Comprehensive Transcriptome Profiling Uncovers Molecular Mechanisms and Potential Candidate Genes Associated with Heat Stress Response in Chickpea
Abstract
1. Introduction
2. Results
2.1. Generation, Mapping, and Assessment of RNA-Seq Reads
2.2. Distribution of DEGs across Six Chickpea Genotypes Contrasting for Heat Stress Response
2.3. Genes Consistently Displaying Significant Differential Expression across all Six Chickpea Genotypes
2.4. Genes That Show Significant Regulation in Tolerant Genotypes but Are Not Substantially Regulated in the Sensitive Genotypes
2.5. Functional Categorization of DEGs
2.6. Differentially Expressed Transcription Factor (TF) Families
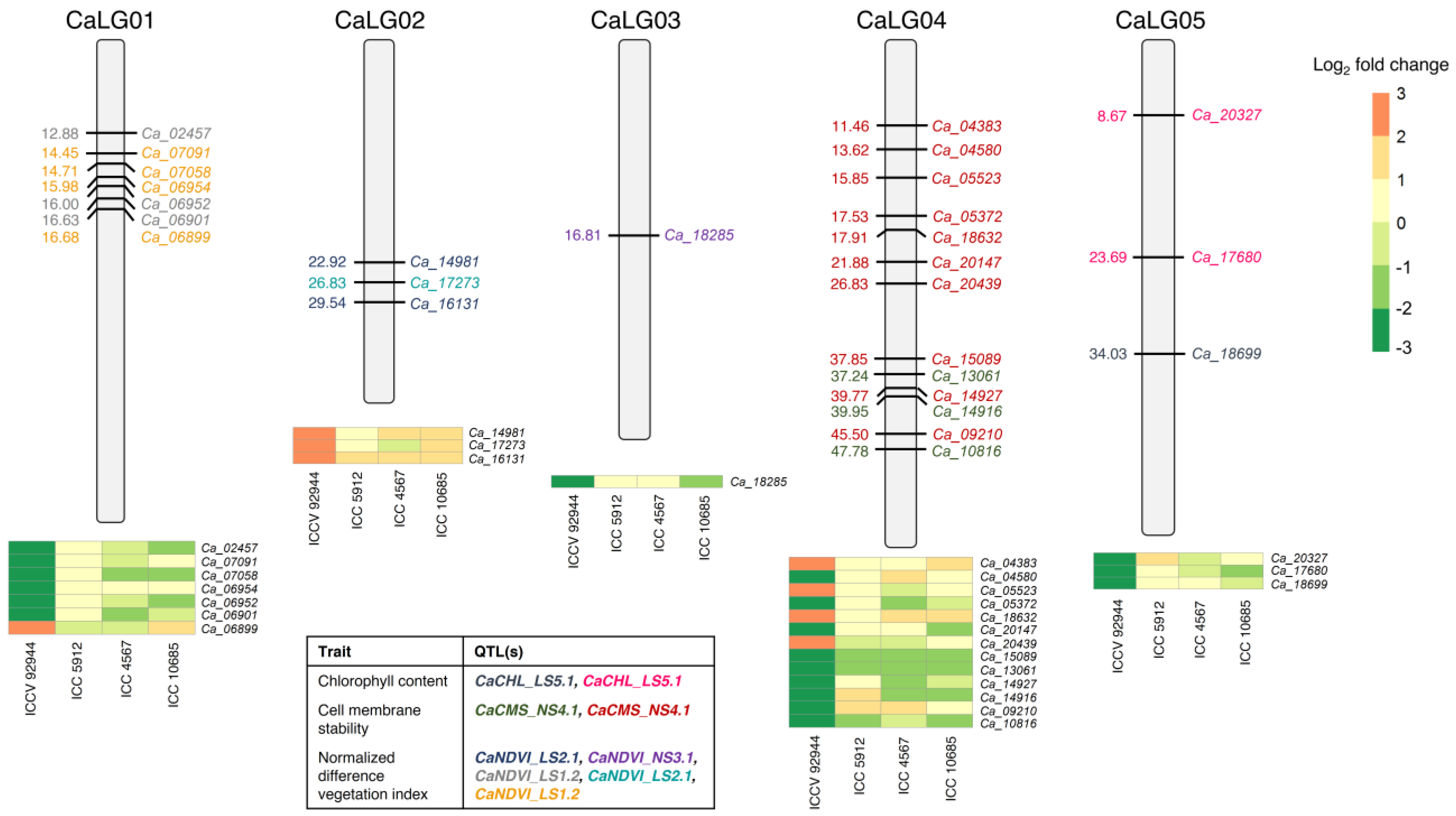
2.7. Candidate Genes Underlying QTLs Governing Heat Tolerance in Chickpea
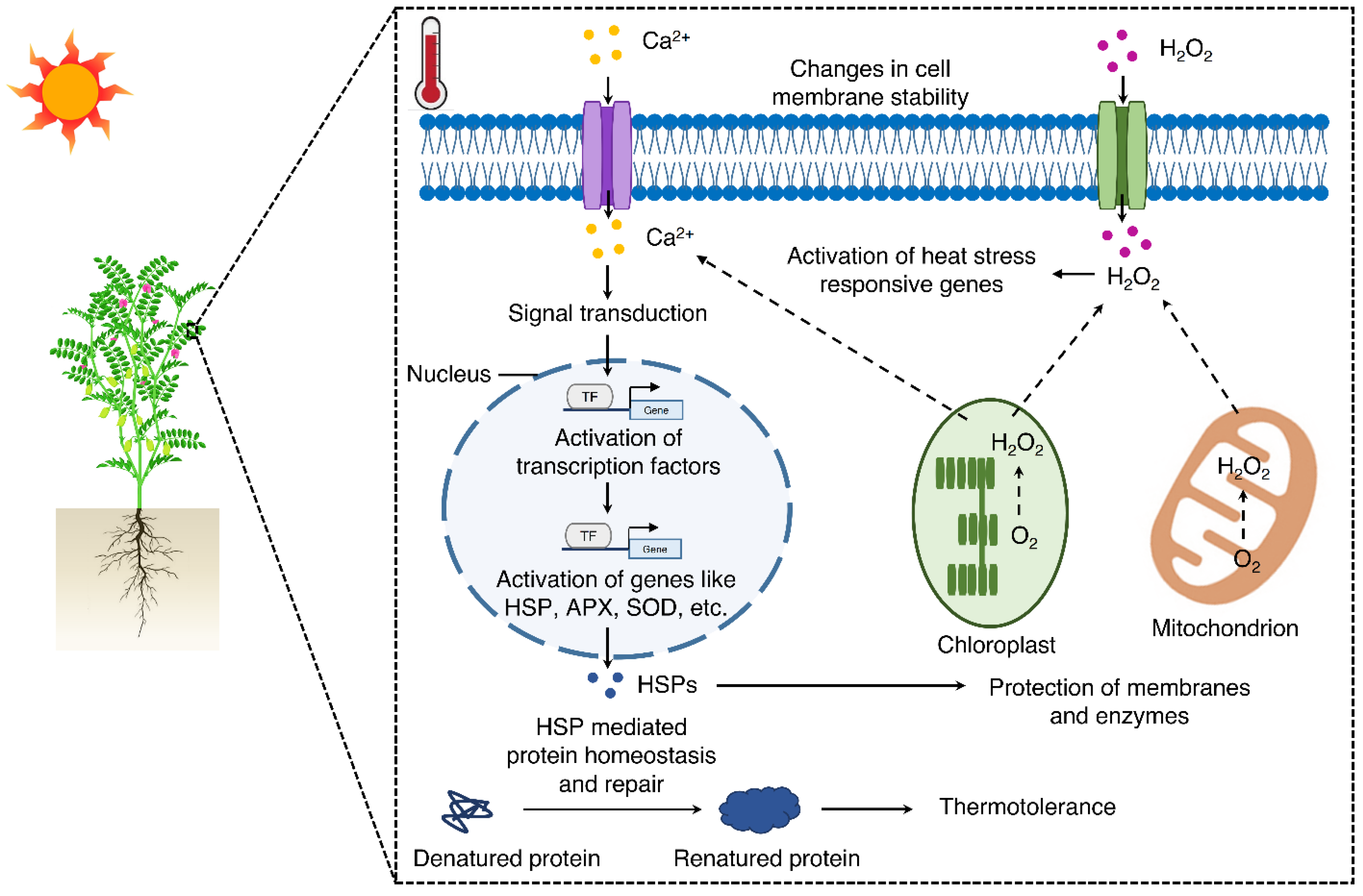
3. Discussion
4. Materials and Methods
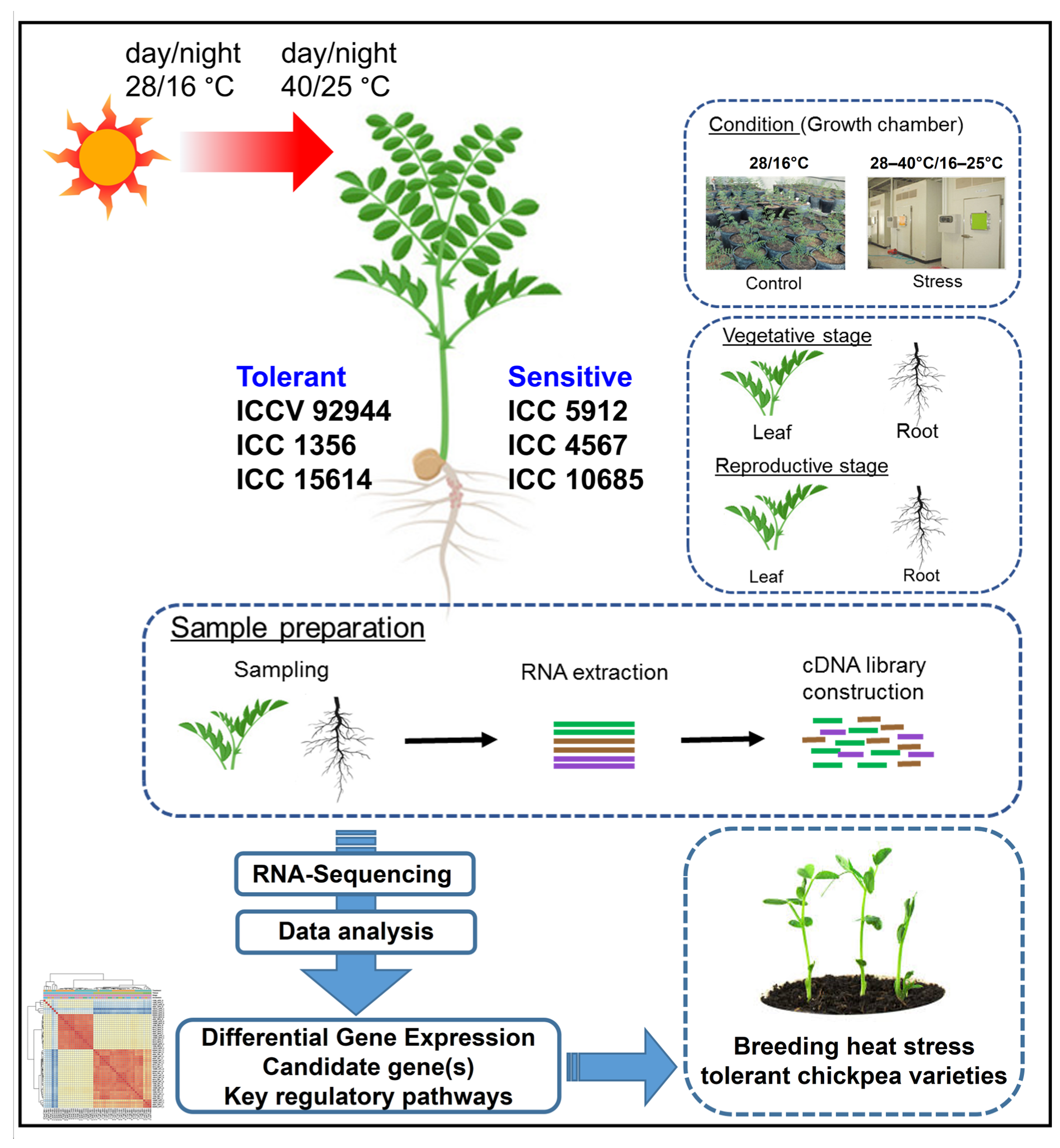
4.1. Plant Material, Growth Conditions, and Heat Stress Treatment Imposition
4.2. RNA Extraction, Illumina Sequencing, and Quality Check of the Sequenced Reads
4.3. Alignment of RNA-Seq Reads to the Chickpea Reference Genome
4.4. Identification of DEGs, GO Enrichment and Pathway Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT 2020. Available online: http://www.fao.org/faostat/en/#data (accessed on 24 March 2022).
- Li, H.; Rodda, M.; Gnanasambandam, A.; Aftab, M.; Redden, R.; Hobson, K.; Rosewarne, G.; Materne, M.; Kaur, S.; Slater, A.T. Breeding for biotic stress resistance in chickpea: Progress and prospects. Euphytica 2015, 204, 257–288. [Google Scholar] [CrossRef]
- Barmukh, R.; Roorkiwal, M.; Garg, V.; Khan, A.; German, L.; Jaganathan, D.; Chitikineni, A.; Kholova, J.; Kudapa, H.; Sivasakthi, K.; et al. Genetic variation in CaTIFY4b contributes to drought adaptation in chickpea. Plant Biotechnol. J. 2022, 20, 1701–1715. [Google Scholar] [CrossRef] [PubMed]
- Barmukh, R.; Roorkiwal, M.; Dixit, G.P.; Bajaj, P.; Kholova, J.; Smith, M.R.; Chitikineni, A.; Bharadwaj, C.; Sheshshayee, M.S.; Rathore, A.; et al. Characterization of ‘QTL-hotspot’ introgression lines reveals physiological mechanisms and candidate genes associated with drought adaptation in chickpea. J. Exp. Bot. 2022, 73, 7255–7272. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Thudi, M.; Nayak, S.N.; Gaur, P.M.; Kashiwagi, J.; Krishnamurthy, L.; Jaganathan, D.; Koppolu, J.; Bohra, A.; Tripathi, S.; et al. Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2014, 127, 445–462. [Google Scholar] [CrossRef]
- Devasirvatham, V.; Gaur, P.M.; Mallikarjuna, N.; Tokachichu, R.N.; Trethowan, R.M.; Tan, D.K.Y. Effect of high temperature on reproductive development of chickpea genotypes under controlled environments. Funct. Plant Biol. 2012, 39, 1009–1018. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Palakurthi, R.; Jha, R.; Valluri, V.; Bajaj, P.; Chitikineni, A.; Singh, N.P.; Varshney, R.K.; Thudi, M. Major QTLs and potential candidate genes for heat stress tolerance identified in chickpea (Cicer arietinum L.). Front. Plant Sci. 2021, 12, 655103. [Google Scholar] [CrossRef]
- Wang, J.; Gan, Y.T.; Clarke, F.; McDonald, C.L. Response of chickpea yield to high temperature stress during reproductive development. Crop Sci. 2006, 46, 2171–2178. [Google Scholar] [CrossRef]
- De Storme, N.; Geelen, D. The impact of environmental stress on male reproductive development in plants: Biological processes and molecular mechanisms. Plant Cell Environ. 2013, 37, 1–18. [Google Scholar] [CrossRef]
- Smith, A.R.; Zhao, D. Sterility caused by floral organ degeneration and abiotic stresses in Arabidopsis and cereal grains. Front Plant Sci. 2016, 7, 1503. [Google Scholar] [CrossRef]
- Krishnamurthy, L.; Gaur, P.M.; Basu, P.S.; Chaturvedi, S.K.; Tripathi, S.; Vadez, V.; Rathore, A.; Varshney, R.K. Large genetic variation for heat tolerance in the reference collection of chickpea (Cicer arietinum L.) germplasm. Plant Genet. Resour. 2011, 9, 59–69. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Dronavalli, N.; Gowda, C.L.L.; Singh, S. Identification and evaluation of chickpea germplasm for tolerance to heat stress. Crop Sci. 2011, 51, 2079–2094. [Google Scholar] [CrossRef]
- Paul, P.J.; Samineni, S.; Sajja, S.B.; Rathore, A.; Das, R.R.; Chaturvedi, S.K.; Lavanya, G.R.; Varshney, R.K.; Gaur, P.M. Capturing genetic variability and selection of traits for heat tolerance in a chickpea recombinant inbred line (RIL) population under field conditions. Euphytica 2018, 214, 27. [Google Scholar] [CrossRef]
- Varshney, R.K.; Song, C.; Saxena, R.K.; Azam, S.; Yu, S.; Sharpe, A.; Cannon, S.; Baek, J.; Rosen, B.D.; Taran, B.; et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 2013, 31, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Misra, G.; Patel, R.K.; Priya, P.; Jhanwar, S.; Khan, A.W.; Shah, N.; Singh, V.K.; Garg, R.; Ganga, J.; et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 2013, 74, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Mir, R.R.; Bhatia, S.; Thudi, M.; Hu, Y.; Azam, S.; Zhang, Y.; Jaganathan, A.; You, F.M.; Gao, J.; et al. Integrated physical, genetic and genome map of chickpea (Cicer arietinum L.). Funct. Integr. Genomics 2014, 14, 59–73. [Google Scholar] [CrossRef]
- Varshney, R.K.; Roorkiwal, M.; Sun, S.; Bajaj, P.; Chitikineni, A.; Thudi, M.; Singh, N.P.; Du, X.; Upadhyaya, H.D.; Khan, A.W.; et al. A chickpea genetic variation map based on the sequencing of 3,366 genomes. Nature 2021, 599, 622–627. [Google Scholar] [CrossRef]
- Varshney, R.K.; Thudi, M.; Roorkiwal, M.; He, W.; Upadhyaya, H.D.; Yang, W.; Bajaj, P.; Cubry, P.; Rathore, A.; Jian, J.; et al. Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 2019, 51, 857–864. [Google Scholar] [CrossRef]
- Palit, P.; Ghosh, R.; Tolani, P.; Tarafdar, A.; Chitikineni, A.; Bajaj, P.; Sharma, M.; Kudapa, H.; Varshney, R.K. Molecular and physiological alterations in chickpea under elevated CO2 concentrations. Plant Cell Physiol. 2020, 61, 1449–1463. [Google Scholar] [CrossRef]
- Kudapa, H.; Azam, S.; Sharpe, A.G.; Taran, B.; Li, R.; Deonovic, B.; Cameron, C.; Farmer, A.D.; Cannon, S.B.; Varshney, R.K. Comprehensive transcriptome assembly of chickpea (Cicer arietinum L.) using sanger and next generation sequencing platforms: Development and applications. PLoS ONE 2014, 9, e86039. [Google Scholar] [CrossRef]
- Hiremath, P.J.; Kumar, A.; Penmetsa, R.V.; Farmer, A.; Schlueter, J.A.; Chamarthi, S.K.; Whaley, A.M.; Carrasquilla-Garcia, N.; Gaur, P.M.; Upadhyaya, H.D.; et al. Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 2012, 10, 716–732. [Google Scholar] [CrossRef]
- Varshney, R.K.; Hiremath, P.J.; Lekha, P.; Kashiwagi, J.; Balaji, J.; Deokar, A.A.; Vadez, V.; Xiao, Y.; Srinivasan, R.; Gaur, P.M.; et al. A comprehensive resource of drought- and salinity-responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.). BMC Genom. 2009, 10, 523. [Google Scholar] [CrossRef]
- Barmukh, R.; Soren, K.R.; Madugula, P.; Gangwar, P.; Shanmugavadivel, P.S.; Bharadwaj, C.; Konda, A.K.; Chaturvedi, S.K.; Bhandari, A.; Rajain, K.; et al. Construction of a high-density genetic map and QTL analysis for yield, yield components and agronomic traits in chickpea (Cicer arietinum L.). PLoS ONE 2021, 16, e0251669. [Google Scholar] [CrossRef]
- Thudi, M.; Bohra, A.; Nayak, S.N.; Varghese, N.; Shah, T.M.; Penmetsa, R.V.; Thirunavukkarasu, N.; Gudapati, S.; Gaur, P.M.; Kulwal, P.; et al. Novel SSR markers from BAC-end sequences, DArT arrays and a comprehensive genetic map with 1,291 marker loci for chickpea (Cicer arietinum L.). PLoS ONE 2011, 6, e27275. [Google Scholar] [CrossRef] [PubMed]
- Kale, S.M.; Jaganathan, D.; Ruperao, P.; Chen, C.; Punna, R.; Kudapa, H.; Thudi, M.; Roorkiwal, M.; Katta, M.A.; Doddamani, D.; et al. Prioritization of candidate genes in "QTL-hotspot" region for drought tolerance in chickpea (Cicer arietinum L.). Sci. Rep. 2015, 19, 15296. [Google Scholar] [CrossRef] [PubMed]
- Kudapa, H.; Garg, V.; Chitikineni, A.; Varshney, R.K. The RNA-Seq based high resolution gene expression atlas of chickpea (Cicer arietinum L.) reveals dynamic spatio-temporal changes associated with growth and development. Plant Cell Environ. 2018, 41, 2209–2225. [Google Scholar] [CrossRef]
- Sabbavarapu, M.M.; Sharma, M.; Chamarthi, S.K.; Swapna, N.; Rathore, A.; Thudi, M.; Gaur, P.M.; Pande, S.; Singh, S.; Kaur, L.; et al. Molecular mapping of QTLs for resistance to Fusarium wilt (race 1) and Ascochyta blight in chickpea (Cicer arietinum L.). Euphytica 2013, 193, 121–133. [Google Scholar] [CrossRef]
- Garg, V.; Khan, A.W.; Kudapa, H.; Kale, S.M.; Chitikineni, A.; Qiwei, S.; Sharma, M.; Li, C.; Zhang, B.; Xin, L.; et al. Integrated transcriptome, small RNA and degradome sequencing approaches provide insights into Ascochyta blight resistance in chickpea. Plant Biotechnol. J. 2019, 17, 914–931. [Google Scholar] [CrossRef] [PubMed]
- Barmukh, R.K.; Roorkiwal, M.; Jaba, J.; Chitikineni, A.; Mishra, S.; Sagurthi, S.; Munghate, R.; Sharma, H.C.; Varshney, R.K. Development of a dense genetic map and QTL analysis for pod borer Helicoverpa armigera (Hübner) resistance component traits in chickpea (Cicer arietinum L.). Plant Genome. 2021, 14, e20071. [Google Scholar] [CrossRef] [PubMed]
- Kaashyap, M.; Ford, R.; Kudapa, H.; Jain, M.; Edwards, D.; Varshney, R.; Mantri, N. Differential regulation of genes involved in root morphogenesis and cell wall modification is associated with salinity tolerance in chickpea. Sci. Rep. 2018, 8, 4855. [Google Scholar] [CrossRef]
- Soren, K.R.; Madugula, P.M.; Kumar, N.; Barmukh, R.; Sengar, M.S.; Bharadwaj, C.; Sharma, P.C.; Singh, S.; Bhandari, A.; Singh, J.; et al. Genetic dissection and identification of candidate genes for salinity tolerance using Axiom®CicerSNP Array in chickpea. Intl. J. Mol. Sci. 2020, 21, 5058. [Google Scholar] [CrossRef]
- Periera, A.M.; Periera, L.G.; Coimbra, S. Arabinogalactan proteins:rising attention from plant biologists. Plant Reprod. 2015, 28, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xu, X.; Shi, Y.; Xu, J.; Huang, B. Transgenic tobacco plants overexpressing a grass PpEXP1 gene exhibit enhanced tolerance to heat stress. PLoS ONE 2014, 9, e100792. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.C.; Molnar, P.; Schwab, W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 2009, 60, 3011–3022. [Google Scholar] [CrossRef]
- Bourgine, B.; Guihur, A. Heat shock signaling in land plants: From plasma membrane sensing to the transcription of small heat shock proteins. Front Plant Sci. 2021, 12, 710801. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Cai, G.; Xu, N.; Zhang, L.; Sun, X.; Guan, J.; Meng, Q. Novel DnaJ protein facilitates thermotolerance of transgenic tomatoes. Int. J. Mol. Sci. 2019, 20, 367. [Google Scholar] [CrossRef]
- Li, Y.J.; Yu, Y.; Liu, X.; Zhang, X.S.; Su, Y.H. The Arabidopsis MATERNAL EFFECT EMBRYO ARREST45 protein modulates maternal auxin biosynthesis and controls seed size by inducing AINTEGUMENTA. Plant Cell 2021, 33, 1907–1926. [Google Scholar] [CrossRef]
- Shao, H.; Dixon, R.A.; Wang, X. Crystal structure of vestitone reductase from Alfalfa (Medicago sativa L.). J. Mol. Biol. 2017, 369, 265–276. [Google Scholar] [CrossRef]
- Mody, T.; Bonnot, T.; Nagel, D.H. Interaction between the circadian clock and regulators of heat stress responses in plants. Genes 2020, 11, 156. [Google Scholar] [CrossRef]
- Boron, A.K.; Orden, J.V.; Markakis, M.N.; Mouille, G.; Adriaensen, D.; Verbelen, J.P.; Hofte, H.; Vissenberg, K. Proline-rich protein-like PRPL1 controls elongation of root hairs in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 5485–5495. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.; Espley, R.V.; Allan, A.C. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Hazratkulova, S.; Sharma, R.; Alikulov, S.; Islomov, S.; Yuldashev, T.; Ziyaev, Z.; Khalikulov, Z.; Ziyadullaev, Z.; Turok, J. Analysis of genotypic variation for normalized difference vegetation index and its relationship with grain yield in winter wheat under terminal heat stress. Plant Breed. 2012, 131, 716–721. [Google Scholar] [CrossRef]
- Casaretto, J.A.; El-kereamy, A.; Zeng, B.; Stiegelmeyer, S.M.; Chen, X.; Bi, Y.-M.; Rothstein, S.J. Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genomics 2016, 17, 312. [Google Scholar] [CrossRef] [PubMed]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Lal, M.K.; Tiwari, R.K.; Gahlaut, V.; Mangal, V.; Kumar, A.; Singh, M.P.; Paul, V.; Kumar, S.; Singh, B.; Zinta, G. Physiological and molecular insights on wheat responses to heat stress. Plant Cell Rep. 2021, 41, 501–518. [Google Scholar] [CrossRef]
- Tiwari, Y.K.; Yadav, S.K. High temperature stress tolerance in maize (Zea mays L.): Physiological and molecular mechanisms. J. Plant Biol. 2019, 62, 93–102. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing future crops: Genomics-assisted breeding comes of age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bohra, A.; Roorkiwal, M.; Barmukh, R.; Cowling, W.A.; Chitikineni, A.; Lam, H.M.; Hickey, L.T.; Croser, J.S.; Bayer, P.E.; et al. Fast-forward breeding for a food-secure world. Trends Genet. 2021, 37, 1124–1136. [Google Scholar] [CrossRef]
- González-Schain, N.; Dreni, L.; Lawas, L.M.F.; Galbiati, M.; Colombo, L.; Heuer, S.; Jagadish, K.S.V.; Kater, M.M. Genome-wide transcriptome analysis during anthesis reveals new insights into the molecular basis of heat stress responses in tolerant and sensitive rice varieties. Plant Cell Physiol. 2016, 57, 57–68. [Google Scholar] [CrossRef]
- Rangan, P.; Furtado, A.; Henry, R. Transcriptome profiling of wheat genotypes under heat stress during grain-filling. J. Cereal Sci. 2020, 91, 102895. [Google Scholar] [CrossRef]
- Sun, M.; Lin, C.; Zhang, A.; Wang, X.; Yan, H.; Khan, I.; Wu, B.; Feng, G.; Nie, G.; Zhang, X.; et al. Transcriptome sequencing revealed the molecular mechanism of response of pearl millet root to heat stress. J. Agron. Crop Sci. 2021, 207, 768–773. [Google Scholar] [CrossRef]
- Jin, J.; Yang, L.; Fan, D.; Liu, X.; Hao, Q. Comparative transcriptome analysis uncovers different heat stress responses in heat-resistant and heat-sensitive jujube cultivars. PLoS ONE 2020, 15, e0235763. [Google Scholar] [CrossRef]
- Li, T.; Xu, X.; Li, Y.; Wang, H.; Li, Z.; Li, Z. Comparative transcriptome analysis reveals differential transcription in heat-susceptible and heat-tolerant pepper (Capsicum annum L.) cultivars under heat stress. J. Plant Biol. 2015, 58, 411–424. [Google Scholar] [CrossRef]
- Dong, Q.; Wallrad, L.; Almutairi, B.O.; Kudla, J. Ca2+ signaling in plant responses to abiotic stresses. J. Integr. Plant Biol. 2022, 64, 287–300. [Google Scholar]
- Demidchik, V.; Shabala, S.; Isayenkov, S.; Cuin, T.A.; Pottosin, I. Calcium transport across plant membranes: Mechanisms and functions. New Phytol. 2018, 220, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Saidi, Y.; Finka, A.; Muriset, M.; Bromberg, Z.; Weiss, Y.G.; Maathuis, F.J.M.; Goloubinoff, P. The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 2009, 21, 2829–2843. [Google Scholar] [CrossRef]
- Gilroy, S.; Suzuki, N.; Miller, G.; Choi, W.-G.; Toyota, M.; Devireddy, A.R.; Mittler, R. A tidal wave of signals: Calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 2014, 19, 623–630. [Google Scholar] [CrossRef]
- Sinha, A.K.; Jaggi, M.; Raghuram, B.; Tuteja, N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 2011, 6, 196–203. [Google Scholar] [CrossRef]
- Ding, H.; He, J.; Wu, Y.; Wu, X.; Ge, C.; Wang, Y.; Zhong, S.; Peiter, E.; Liang, J.; Xu, W. The tomato mitogen-activated protein kinase SlMPK1 is as a negative regulator of the high-temperature stress response. Plant Physiol. 2018, 177, 633–651. [Google Scholar] [CrossRef]
- Liao, C.; Zheng, Y.; Guo, Y. MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 2017, 216, 163–177. [Google Scholar] [CrossRef]
- Chauhan, H.; Khurana, N.; Nijhavan, A.; Khurana, J.P.; Khurana, P. The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 2012, 35, 1912–1931. [Google Scholar] [CrossRef]
- Timperio, A.M.; Egidi, M.G.; Zolla, L. Proteomics applied on plant abiotic stresses: Role of heat shock proteins (HSP). J Proteomics 2008, 71, 391–411. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Qu, A.L.; Ding, Y.F.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kaushal, N.; Nayyar, H.; Gaur, P.M. Abscisic acid induces heat tolerance in chickpea (Cicer arietinum L.) seedlings by facilitated accumulation of osmoprotectants. Acta Physiol. Plant. 2012, 34, 1651–1658. [Google Scholar] [CrossRef]
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Bürger, M.; Wang, Y.; Chory, J. Two interacting ethylene response factors regulate heat stress response. Plant Cell 2021, 33, 338–357. [Google Scholar] [CrossRef] [PubMed]
- Katta, M.A.; Khan, A.W.; Doddamani, D.; Thudi, M.; Varshney, R.K. NGS-QCbox and raspberry for parallel, automated and rapid quality control analysis of large-scale next generation sequencing (Illumina) data. PLoS ONE 2015, 10, e0139868. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]


| S. No. | Sample ID | Accession | Stage | Tissue | Treat-ment | Total Reads | Filtered Reads | Filtered Reads (%) | Mapped Reads | Mapped Reads (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 92944_BFL_C | ICCV 92944 | Vegetative | Leaf | Control | 10,535,724 | 10,396,767 | 98.68 | 10,200,059 | 98.11 |
| 2 | 92944_BFL_S | Vegetative | Leaf | Stress | 9,748,624 | 9,586,491 | 98.34 | 9,420,776 | 98.27 | |
| 3 | 92944_BFR_C | Vegetative | Root | Control | 9,219,200 | 9,109,880 | 98.81 | 8,903,056 | 97.73 | |
| 4 | 92944_BFR_S | Vegetative | Root | Stress | 7,212,904 | 7,145,108 | 99.06 | 6,941,474 | 97.15 | |
| 5 | 92944_AFL_C | Reproductive | Leaf | Control | 12,186,436 | 12,019,055 | 98.63 | 11,785,230 | 98.05 | |
| 6 | 92944_AFL_S | Reproductive | Leaf | Stress | 10,763,618 | 10,651,458 | 98.96 | 10,462,620 | 98.23 | |
| 7 | 92944_AFR_C | Reproductive | Root | Control | 8250,522 | 8,127,931 | 98.51 | 7,808,259 | 96.07 | |
| 8 | 92944_AFR_S | Reproductive | Root | Stress | 5,524,034 | 5,385,805 | 97.50 | 5,014,912 | 93.11 | |
| 9 | 1356_BFL_C | ICC 1356 | Vegetative | Leaf | Control | 13,092,636 | 12,902,779 | 98.55 | 12,623,893 | 97.84 |
| 10 | 1356_BFL_S | Vegetative | Leaf | Stress | 7,848,062 | 7,762,543 | 98.91 | 7,631,234 | 98.31 | |
| 11 | 1356_BFR_C | Vegetative | Root | Control | 8,840,168 | 8,731,055 | 98.77 | 8,530,613 | 97.70 | |
| 12 | 1356_BFR_S | Vegetative | Root | Stress | 7,331,290 | 7,249,290 | 98.88 | 7,071,163 | 97.54 | |
| 13 | 1356_AFL_C | Reproductive | Leaf | Control | 13,595,606 | 13,428,551 | 98.77 | 13,186,265 | 98.20 | |
| 14 | 1356_AFL_S | Reproductive | Leaf | Stress | 14,108,618 | 13,902,500 | 98.54 | 13,657,584 | 98.24 | |
| 15 | 1356_AFR_C | Reproductive | Root | Control | 9,227,816 | 9,070,316 | 98.29 | 8,845,298 | 97.52 | |
| 16 | 1356_AFR_S | Reproductive | Root | Stress | 11,544,052 | 11,227,552 | 97.26 | 11,026,008 | 98.20 | |
| 17 | 15614_BFL_C | ICC 15614 | Vegetative | Leaf | Control | 10,665,666 | 10,509,803 | 98.54 | 10,299,447 | 98.00 |
| 18 | 15614_BFL_S | Vegetative | Leaf | Stress | 9,627,154 | 9,487,221 | 98.55 | 9,310,571 | 98.14 | |
| 19 | 15614_BFR_C | Vegetative | Root | Control | 9,227,608 | 8,942,970 | 96.92 | 8,672,250 | 96.97 | |
| 20 | 15614_BFR_S | Vegetative | Root | Stress | 9,642,178 | 9,537,244 | 98.91 | 9,229,981 | 96.78 | |
| 21 | 15614_AFL_C | Reproductive | Leaf | Control | 8,068,362 | 7,934,679 | 98.34 | 7,776,680 | 98.01 | |
| 22 | 15614_AFL_S | Reproductive | Leaf | Stress | 7,523,960 | 7,432,553 | 98.79 | 7,246,601 | 97.50 | |
| 23 | 15614_AFR_C | Reproductive | Root | Control | 4,552,154 | 4,432,821 | 97.38 | 4,332,522 | 97.74 | |
| 24 | 15614_AFR_S | Reproductive | Root | Stress | 8,145,728 | 8,044,853 | 98.76 | 7,394,860 | 91.92 | |
| 25 | 5912_BFL_C | ICC 5912 | Vegetative | Leaf | Control | 7,969,184 | 7,869,032 | 98.74 | 7,618,997 | 96.82 |
| 26 | 5912_BFL_S | Vegetative | Leaf | Stress | 7,941,190 | 7,856,097 | 98.93 | 7,716,168 | 98.22 | |
| 27 | 5912_BFR_C | Vegetative | Root | Control | 10,438,074 | 10,085,541 | 96.62 | 9,785,839 | 97.03 | |
| 28 | 5912_BFR_S | Vegetative | Root | Stress | 8,414,738 | 8,336,666 | 99.07 | 8,141,952 | 97.66 | |
| 29 | 5912_AFL_C | Reproductive | Leaf | Control | 10,500,550 | 10,363,949 | 98.70 | 10,182,627 | 98.25 | |
| 30 | 5912_AFL_S | Reproductive | Leaf | Stress | 5,180,758 | 5,117,767 | 98.78 | 4,961,472 | 96.95 | |
| 31 | 5912_AFR_C | Reproductive | Root | Control | 6,801,290 | 6,728,029 | 98.92 | 6,557,444 | 97.46 | |
| 32 | 5912_AFR_S | Reproductive | Root | Stress | 11,591,808 | 11,410,935 | 98.44 | 11,130,213 | 97.54 | |
| 33 | 4567_BFL_C | ICC4567 | Vegetative | Leaf | Control | 12,433,794 | 12,216,153 | 98.25 | 11,994,301 | 98.18 |
| 34 | 4567_BFL_S | Vegetative | Leaf | Stress | 10,425,446 | 10,288,128 | 98.68 | 10,068,543 | 97.87 | |
| 35 | 4567_BFR_C | Vegetative | Root | Control | 9,393,346 | 9,167,255 | 97.59 | 8,885,865 | 96.93 | |
| 36 | 4567_BFR_S | Vegetative | Root | Stress | 5,773,594 | 5,725,855 | 99.17 | 5,541,685 | 96.78 | |
| 37 | 4567_AFL_C | Reproductive | Leaf | Control | 12,591,206 | 12,398,886 | 98.47 | 12,166,369 | 98.12 | |
| 38 | 4567_AFL_S | Reproductive | Leaf | Stress | 9,909,176 | 9,778,957 | 98.69 | 9,596,564 | 98.13 | |
| 39 | 4567_AFR_C | Reproductive | Root | Control | 13,244,230 | 13,065,481 | 98.65 | 12,701,915 | 97.22 | |
| 40 | 4567_AFR_S | Reproductive | Root | Stress | 7,681,168 | 7,497,257 | 97.61 | 7,175,938 | 95.71 | |
| 41 | 10685_BFL_C | ICC 10685 | Vegetative | Leaf | Control | 10,826,998 | 10,674,415 | 98.59 | 10,455,869 | 97.95 |
| 42 | 10685_BFL_S | Vegetative | Leaf | Stress | 11,806,122 | 11,612,804 | 98.36 | 11,332,255 | 97.58 | |
| 43 | 10685_BFR_C | Vegetative | Root | Control | 11,425,388 | 11,082,467 | 97.00 | 10,756,797 | 97.06 | |
| 44 | 10685_BFR_S | Vegetative | Root | Stress | 7,308,052 | 7,241,501 | 99.09 | 7,058,819 | 97.48 | |
| 45 | 10685_AFL_C | Reproductive | Leaf | Control | 12,762,970 | 12,546,086 | 98.30 | 12,296,546 | 98.01 | |
| 46 | 10685_AFL_S | Reproductive | Leaf | Stress | 10,978,522 | 10,853,672 | 98.86 | 10,661,986 | 98.23 | |
| 47 | 10685_AFR_C | Reproductive | Root | Control | 7,721,326 | 7,641,066 | 98.96 | 7,464,657 | 97.69 | |
| 48 | 10685_AFR_S | Reproductive | Root | Stress | 9,251,526 | 9,124,015 | 98.62 | 8,884,704 | 97.38 | |
| Total | 458,852,576 | 451,701,239 | 98.45 | 440,508,881 | 97.41 |
| Gene ID | XLOC ID | Log2 Fold Change (Treatment/Control) | Gene Description | |||||
|---|---|---|---|---|---|---|---|---|
| ICCV 92944 | ICC 1356 | ICC 15614 | ICC 5912 | ICC 4567 | ICC 10685 | |||
| Leaf sample—before flowering | ||||||||
| Ca_14776 | XLOC_001145 | −2.161 | −2.165 | −2.125 | −1.702 | −1.651 | 1.532 | pathogenesis-related protein 10 |
| Ca_07845 | XLOC_009218 | −2.956 | 2.458 | −2.134 | −0.806 | 1.402 | −0.163 | adenylate isopentenyltransferase |
| Leaf sample—after flowering | ||||||||
| Ca_00611 | XLOC_000503 | 3.140 | −3.321 | 2.981 | −0.057 | 0.576 | 1.887 | basic 7S globulin |
| Ca_00716, Ca_00717 | XLOC_007162 | −2.600 | 2.662 | 2.696 | 0.615 | 1.725 | −0.349 | IST1-like protein isoform X2 |
| Ca_05434 | XLOC_010012 | −2.650 | 2.103 | 4.128 | −0.786 | 1.774 | −0.593 | protein trichome birefringence-like 36 |
| Ca_10969 | XLOC_012559 | −2.618 | 2.066 | 2.539 | 1.526 | 1.894 | 0.050 | selenoprotein H |
| Ca_11344 | XLOC_014356 | −2.875 | 2.951 | 2.332 | 0.728 | 1.912 | NA | nuclear transcription factor Y subunit C-4 |
| Ca_25834 | XLOC_017871 | 3.984 | 2.594 | 2.935 | 0.960 | 0.517 | 1.430 | glycine-rich cell wall structural 1-like |
| Ca_05233 | XLOC_019232 | −2.424 | 2.341 | 2.077 | 1.273 | 0.213 | −0.392 | beta-xylosidase/alpha-L-arabinofuranosidase 2 |
| Ca_06616 | XLOC_022893 | −2.089 | 2.078 | 2.323 | 0.228 | 1.379 | −0.698 | DUF936 family protein |
| Ca_01970 | XLOC_025377 | −2.259 | 2.122 | 2.621 | 1.089 | 1.683 | 0.103 | chalcone-flavanone isomerase family |
| Ca_27831 | XLOC_030374 | −2.519 | 2.197 | 2.352 | 0.991 | 1.566 | 1.025 | CTP synthase-like isoform X1 |
| Ca_00464 | XLOC_000429 | 2.041 | −3.878 | −3.219 | 0.674 | NA | −0.680 | serine carboxypeptidase-like 11 |
| Ca_06899 | XLOC_002705 | 2.490 | −2.172 | −2.465 | −0.001 | −0.193 | 1.441 | NAC family transcription factor 4 |
| Ca_09600 | XLOC_018907 | 2.256 | −2.235 | −2.951 | −0.953 | −0.489 | −0.075 | transcription factor TGA4-like isoform X1 |
| Ca_02987 | XLOC_021039 | 2.420 | −9.719 | −4.680 | 0.607 | −0.407 | 1.176 | ABA-responsive protein ABR18-like |
| Root sample—before flowering | ||||||||
| Ca_14147 | XLOC_000968 | 4.027 | 2.467 | 2.073 | 1.012 | NA | 1.305 | cysteine-rich repeat secretory protein 38-like |
| Ca_03546 | XLOC_009482 | 3.704 | 3.129 | 2.302 | 1.785 | 1.538 | 0.734 | peroxidase 5-like |
| Ca_06976, Ca_06977 | XLOC_001062 | −2.850 | −2.721 | −2.895 | 0.538 | 0.289 | −0.728 | UDP-glycosyltransferase 79B30 |
| Ca_06902 | XLOC_002703 | −3.674 | 2.910 | −2.009 | 0.407 | −1.651 | 1.136 | PREDICTED: uncharacterized protein LOC101506036 |
| Ca_14738 | XLOC_002759 | −2.465 | −2.426 | −2.305 | −1.309 | −1.193 | NA | E3 ubiquitin- ligase LIN-1 |
| Ca_19160 | XLOC_003847 | 2.254 | −2.392 | −3.952 | −1.107 | −0.690 | −1.771 | arabinogalactan peptide 21-like |
| Ca_18792 | XLOC_006264 | −2.067 | −2.650 | −3.088 | −0.820 | −1.456 | −0.846 | dirigent protein 9-like |
| Ca_16612 | XLOC_010291 | −2.866 | −2.265 | −3.618 | −1.585 | −1.241 | −0.737 | probable pectate lyase 1 |
| Ca_23747 | XLOC_013214 | −2.464 | −2.961 | −2.093 | −1.327 | NA | 0.074 | probable pectinesterase/pectinesterase inhibitor 25 |
| Ca_22983 | XLOC_026437 | −2.194 | 3.352 | −2.720 | 1.481 | −1.969 | 1.948 | hypothetical protein glysoja_025159 |
| Root sample—after flowering | ||||||||
| Ca_11978 | XLOC_007090 | −2.016 | −2.800 | 2.140 | −0.071 | −0.706 | −0.682 | ras-related protein RABE1c-like |
| Ca_18112 | XLOC_003876 | 2.567 | 2.437 | −2.526 | 1.302 | −1.692 | 1.597 | mitochondrial uncoupling protein 5 |
| Ca_09783 | XLOC_005932 | 2.229 | −2.034 | −2.205 | −0.125 | NA | −0.189 | OBERON-like protein |
| Ca_05504 | XLOC_009968 | −2.783 | −2.225 | −2.072 | −0.018 | −0.801 | −0.283 | glutamine--tRNA ligase |
| Ca_24258 | XLOC_022284 | −3.033 | 2.178 | −3.424 | 0.380 | NA | 0.127 | NA |
| NA, not available | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudapa, H.; Barmukh, R.; Garg, V.; Chitikineni, A.; Samineni, S.; Agarwal, G.; Varshney, R.K. Comprehensive Transcriptome Profiling Uncovers Molecular Mechanisms and Potential Candidate Genes Associated with Heat Stress Response in Chickpea. Int. J. Mol. Sci. 2023, 24, 1369. https://doi.org/10.3390/ijms24021369
Kudapa H, Barmukh R, Garg V, Chitikineni A, Samineni S, Agarwal G, Varshney RK. Comprehensive Transcriptome Profiling Uncovers Molecular Mechanisms and Potential Candidate Genes Associated with Heat Stress Response in Chickpea. International Journal of Molecular Sciences. 2023; 24(2):1369. https://doi.org/10.3390/ijms24021369
Chicago/Turabian StyleKudapa, Himabindu, Rutwik Barmukh, Vanika Garg, Annapurna Chitikineni, Srinivasan Samineni, Gaurav Agarwal, and Rajeev K. Varshney. 2023. "Comprehensive Transcriptome Profiling Uncovers Molecular Mechanisms and Potential Candidate Genes Associated with Heat Stress Response in Chickpea" International Journal of Molecular Sciences 24, no. 2: 1369. https://doi.org/10.3390/ijms24021369
APA StyleKudapa, H., Barmukh, R., Garg, V., Chitikineni, A., Samineni, S., Agarwal, G., & Varshney, R. K. (2023). Comprehensive Transcriptome Profiling Uncovers Molecular Mechanisms and Potential Candidate Genes Associated with Heat Stress Response in Chickpea. International Journal of Molecular Sciences, 24(2), 1369. https://doi.org/10.3390/ijms24021369







