VanGompel, M.J.W.; Leib, D.; et al. The Evf2 Ultraconserved Enhancer lncRNA Functionally and Spatially Organizes Megabase Distant Genes in the Developing Forebrain. Mol. Cell 2018, 71, 956–972.e959. [Google Scholar] [CrossRef]
- Foliaki, S.T.; Schwarz, B.; Groveman, B.R.; Walters, R.O.; Ferreira, N.C.; Orru, C.D.; Smith, A.; Wood, A.; Schmit, O.M.; Freitag, P.; et al. Neuronal excitatory-to-inhibitory balance is altered in cerebral organoid models of genetic neurological diseases. Mol. Brain 2021, 14, 156. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.T.; Chang, Y.G.; Chern, Y. Insights into GABAAergic system alteration in Huntington’s disease. Open Biol. 2018, 8, 180165. [Google Scholar] [CrossRef] [PubMed]
- Garret, M.; Du, Z.; Chazalon, M.; Cho, Y.H.; Baufreton, J. Alteration of GABAergic neurotransmission in Huntington’s disease. CNS Neurosci. Ther. 2018, 24, 292–300. [Google Scholar] [CrossRef]
- Deidda, G.; Bozarth, I.F.; Cancedda, L. Modulation of GABAergic transmission in development and neurodevelopmental disorders: Investigating physiology and pathology to gain therapeutic perspectives. Front. Cell Neurosci. 2014, 8, 119. [Google Scholar] [CrossRef]
- Si, W.; Huang, Z.; Li, X.; Zhao, L.; Ji, Y.; Li, H.; Liu, X.; Ye, S.; Chen, D.; Liu, H.; et al. Super-enhancer-driven Sorting Nexin 5 expression promotes dopaminergic neuronal ferroptosis in Parkinson’s disease models. Biochem. Biophys. Res. Commun. 2021, 567, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Onoguchi, M.; Hirabayashi, Y.; Koseki, H.; Gotoh, Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc. Natl. Acad. Sci. USA 2012, 109, 16939–16944. [Google Scholar] [CrossRef] [PubMed]
- Schuurmans, C.; Armant, O.; Nieto, M.; Stenman, J.M.; Britz, O.; Klenin, N.; Brown, C.; Langevin, L.M.; Seibt, J.; Tang, H.; et al. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004, 23, 2892–2902. [Google Scholar] [CrossRef]
- Chang, H.C.; Huang, H.C.; Juan, H.F.; Hsu, C.L. Investigating the role of super-enhancer RNAs underlying embryonic stem cell differentiation. BMC Genom. 2019, 20, 896. [Google Scholar] [CrossRef]
- Thiriet, M. Cardiovascular Disease: An Introduction. In Vasculopathies; Biomathematical and Biomechanical Modeling of the Circulatory and Ventilatory Systems; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–90. [Google Scholar]
- Ounzain, S.; Pezzuto, I.; Micheletti, R.; Burdet, F.; Sheta, R.; Nemir, M.; Gonzales, C.; Sarre, A.; Alexanian, M.; Blow, M.J.; et al. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J. Mol. Cell Cardiol. 2014, 76, 55–70. [Google Scholar] [CrossRef]
- Ounzain, S.; Pedrazzini, T. Super-enhancer lncs to cardiovascular development and disease. Biochim. Biophys. Acta 2016, 1863, 1953–1960. [Google Scholar] [CrossRef]
- Ounzain, S.; Pedrazzini, T. The promise of enhancer-associated long noncoding RNAs in cardiac regeneration. Trends Cardiovasc. Med. 2015, 25, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.J.; Kim, S.H.; Kim, E.K.; Han, J.W.; Shin, K.-H.; Hu, H.; Kim, K.S.; Choi, Y.D.; Kim, S.; Lee, Y.H.; et al. Prognostic implications of polycomb proteins ezh2, suz12, and eed1 and histone modification by H3K27me3 in sarcoma. BMC Cancer 2018, 18, 158. [Google Scholar] [CrossRef] [PubMed]
- Ounzain, S.; Micheletti, R.; Arnan, C.; Plaisance, I.; Cecchi, D.; Schroen, B.; Reverter, F.; Alexanian, M.; Gonzales, C.; Ng, S.Y.; et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell Cardiol. 2015, 89, 98–112. [Google Scholar] [CrossRef]
- Yang, X.H.; Nadadur, R.D.; Hilvering, C.R.E.; Bianchi, V.; Werner, M.; Mazurek, S.R.; Gadek, M.; Shen, K.M.; Goldman, J.A.; Tyan, L.; et al. Transcription-factor-dependent enhancer transcription defines a gene regulatory network for cardiac rhythm. eLife 2017, 6, e31683. [Google Scholar] [CrossRef]
- Arnolds, D.E.; Liu, F.; Fahrenbach, J.P.; Kim, G.H.; Schillinger, K.J.; Smemo, S.; McNally, E.M.; Nobrega, M.A.; Patel, V.V.; Moskowitz, I.P. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J. Clin. Investig. 2012, 122, 2509–2518. [Google Scholar] [CrossRef]
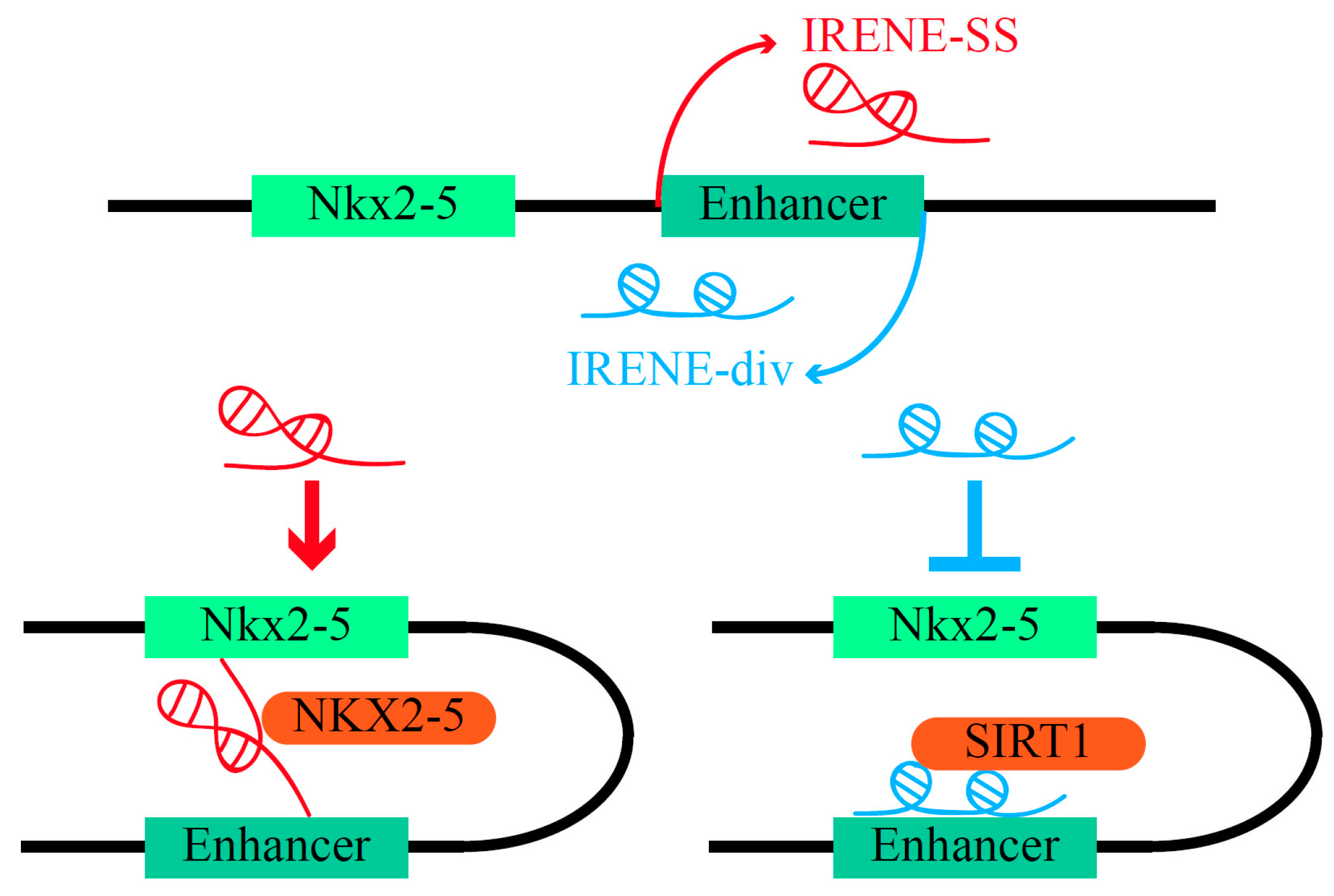
- Salamon, I.; Serio, S.; Bianco, S.; Pagiatakis, C.; Crasto, S.; Chiariello, A.M.; Conte, M.; Cattaneo, P.; Fiorillo, L.; Felicetta, A.; et al. Divergent Transcription of the Nkx2-5 Locus Generates Two Enhancer RNAs with Opposing Functions. iScience 2020, 23, 101539. [Google Scholar] [CrossRef]
- McElhinney, D.B.; Geiger, E.; Blinder, J.; Benson, W.; Goldmuntz, E. NKX2.5 Mutations in Patients with Congenital Heart Disease. J. Am. Coll. Cardiol. 2003, 42, 1650–1655. [Google Scholar] [CrossRef]
- Schott, J.-J.; Benson, D.W.; Basson, C.T.; William Pease, G.; Silberbach, M.; Moak, J.P.; Maron, B.J.; Seidman, C.E.; Seidman, J.G. Congenital Heart Disease Caused by Mutations in the Transcription Factor NKX2-5. Science 1998, 281, 108–111. [Google Scholar] [CrossRef]
- Micheletti, R.; Plaisance, I.; Abraham, B.J.; Sarre, A.; Ting, C.-C.; Alexanian, M.; Maric, D.; Maison, D.; Nemir, M.; Young, R.A.; et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017, 9, eaai9118. [Google Scholar] [CrossRef]
- Prabhu, S.D.; Frangogiannis, N.G. The Biological Basis for Cardiac Repair After Myocardial Infarction: From Inflammation to Fibrosis. Circ. Res. 2016, 119, 91–112. [Google Scholar] [CrossRef]
- Mirtschink, P.; Bischof, C.; Pham, M.-D.; Sharma, R.; Khadayate, S.; Rossi, G.; Fankhauser, N.; Traub, S.; Sossalla, S.; Hagag, E.; et al. Inhibition of the Hypoxia-Inducible Factor 1α–Induced Cardiospecific HERNA1 Enhance-Templated RNA Protects From Heart Disease. Circulation 2019, 139, 2778–2792. [Google Scholar] [CrossRef] [PubMed]
- Mirtschink, P.; Krek, W. Hypoxia-driven glycolytic and fructolytic metabolic programs: Pivotal to hypertrophic heart disease. Biochim. Biophys. Acta 2016, 1863, 1822–1828. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013, 152, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Alexanian, M.; Maric, D.; Jenkinson, S.P.; Mina, M.; Friedman, C.E.; Ting, C.C.; Micheletti, R.; Plaisance, I.; Nemir, M.; Maison, D.; et al. A transcribed enhancer dictates mesendoderm specification in pluripotency. Nat. Commun. 2017, 8, 1806. [Google Scholar] [CrossRef] [PubMed]
- Cao, H. Adipocytokines in obesity and metabolic disease. J. Endocrinol. 2014, 220, T47–T59. [Google Scholar] [CrossRef]
- O’Rahilly, S. Human genetics illuminates the paths to metabolic disease. Nature 2009, 462, 307–314. [Google Scholar] [CrossRef]
- den Boer, M.; Voshol, P.J.; Kuipers, F.; Havekes, L.M.; Romijn, J.A. Hepatic steatosis: A mediator of the metabolic syndrome. Lessons from animal models. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 644–649. [Google Scholar] [CrossRef]
- Lo, K.A.; Huang, S.; Walet, A.C.E.; Zhang, Z.-c.; Leow, M.K.-S.; Liu, M.; Sun, L. Adipocyte Long-Noncoding RNA Transcriptome Analysis of Obese Mice Identified Lnc-Leptin, Which Regulates Leptin. Diabetes 2018, 67, 1045–1056. [Google Scholar] [CrossRef]
- Qadir, M.I.; Ahmed, Z. lep Expression and Its Role in Obesity and Type-2 Diabetes. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 47–51. [Google Scholar] [CrossRef]
- Zhang, Y.; Proenca, R.; Maffei, M.; Barone, M.; Leopold, L.; Friedman, J.M. Positional cloning of the mouse obese gene and its human homologue. Nature 1994, 372, 425–432. [Google Scholar] [CrossRef]
- Sureshchandra, S.; Wilson, R.M.; Rais, M.; Marshall, N.E.; Purnell, J.Q.; Thornburg, K.L.; Messaoudi, I. Maternal Pregravid Obesity Remodels the DNA Methylation Landscape of Cord Blood Monocytes Disrupting Their Inflammatory Program. J. Immunol. 2017, 199, 2729–2744. [Google Scholar] [CrossRef] [PubMed]
- Benhammou, J.N.; Ko, A.; Alvarez, M.; Kaikkonen, M.U.; Rankin, C.; Garske, K.M.; Padua, D.; Bhagat, Y.; Kaminska, D.; Kärjä, V.; et al. Novel Lipid Long Intervening Noncoding RNA, Oligodendrocyte Maturation-Associated Long Intergenic Noncoding RNA, Regulates the Liver Steatosis Gene Stearoyl-Coenzyme A Desaturase As an Enhancer RNA. Hepatol. Commun. 2019, 3, 1356–1372. [Google Scholar] [CrossRef] [PubMed]
- Kotronen, A.; Seppanen-Laakso, T.; Westerbacka, J.; Kiviluoto, T.; Arola, J.; Ruskeepaa, A.L.; Oresic, M.; Yki-Jarvinen, H. Hepatic stearoyl-CoA desaturase (SCD)-1 activity and diacylglycerol but not ceramide concentrations are increased in the nonalcoholic human fatty liver. Diabetes 2009, 58, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Li, S.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.H.; et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1alpha. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef]
- Rius-Perez, S.; Torres-Cuevas, I.; Millan, I.; Ortega, A.L.; Perez, S. PGC-1alpha, Inflammation, and Oxidative Stress: An Integrative View in Metabolism. Oxid. Med. Cell. Longev. 2020, 2020, 1452696. [Google Scholar] [CrossRef]
- Abhimanyu Garg, M.D. Acquired and Inherited Lipodystrophies. N. Engl. J. Med. 2004, 350, 1220–1234. [Google Scholar] [CrossRef]
- Akinci, B.; Sahinoz, M.; Oral, E. Lipodystrophy Syndromes: Presentation and Treatment; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Czapiewski, R.; Batrakou, D.G.; de Las Heras, J.I.; Carter, R.N.; Sivakumar, A.; Sliwinska, M.; Dixon, C.R.; Webb, S.; Lattanzi, G.; Morton, N.M.; et al. Genomic loci mispositioning in Tmem120a knockout mice yields latent lipodystrophy. Nat. Commun. 2022, 13, 321. [Google Scholar] [CrossRef]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J., Jr.; Liu, X.S.; et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef]
- Kahn, B.B.; McGraw, T.E. Rosiglitazone, PPARγ, and Type 2 Diabetes. N. Engl. J. Med. 2010, 363, 2667–2669. [Google Scholar] [CrossRef]
- Step, S.E.; Lim, H.W.; Marinis, J.M.; Prokesch, A.; Steger, D.J.; You, S.H.; Won, K.J.; Lazar, M.A. Anti-diabetic rosiglitazone remodels the adipocyte transcriptome by redistributing transcription to PPARgamma-driven enhancers. Genes Dev. 2014, 28, 1018–1028. [Google Scholar] [CrossRef]
- Ye, J. Mechanisms of insulin resistance in obesity. Front. Med. 2013, 7, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Degirmenci, U.; Li, J.; Lim, Y.C.; Siang, D.T.C.; Lin, S.; Liang, H.; Sun, L. Silencing an insulin-induced lncRNA, LncASIR, impairs the transcriptional response to insulin signalling in adipocytes. Sci. Rep. 2019, 9, 5608. [Google Scholar] [CrossRef] [PubMed]

| Identified eRNA | Disease Type | Target Gene | Experimental or Clinical Resources | Reference |
|---|---|---|---|---|
| CCAT1 | CRC, PRAD, BRCA and ESCA | MYC | Patient and/or surgical samples and cell lines | [72,73,74,78] |
| MMP9 and CCL2 eRNA | BRCA, ESCA and PAAD | Brd4 | Cell lines (SW480) | [86] |
| FOXP4-AS1 | HCC, CRC, PRAD and OV | FOXP4 | Patient and/or surgical samples and cell lines (PC-3, DU145, VCaP, LNCaP, RWPE-1, MHCC-97H, HepG2, LM3, SMMC-7721, DLD-1, HT-29, HCT116, SW480, Lovo) | [91,92,93] |
| BIR3C | Gastric cancer | cIAP2 | Cell lines (AGS, MKN28, MKN45) | [94] |
| WAKMAR2 | Breast cancer | IL27RA, RAC2, FABP7, IGLV1-51, IGHA1 and IGHD | Patient samples and cell lines (MB-231, MCF7) | [95] |
| TBX5-AS1 | Lung cancer non-small cells | PI3K and AKT | Patient samples and cell lines (16HBE, A549, H1299, NCI-H520) | [96,97] |
| KLK3e | PRAD | PSA | Cell lines (LNCaP, VCaP, COS-7) | [98] |
| NET1e | PRAD, STAD, LIHC, KIRC and KIRP | Downstream miRNAs (let-7e, miR-34, miR-98, miR-107, etc.) | Patient samples | [100] |
| Bdnf-Enhg1 and Bdnf-Enhg2 | AD | Bdnf | Mice models | [112] |
| Evf2 | HD and epilepsy | Dlx5/6 | Rabbit models and cell lines (mouse EL250, PL253 and PL452) | [120] |
| utNgn1 | Neurogenesis | Neurog1 | Mice models and ESC lines | [126] |
| IRENEs | CHD | Nkx2-5 | Mice models and cell lines (human RUES2 cells and iPSC-derived CMs, mouse HL-1) | [137] |
| Wisper | AOS and CM | Wisp2 | Patient samples and mice models | [140] |
| HERNA1 | AS and HCM | synaptotagmin XVII and SMG1 | Patient samples, mice models and cell lines (HEK-293T cells) | [142] |
| Bvht | CVD | Mesp1 | ESC lines | [144] |
| Meteor | CVD | Eomes, T, Gsc, Gata4 and Isl1 | Mice models and ESC lines | [145] |
| Lnc-leptin | Obesity | Lep | Mice models | [149] |
| OLMALINC | Hepatic steatosis and NAFLD | SCD | Patient samples and cell lines (HepG2 and Fa2N4) | [153] |
| LncASIR | T2D | PI3K, Fabp4, Glut4 and Srebp1c | Mice models | [164] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhang, C.; Wang, Y.; Liu, X.; Zhang, Z. Enhancer RNA (eRNA) in Human Diseases. Int. J. Mol. Sci. 2022, 23, 11582. https://doi.org/10.3390/ijms231911582
Wang Y, Zhang C, Wang Y, Liu X, Zhang Z. Enhancer RNA (eRNA) in Human Diseases. International Journal of Molecular Sciences. 2022; 23(19):11582. https://doi.org/10.3390/ijms231911582
Chicago/Turabian StyleWang, Yunzhe, Chenyang Zhang, Yuxiang Wang, Xiuping Liu, and Zhao Zhang. 2022. "Enhancer RNA (eRNA) in Human Diseases" International Journal of Molecular Sciences 23, no. 19: 11582. https://doi.org/10.3390/ijms231911582
APA StyleWang, Y., Zhang, C., Wang, Y., Liu, X., & Zhang, Z. (2022). Enhancer RNA (eRNA) in Human Diseases. International Journal of Molecular Sciences, 23(19), 11582. https://doi.org/10.3390/ijms231911582




