mTOR Complex 1 Content and Regulation Is Adapted to Animal Longevity
Abstract
:1. Introduction
2. The Structure and Regulation of the mTORC1
3. mTORC1 and Longevity
4. mTORC1 and Ageing and the Development of Age-Related Diseases
5. mTORC1 and Nutritional Interventions in Ageing
5.1. Rapamycin Treatment
| Species | Sex | Intervention | mTORC1 | Tissue | Phenotype | Longevity | Ref. |
|---|---|---|---|---|---|---|---|
| Worm | - | Rapamycin A | Reduced * | Whole | n.d. | +21% | [55] |
| Fly | M/F | Rapamycin D | Reduced # | Whole, abdomen, thorax and head | n.d. | +18% | [50] |
| Fly | F | Rapamycin D | Reduced # | Intestine | n.d. | +8% | [51] |
| Fly | M/F | Rapamycin D | Reduced | n.d. | n.d. | +33% | [54] |
| Mouse | M/F | Rapamycin A2 | Reduced # | Heart, liver, kidney and intestine | n.d. | n.d. | [53] |
| Mouse | M | Rapamycin B3 | Reduced # | Liver | Delayed ageing | n.d. | [36] |
| Mouse | F | Rapamycin B4 | Reduced | n.d. | n.d. | +4% | [57] |
| Mouse | M | Rapamycin B4 | Reduced | n.d. | n.d. | +5% | [57] |
| Mouse | F | Rapamycin B4 | Reduced | n.d. | Delayed ageing | +9–14% | [58] |
| Mouse | M | Rapamycin B4 | Reduced | n.d. | Delayed ageing | +14% | [58] |
| Mouse | M | Rapamycin B4 | Reduced * | n.d. | n.d. | +60% | [37] |
| Mouse | F | Rapamycin D1 | Reduced | n.d. | Reduced cancer | +9% | [62] |
| Mouse | F | Rapamycin D2 | Reduced | n.d. | n.d. | +9–15% | [56] |
| Mouse | M | Rapamycin D2 | Reduced | n.d. | n.d. | +15–18% | [56] |
| Mouse | F | Rapamycin C2/D4 | Reduced # | Visceral fat | n.d. | +14% | [52] |
| Mouse | M | Rapamycin C2/D4 | Reduced # | Visceral fat | n.d. | +9% | [52] |
| Mouse | F | Rapamycin D4 | Reduced | n.d. | n.d. | +12% | [57] |
| Mouse | M | Rapamycin D4 | Reduced | n.d. | n.d. | +9% | [57] |
5.2. Calorie and Amino Acid Restriction
| Species | Sex | Intensity | mTORC1 | Tissue | Phenotype | Longevity | Ref. |
|---|---|---|---|---|---|---|---|
| Mouse | M/F | CR (50%) A1 | Reduced | - | n.d. | n.d. | [76] |
| Rat | F | CR (40%) B1 | Reduced * | Liver | n.d. | n.d. | [73] |
| Human | F | CR (−1000 kcal) B3 | Reduced | Amygdala | Improved function and metabolic health | n.d. | [77] |
| Rat | F | CR (40%) B3 | Reduced * | Skeletal muscle | Delayed ageing | n.d. | [38] |
| Humans | F/M | CR (30%) B4 | Reduced | - | Improved memory and metabolic health | n.d. | [79] |
| Mouse | M | CR (30%) C1 | Reduced * | Hippocampus (neurons) | n.d. | n.d. | [69] |
| Mouse | M | CR (30%) C1 | Reduced * | Hippocampus (neurons) | Improved learning and memory | n.d. | [70] |
| Mouse | M | CR (30%) C1 | Reduced * | Hippocampus (astrocytes) | n.d. | n.d. | [71] |
| Mouse | F | PR (100%) A1 | Reduced # | Liver | n.d. | n.d. | [74] |
| Mouse | F | PR (7–21%) D1 | Reduced # | Liver, heart skeletal muscle, adipose tissue | n.d. | n.d. | [80] |
| Mouse | F | MetR (80%) B1 | Reduced | Hippocampus, cortex | Improved learning and memory | n.d. | [78] |
| Mouse | M | MetR (80%) B2 | Reduced * | Liver | Improved metabolic health | n.d. | [72] |
| Mouse | M | MetR (80%) B4 | Reduced * | Kidney | Delayed ageing | n.d. | [75] |
| Mouse | M/F | MetR (80%) D1 | Reduced * | n.d. | n.d. | +25% | [75] |
| Mouse | M | BCAAR (70%) C1/C3/D1 | Reduced | Heart, liver | Delayed ageing | +12.3 | [39] |
| Mouse | F | BCAAR (70%) D1/D3 | Reduced | n.d. | Improved metabolic health | n.d. | [39] |
| Mouse | M | LeuR (80%) B2 | Reduced * | Liver | n.d. | n.d. | [72] |
| Mouse | M | LeuR (0%) B2 | Reduced * | Liver | n.d. | n.d. | [85] |
6. mTORC1 and Nutritional Interventions in Pathological Processes
| Species | Sex | Disease | Intervention | mTORC1 | Tissue | Phenotype | Ref. |
|---|---|---|---|---|---|---|---|
| Mouse | M | AD | Rapamycin B1 | Reduced | Cortex | Improved health | [45] |
| Mouse | M | AD | Rapamycin B2 | Reduced | Brain | Improved health | [45] |
| Mouse | M | AD | Rapamycin C1 | Reduced | Brain | Improved health | [45] |
| Mouse | M | AD | CR (40%) B1 | Reduced | Hippocampus, cortex miand dentate gyrus | Improved health | [87] |
| Mouse | MF | AD | CR (30%) B1 | Reduced | Hippocampus | Improved health | [88] |
| Mouse | F | AD | CR (30%) C1 | Reduced | Hippocampus, neocortex | Improved health | [90] |
| Mouse | F | AD | CR (30%) C1 | Reduced | Hippocampus, cortex, entorhinal cortex | Improved health | [89] |
| Mouse | F | Cancer | Rapamycin B1 | Reduced | Tumour | Reduced tumour size | [95] |
| Mouse | F | Cancer | Rapamycin B1 | Reduced | n.d. | Reduced cancer incidence | [100] |
| Mouse | F | Cancer | DR (50%) A1 | Reduced | Tumour | Reduced tumour size | [96] |
| Rat | F | Cancer | DR (40%) B1 | Reduced * | Tumour | Reduced cancer incidence | [73] |
| Mouse | F | Cancer | PR (7–21%) D1 | Reduced # | Tumour | Reduced tumour size | [80] |
| Rat | M | Diabetes | PR (25%) A2 | Reduced # | Kidney | Improved health | [101] |
| Mouse | M/F | Progeria | mTOR (Δ/+) | Reduced * | Fibroblasts | Improved health | [99] |
| Mouse | M/F | Progeria | BCAAR D2 | Reduced | n.d. | Improved health | [39] |
7. Clinical Trials Aimed at Delaying the Ageing Process
8. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, S.C.; Rabinovitch, P.S.; Kaeberlein, M. mTOR is a key modulator of ageing and age-related disease. Nature 2013, 493, 338–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valvezan, A.J.; Manning, B.D. Molecular logic of mTORC1 signalling as a metabolic rheostat. Nat. Metab. 2019, 1, 321–333. [Google Scholar] [CrossRef]
- Chellappa, K.; Brinkman, J.A.; Mukherjee, S.; Morrison, M.; Alotaibi, M.I.; Carbajal, K.A.; Alhadeff, A.L.; Perron, I.J.; Yao, R.; Purdy, C.S.; et al. Hypothalamic mTORC2 is essential for metabolic health and longevity. Aging Cell 2019, 18, e13014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barja, G. Towards a unified mechanistic theory of aging. Exp. Gerontol. 2019, 124, 110627. [Google Scholar] [CrossRef]
- Kapahi, P.; Chen, D.; Rogers, A.N.; Katewa, S.D.; Li, P.W.-L.; Thomas, E.L.; Kockel, L. With TOR, less is more: A key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010, 11, 453–465. [Google Scholar] [CrossRef] [Green Version]
- Khayati, K.; Antikainen, H.; Bonder, E.M.; Weber, G.F.; Kruger, W.D.; Jakubowski, H.; Dobrowolski, R. The amino acid metabolite homocysteine activates mTORC1 to inhibit autophagy and form abnormal proteins in human neurons and mice. FASEB J. 2017, 31, 598–609. [Google Scholar] [CrossRef] [Green Version]
- Papadopoli, D.; Boulay, K.; Kazak, L.; Pollak, M.; Mallette, F.; Topisirovic, I.; Hulea, L. mTOR as a central regulator of lifespan and aging. F1000Research 2019, 8, 998. [Google Scholar] [CrossRef]
- Weichhart, T. mTOR as regulator of lifespan, aging, and cellular senescence: A mini-review. Gerontology 2018, 64, 127–134. [Google Scholar] [CrossRef]
- Chao, L.H.; Avruch, J. Cryo-EM insight into the structure of mTOR complex 1 and its interactions with Rheb and substrates. F1000Research 2019, 8, 14. [Google Scholar] [CrossRef]
- Peterson, T.R.; Laplante, M.; Thoreen, C.C.; Sancak, Y.; Kang, S.A.; Kuehl, W.M.; Gray, N.S.; Sabatini, D.M. DEPTOR Is an mTOR Inhibitor Frequently Overexpressed in Multiple Myeloma Cells and Required for Their Survival. Cell 2009, 137, 873–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Chen, X.; Liu, M.; Xu, Y. The structure of mTOR complexes at a glance. Precis. Cancer Med. 2018, 1, 7. [Google Scholar] [CrossRef]
- Aylett, C.H.S.; Sauer, E.; Imseng, S.; Boehringer, D.; Hall, M.N.; Ban, N.; Maier, T. Architecture of human mTOR complex 1. Science 2016, 351, 48–52. [Google Scholar] [CrossRef]
- Mota-Martorell, N.; Jove, M.; Pradas, I.; Berdún, R.; Sanchez, I.; Naudi, A.; Gari, E.; Barja, G.; Pamplona, R. Gene expression and regulatory factors of the mechanistic target of rapamycin (mTOR) complex 1 predict mammalian longevity. Geroscience 2020, 42, 1157–1173. [Google Scholar] [CrossRef]
- Li, X.; Yan, X. Sensors for the mTORC1 pathway regulated by amino acids. J. Zhejiang Univ. B 2019, 20, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Melick, C.H.; Jewell, J.L. Regulation of mTORC1 by upstream stimuli. Genes 2020, 11, 989. [Google Scholar] [CrossRef] [PubMed]
- Sekulić, A.; Hudson, C.C.; Homme, J.L.; Yin, P.; Otterness, D.M.; Karnitz, L.M.; Abraham, R.T. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 2000, 60, 3504–3513. [Google Scholar] [PubMed]
- Lin, J.; Fang, Y.; Zhang, M.; Wang, X.; Li, L.; He, M.; Xue, A.; Zhu, K.; Shen, Y.; Li, B. Phosphorylation of PRAS40 contributes to the activation of the PI3K/AKT/mTOR signaling pathway and the inhibition of autophagy following status epilepticus in rats. Exp. Ther. Med. 2020, 20, 3625–3632. [Google Scholar] [CrossRef]
- Wang, L.; Harris, T.E.; Lawrence, J.C. Regulation of proline-rich Akt Substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated Phosphorylation. J. Biol. Chem. 2008, 283, 15619–15627. [Google Scholar] [CrossRef] [Green Version]
- Lv, D.; Guo, L.; Zhang, T.; Huang, L. PRAS40 signaling in tumor. Oncotarget 2017, 8, 69076–69085. [Google Scholar] [CrossRef] [Green Version]
- Deleyto-Seldas, N.; Efeyan, A. The mTOR–autophagy axis and the control of metabolism. Front. Cell Dev. Biol. 2021, 9, 1519. [Google Scholar] [CrossRef] [PubMed]
- Sharp, Z.D.; Bartke, A. Evidence for Down-Regulation of Phosphoinositide 3-Kinase/Akt/Mammalian Target of Rapamycin (PI3K/Akt/mTOR)-Dependent Translation Regulatory Signaling Pathways in Ames Dwarf Mice. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2005, 60, 293–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Lewis, K.N.; Tulsian, R.; Astafev, A.A.; Buffenstein, R.; Kondratov, R.V. It’s about time; divergent circadian clocks in livers of mice and naked mole-rats. FASEB J. 2021, 35, e21590. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Gladyshev, V.N. Molecular signatures of longevity: Insights from cross-species comparative studies. Semin. Cell Dev. Biol. 2017, 70, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Y.; Simon, M.; Zhao, Y.; Ablaeva, J.; Corson, N.; Choi, Y.; Yamada, K.Y.H.; Schork, N.J.; Hood, W.R.; Hill, G.E.; et al. Comparative transcriptomics reveals circadian and pluripotency networks as two pillars of longevity regulation. Cell Metab. 2022, 34, 836–856.e5. [Google Scholar] [CrossRef]
- Passtoors, W.M.; Beekman, M.; Deelen, J.; van der Breggen, R.; Maier, A.B.; Guigas, B.; Derhovanessian, E.; van Heemst, D.; de Craen, A.J.M.; Gunn, D.A.; et al. Gene expression analysis of mTOR pathway: Association with human longevity. Aging Cell 2013, 12, 24–31. [Google Scholar] [CrossRef] [Green Version]
- Kapahi, P.; Zid, B.M.; Harper, T.; Koslover, D.; Sapin, V.; Benzer, S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004, 14, 885–890. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.-M.; Diaz, V.; Walsh, M.E.; Zhang, Y. Moderate lifelong overexpression of tuberous sclerosis complex 1 (TSC1) improves health and survival in mice. Sci. Rep. 2017, 7, 834. [Google Scholar] [CrossRef] [Green Version]
- Selman, C.; Tullet, J.M.A.; Wieser, D.; Irvine, E.; Lingard, S.J.; Choudhury, A.I.; Claret, M.; Al-Qassab, H.; Carmignac, D.; Ramadani, F.; et al. Ribosomal protein S6 Kinase 1 signaling regulates mammalian life span. Science 2009, 326, 140–144. [Google Scholar] [CrossRef] [Green Version]
- Shima, H. Disruption of the p70s6k/p85s6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998, 17, 6649–6659. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Ou, Y.; Li, Y.; Hu, S.; Shao, L.-W.; Liu, Y. Metformin extends C. elegans lifespan through lysosomal pathway. Elife 2017, 6, e31268. [Google Scholar] [CrossRef]
- Vellai, T.; Takacs-Vellai, K.; Zhang, Y.; Kovacs, A.L.; Orosz, L.; Müller, F. Influence of TOR kinase on lifespan in C. elegans. Nature 2003, 426, 620. [Google Scholar] [CrossRef] [PubMed]
- Lamming, D.W.; Ye, L.; Katajisto, P.; Goncalves, M.D.; Saitoh, M.; Stevens, D.M.; Davis, J.G.; Salmon, A.B.; Richardson, A.; Ahima, R.S.; et al. Rapamycin-Induced Insulin Resistance Is Mediated by mTORC2 Loss and Uncoupled from Longevity. Science 2012, 335, 1638–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.J.; Liu, J.; Chen, E.B.; Wang, J.J.; Cao, L.; Narayan, N.; Fergusson, M.M.; Rovira, I.I.; Allen, M.; Springer, D.A.; et al. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 2013, 4, 913–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeffer, C.A.; Tang, W.; Wong, H.; Santillan, A.; Patterson, R.J.; Martinez, L.A.; Tejada-Simon, M.V.; Paylor, R.; Hamilton, S.L.; Klann, E. Removal of FKBP12 enhances mTOR-Raptor interactions, LTP, memory, and perseverative/repetitive behavior. Neuron 2008, 60, 832–845. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Cisuelo, V.; Gómez, J.; García-Junceda, I.; Naudí, A.; Cabré, R.; Mota-Martorell, N.; López-Torres, M.; González-Sánchez, M.; Pamplona, R.; Barja, G. Rapamycin reverses age-related increases in mitochondrial ROS production at complex I, oxidative stress, accumulation of mtDNA fragments inside nuclear DNA, and lipofuscin level, and increases autophagy, in the liver of middle-aged mice. Exp. Gerontol. 2016, 83, 130–138. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Y.; Liu, Y.; Zheng, P. mTOR Regulation and Therapeutic Rejuvenation of Aging Hematopoietic Stem Cells. Sci. Signal. 2009, 2, ra75. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-N.; Liao, Y.-H.; Tsai, S.-C.; Thompson, L.V. Age-dependent effects of caloric restriction on mTOR and ubiquitin-proteasome pathways in skeletal muscles. Geroscience 2019, 41, 871–880. [Google Scholar] [CrossRef]
- Richardson, N.E.; Konon, E.N.; Schuster, H.S.; Mitchell, A.T.; Boyle, C.; Rodgers, A.C.; Finke, M.; Haider, L.R.; Yu, D.; Flores, V.; et al. Lifelong restriction of dietary branched-chain amino acids has sex-specific benefits for frailty and life span in mice. Nat. Aging 2021, 1, 73–86. [Google Scholar] [CrossRef]
- Tang, H.; Inoki, K.; Brooks, S.V.; Okazawa, H.; Lee, M.; Wang, J.; Kim, M.; Kennedy, C.L.; Macpherson, P.C.D.; Ji, X.; et al. mTORC1 underlies age-related muscle fiber damage and loss by inducing oxidative stress and catabolism. Aging Cell 2019, 18, e12943. [Google Scholar] [CrossRef] [Green Version]
- Cabré, R.; Naudí, A.; Dominguez-Gonzalez, M.; Ayala, V.; Jové, M.; Mota-Martorell, N.; Piñol-Ripoll, G.; Gil-Villar, M.P.; Rué, M.; Portero-Otín, M.; et al. Sixty years old is the breakpoint of human frontal cortex aging. Free Radic. Biol. Med. 2017, 103, 14–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabré, R.; Naudí, A.; Dominguez-Gonzalez, M.; Jové, M.; Ayala, V.; Mota-Martorell, N.; Pradas, I.; Nogueras, L.; Rué, M.; Portero-Otín, M.; et al. Lipid Profile in Human Frontal Cortex Is Sustained Throughout Healthy Adult Life Span to Decay at Advanced Ages. J. Gerontol.—Ser. A Biol. Sci. Med. Sci. 2018, 73, 703–710. [Google Scholar] [CrossRef]
- Perluigi, M.; Di Domenico, F.; Barone, E.; Butterfield, D.A. mTOR in Alzheimer disease and its earlier stages: Links to oxidative damage in the progression of this dementing disorder. Free Radic. Biol. Med. 2021, 169, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Jové, M.; Mota-Martorell, N.; Torres, P.; Ayala, V.; Portero-Otin, M.; Ferrer, I.; Pamplona, R. The Causal Role of Lipoxidative Damage in Mitochondrial Bioenergetic Dysfunction Linked to Alzheimer’s Disease Pathology. Life 2021, 11, 388. [Google Scholar] [CrossRef]
- Van Skike, C.E.; Hussong, S.A.; Hernandez, S.F.; Banh, A.Q.; DeRosa, N.; Galvan, V. mTOR Attenuation with Rapamycin Reverses Neurovascular Uncoupling and Memory Deficits in Mice Modeling Alzheimer’s Disease. J. Neurosci. 2021, 41, 4305–4320. [Google Scholar] [CrossRef]
- Davies, J.; Zachariades, E.; Rogers-Broadway, K.-R.; Kartiers, E. Elucidating the role of DEPTOR in Alzheimer’s disease. Int. J. Mol. Med. 2014, 34, 1195–1200. [Google Scholar] [CrossRef] [Green Version]
- Ham, D.J.; Börsch, A.; Lin, S.; Thürkauf, M.; Weihrauch, M.; Reinhard, J.R.; Delezie, J.; Battilana, F.; Wang, X.; Kaiser, M.S.; et al. The neuromuscular junction is a focal point of mTORC1 signaling in sarcopenia. Nat. Commun. 2020, 11, 4510. [Google Scholar] [CrossRef]
- Joseph, G.A.; Wang, S.X.; Jacobs, C.E.; Zhou, W.; Kimble, G.C.; Tse, H.W.; Eash, J.K.; Shavlakadze, T.; Glass, D.J. Partial Inhibition of mTORC1 in Aged Rats Counteracts the Decline in Muscle Mass and Reverses Molecular Signaling Associated with Sarcopenia. Mol. Cell. Biol. 2019, 39, e00141-19. [Google Scholar] [CrossRef] [Green Version]
- Romero, D.F.; Buchinsky, F.J.; Rucinski, B.; Cvetkovic, M.; Bryer, H.P.; Liang, X.G.; Ma, Y.F.; Jee, W.S.S.; Epstein, S. Rapamycin: A bone sparing immunosuppressant? J. Bone Miner. Res. 2009, 10, 760–768. [Google Scholar] [CrossRef]
- Bjedov, I.; Toivonen, J.M.; Kerr, F.; Slack, C.; Jacobson, J.; Foley, A.; Partridge, L. Mechanisms of Life Span Extension by Rapamycin in the Fruit Fly Drosophila melanogaster. Cell Metab. 2010, 11, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.-X.; Regan, J.C.; Eßer, J.; Drews, L.F.; Weinseis, T.; Stinn, J.; Hahn, O.; Miller, R.A.; Grönke, S.; Partridge, L. A TORC1-histone axis regulates chromatin organisation and non-canonical induction of autophagy to ameliorate ageing. Elife 2021, 10, e62233. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leontieva, O.V.; Paszkiewicz, G.M.; Blagosklonny, M.V. Mechanistic or mammalian target of rapamycin (mTOR) may determine robustness in young male mice at the cost of accelerated aging. Aging 2012, 4, 899–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moskalev, A.A.; Shaposhnikov, M.V. Pharmacological Inhibition of Phosphoinositide 3 and TOR Kinases Improves Survival of Drosophila melanogaster. Rejuvenation Res. 2010, 13, 246–247. [Google Scholar] [CrossRef]
- Peng, H.-H.; Wu, C.-Y.; Hsiao, Y.-C.; Martel, J.; Ke, P.-Y.; Chiu, C.-Y.; Liau, J.-C.; Chang, I.-T.; Su, Y.-H.; Ko, Y.-F.; et al. Ganoderma lucidum stimulates autophagy-dependent longevity pathways in Caenorhabditis elegans and human cells. Aging 2021, 13, 13474–13495. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.A.; Harrison, D.E.; Astle, C.M.; Baur, J.A.; Boyd, A.R.; de Cabo, R.; Fernandez, E.; Flurkey, K.; Javors, M.A.; Nelson, J.F.; et al. Rapamycin, But Not Resveratrol or Simvastatin, Extends Life Span of Genetically Heterogeneous Mice. J. Gerontol. Ser. A 2011, 66A, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Strong, R.; Miller, R.A.; Bogue, M.; Fernandez, E.; Javors, M.A.; Libert, S.; Marinez, P.A.; Murphy, M.P.; Musi, N.; Nelson, J.F.; et al. Rapamycin-mediated mouse lifespan extension: Late-life dosage regimes with sex-specific effects. Aging Cell 2020, 19. [Google Scholar] [CrossRef]
- Bitto, A.; Ito, T.K.; Pineda, V.V.; LeTexier, N.J.; Huang, H.Z.; Sutlief, E.; Tung, H.; Vizzini, N.; Chen, B.; Smith, K.; et al. Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. Elife 2016, 5, e16351. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [Green Version]
- Nandagopal, N.; Roux, P.P. Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation 2015, 3, e983402. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.S.Y.; Kranzusch, P.J.; Cate, J.H.D. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 2015, 522, 111–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anisimov, V.N.; Zabezhinski, M.A.; Popovich, I.G.; Piskunova, T.S.; Semenchenko, A.V.; Tyndyk, M.L.; Yurova, M.N.; Rosenfeld, S.V.; Blagosklonny, M. V Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle 2011, 10, 4230–4236. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M. Rapamycin and Ageing: When, for How Long, and How Much? J. Genet. Genom. 2014, 41, 459–463. [Google Scholar] [CrossRef] [Green Version]
- Mukhopadhyay, S.; Frias, M.A.; Chatterjee, A.; Yellen, P.; Foster, D.A. The Enigma of Rapamycin Dosage. Mol. Cancer Ther. 2016, 15, 347–353. [Google Scholar] [CrossRef] [Green Version]
- Sanz, A.; Caro, P.; Barja, G. Protein Restriction Without Strong Caloric Restriction Decreases Mitochondrial Oxygen Radical Production and Oxidative DNA Damage in Rat Liver. J. Bioenerg. Biomembr. 2004, 36, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.; Caro, P.; Gomez Sanchez, J.; Barja, G. Effect of Lipid Restriction on Mitochondrial Free Radical Production and Oxidative DNA Damage. Ann. N. Y. Acad. Sci. 2006, 1067, 200–209. [Google Scholar] [CrossRef]
- Sanz, A.; Gómez, J.; Caro, P.; Barja, G. Carbohydrate restriction does not change mitochondrial free radical generation and oxidative DNA damage. J. Bioenerg. Biomembr. 2006, 38, 327–333. [Google Scholar] [CrossRef]
- Pamplona, R.; Barja, G. Mitochondrial oxidative stress, aging and caloric restriction: The protein and methionine connection. Biochim. Biophys. Acta—Bioenerg. 2006, 1757, 496–508. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Dong, W.; Wang, R.; Li, Y.; Xu, B.; Zhang, J.; Zhao, Z.; Wang, Y. Effect of caloric restriction on the SIRT1/mTOR signaling pathways in senile mice. Brain Res. Bull. 2015, 116, 67–72. [Google Scholar] [CrossRef]
- Dong, W.; Wang, R.; Ma, L.-N.; Xu, B.-L.; Zhang, J.-S.; Zhao, Z.-W.; Wang, Y.-L.; Zhang, X. Autophagy involving age-related cognitive behavior and hippocampus injury is modulated by different caloric intake in mice. Int. J. Clin. Exp. Med. 2015, 8, 11843–11853. [Google Scholar]
- Dong, W.; Wang, R.; Ma, L.-N.; Xu, B.-L.; Zhang, J.-S.; Zhao, Z.-W.; Wang, Y.-L.; Zhang, X. Influence of age-related learning and memory capacity of mice: Different effects of a high and low caloric diet. Aging Clin. Exp. Res. 2016, 28, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Lees, E.K.; Banks, R.; Cook, C.; Hill, S.; Morrice, N.; Grant, L.; Mody, N.; Delibegovic, M. Direct comparison of methionine restriction with leucine restriction on the metabolic health of C57BL/6J mice. Sci. Rep. 2017, 7, 9977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Zhu, Z.; Thompson, H.J. Dietary Energy Restriction Modulates the Activity of AMP-Activated Protein Kinase, Akt, and Mammalian Target of Rapamycin in Mammary Carcinomas, Mammary Gland, and Liver. Cancer Res. 2008, 68, 5492–5499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harputlugil, E.; Hine, C.; Vargas, D.; Robertson, L.; Manning, B.D.; Mitchell, J.R. The TSC Complex Is Required for the Benefits of Dietary Protein Restriction on Stress Resistance In Vivo. Cell Rep. 2014, 8, 1160–1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.-Y.; Wang, W.-J.; Liu, J.-Q.; Song, Y.-H.; Li, P.; Sun, X.-F.; Cai, G.-Y.; Chen, X.-M. Methionine restriction delays senescence and suppresses the senescence-associated secretory phenotype in the kidney through endogenous hydrogen sulfide. Cell Cycle 2019, 18, 1573–1587. [Google Scholar] [CrossRef]
- Hine, C.; Harputlugil, E.; Zhang, Y.; Ruckenstuhl, C.; Lee, B.C.; Brace, L.; Longchamp, A.; Treviño-Villarreal, J.H.; Mejia, P.; Ozaki, C.K.; et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 2015, 160, 132–144. [Google Scholar] [CrossRef] [Green Version]
- Jakobsdottir, S.; van Nieuwpoort, I.C.; van Bunderen, C.C.; de Ruiter, M.B.; Twisk, J.W.R.; Deijen, J.B.; Veltman, D.J.; Drent, M.L. Acute and short-term effects of caloric restriction on metabolic profile and brain activation in obese, postmenopausal women. Int. J. Obes. 2016, 40, 1671–1678. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, Y.; Sun, J.; Zhang, Y.; Luo, T.; Li, B.; Jiang, Y.; Shi, Y.; Le, G. Dietary methionine restriction ameliorates the impairment of learning and memory function induced by obesity in mice. Food Funct. 2019, 10, 1411–1425. [Google Scholar] [CrossRef]
- Witte, A.V.; Fobker, M.; Gellner, R.; Knecht, S.; Flöel, A. Caloric restriction improves memory in elderly humans. Proc. Natl. Acad. Sci. USA 2009, 106, 1255–1260. [Google Scholar] [CrossRef] [Green Version]
- Lamming, D.W.; Cummings, N.E.; Rastelli, A.L.; Gao, F.; Cava, E.; Bertozzi, B.; Spelta, F.; Pili, R.; Fontana, L. Restriction of dietary protein decreases mTORC1 in tumors and somatic tissues of a tumor-bearing mouse xenograft model. Oncotarget 2015, 6, 31233–31240. [Google Scholar] [CrossRef] [Green Version]
- Stone, K.P.; Ghosh, S.; Kovalik, J.P.; Orgeron, M.; Wanders, D.; Sims, L.C.; Gettys, T.W. The acute transcriptional responses to dietary methionine restriction are triggered by inhibition of ternary complex formation and linked to Erk1/2, mTOR, and ATF4. Sci. Rep. 2021, 11, 3765. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solon-Biet, S.M.; Cogger, V.C.; Pulpitel, T.; Wahl, D.; Clark, X.; Bagley, E.E.; Gregoriou, G.C.; Senior, A.M.; Wang, Q.-P.; Brandon, A.E.; et al. Branched-chain amino acids impact health and lifespan indirectly via amino acid balance and appetite control. Nat. Metab. 2019, 1, 532–545. [Google Scholar] [CrossRef] [PubMed]
- Zhenyukh, O.; Civantos, E.; Ruiz-Ortega, M.; Sánchez, M.S.; Vázquez, C.; Peiró, C.; Egido, J.; Mas, S. High concentration of branched-chain amino acids promotes oxidative stress, inflammation and migration of human peripheral blood mononuclear cells via mTORC1 activation. Free Radic. Biol. Med. 2017, 104, 165–177. [Google Scholar] [CrossRef]
- Xiao, F.; Huang, Z.; Li, H.; Yu, J.; Wang, C.; Chen, S.; Meng, Q.; Cheng, Y.; Gao, X.; Li, J.; et al. Leucine Deprivation Increases Hepatic Insulin Sensitivity via GCN2/mTOR/S6K1 and AMPK Pathways. Diabetes 2011, 60, 746–756. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Liu, Y.; Sun, M. Autophagy and Alzheimer’s Disease. Cell. Mol. Neurobiol. 2017, 37, 377–388. [Google Scholar] [CrossRef]
- Patel, N.V.; Gordon, M.N.; Connor, K.E.; Good, R.A.; Engelman, R.W.; Mason, J.; Morgan, D.G.; Morgan, T.E.; Finch, C.E. Caloric restriction attenuates Aβ-deposition in Alzheimer transgenic models. Neurobiol. Aging 2005, 26, 995–1000. [Google Scholar] [CrossRef]
- Wu, P.; Shen, Q.; Dong, S.; Xu, Z.; Tsien, J.Z.; Hu, Y. Calorie restriction ameliorates neurodegenerative phenotypes in forebrain-specific presenilin-1 and presenilin-2 double knockout mice. Neurobiol. Aging 2008, 29, 1502–1511. [Google Scholar] [CrossRef]
- Schafer, M.J.; Alldred, M.J.; Lee, S.H.; Calhoun, M.E.; Petkova, E.; Mathews, P.M.; Ginsberg, S.D. Reduction of β-amyloid and γ-secretase by calorie restriction in female Tg2576 mice. Neurobiol. Aging 2015, 36, 1293–1302. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ho, L.; Qin, W.; Rocher, A.B.; Seror, I.; Humala, N.; Maniar, K.; Dolios, G.; Wang, R.; Hof, P.R.; et al. Caloric restriction attenuates β-amyloid neuropathology in a mouse model of Alzheimer’s disease. FASEB J. 2005, 19, 1–18. [Google Scholar] [CrossRef]
- Alachkar, A.; Agrawal, S.; Baboldashtian, M.; Nuseir, K.; Salazar, J.; Agrawal, A. L-methionine enhances neuroinflammation and impairs neurogenesis: Implication for Alzheimer’s disease. J. Neuroimmunol. 2022, 366, 577843. [Google Scholar] [CrossRef] [PubMed]
- Kalani, A.; Chaturvedi, P.; Kalani, K.; Kamat, P.; Chaturvedi, P.; Tyagi, N. A high methionine, low folate and vitamin B 6/B 12 containing diet can be associated with memory loss by epigenetic silencing of netrin-1. Neural Regen. Res. 2019, 14, 1247. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Mei, X.; Shen, Z.; Bi, Y.; Yuan, Y.; Guo, Z.; Wang, H.; Zhao, H.; Zhou, Z.; Wang, C.; et al. Netrin-1 Improves Functional Recovery through Autophagy Regulation by Activating the AMPK/mTOR Signaling Pathway in Rats with Spinal Cord Injury. Sci. Rep. 2017, 7, 42288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, H.; Kong, Q.; Zhang, H.; Wang, J.; Luo, T.; Jiang, Y. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ou, Y.; Luo, R.; Wang, J.; Wang, D.; Guan, J.; Li, Y.; Xia, P.; Chen, P.R.; Liu, Y. SAR1B senses leucine levels to regulate mTORC1 signalling. Nature 2021, 596, 281–284. [Google Scholar] [CrossRef]
- Salvadori, G.; Zanardi, F.; Iannelli, F.; Lobefaro, R.; Vernieri, C.; Longo, V.D. Fasting-mimicking diet blocks triple-negative breast cancer and cancer stem cell escape. Cell Metab. 2021, 33, 2247–2259.e6. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Saqcena, M.; Foster, D.A. Synthetic lethality in KRas-driven cancer cells created by glutamine deprivation. Oncoscience 2015, 2, 807–808. [Google Scholar] [CrossRef]
- Shevchenko, I.; Bazhin, A. V Metabolic Checkpoints: Novel Avenues for Immunotherapy of Cancer. Front. Immunol. 2018, 9, 1816. [Google Scholar] [CrossRef] [Green Version]
- Cabral, W.A.; Tavarez, U.L.; Beeram, I.; Yeritsyan, D.; Boku, Y.D.; Eckhaus, M.A.; Nazarian, A.; Erdos, M.R.; Collins, F.S. Genetic reduction of mTOR extends lifespan in a mouse model of Hutchinson-Gilford Progeria syndrome. Aging Cell 2021, 20, e13457. [Google Scholar] [CrossRef]
- Anisimov, V.N.; Zabezhinski, M.A.; Popovich, I.G.; Piskunova, T.S.; Semenchenko, A.V.; Tyndyk, M.L.; Yurova, M.N.; Antoch, M.P.; Blagosklonny, M.V. Rapamycin Extends Maximal Lifespan in Cancer-Prone Mice. Am. J. Pathol. 2010, 176, 2092–2097. [Google Scholar] [CrossRef]
- Kitada, M.; Ogura, Y.; Suzuki, T.; Sen, S.; Lee, S.M.; Kanasaki, K.; Kume, S.; Koya, D. A very-low-protein diet ameliorates advanced diabetic nephropathy through autophagy induction by suppression of the mTORC1 pathway in Wistar fatty rats, an animal model of type 2 diabetes and obesity. Diabetologia 2016, 59, 1307–1317. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J.L.; Bakula, D.; Scheibye-Knudsen, M. Clinical Trials Targeting Aging. Front. Aging 2022, 3, 820215. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.A.; Demetriades, C. The Multifaceted Role of Nutrient Sensing and mTORC1 Signaling in Physiology and Aging. Front. Aging 2021, 2, 38. [Google Scholar] [CrossRef] [PubMed]
- Kraig, E.; Linehan, L.A.; Liang, H.; Romo, T.Q.; Liu, Q.; Wu, Y.; Benavides, A.D.; Curiel, T.J.; Javors, M.A.; Musi, N.; et al. A randomized control trial to establish the feasibility and safety of rapamycin treatment in an older human cohort: Immunological, physical performance, and cognitive effects. Exp. Gerontol. 2018, 105, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Mannick, J.B.; Teo, G.; Bernardo, P.; Quinn, D.; Russell, K.; Klickstein, L.; Marshall, W.; Shergill, S. Targeting the biology of ageing with mTOR inhibitors to improve immune function in older adults: Phase 2b and phase 3 randomised trials. Lancet Heal. Longev. 2021, 2, e250–e262. [Google Scholar] [CrossRef]
- Mannick, J.B.; Morris, M.; Hockey, H.-U.P.; Roma, G.; Beibel, M.; Kulmatycki, K.; Watkins, M.; Shavlakadze, T.; Zhou, W.; Quinn, D.; et al. TORC1 inhibition enhances immune function and reduces infections in the elderly. Sci. Transl. Med. 2018, 10, eaaq1564. [Google Scholar] [CrossRef] [Green Version]
- Perl, A. Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases. Nat. Rev. Rheumatol. 2016, 12, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Perl, A. mTOR activation is a biomarker and a central pathway to autoimmune disorders, cancer, obesity, and aging. Ann. N. Y. Acad. Sci. 2015, 1346, 33–44. [Google Scholar] [CrossRef] [Green Version]
- Lai, Z.-W.; Kelly, R.; Winans, T.; Marchena, I.; Shadakshari, A.; Yu, J.; Dawood, M.; Garcia, R.; Tily, H.; Francis, L.; et al. Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: A single-arm, open-label, phase 1/2 trial. Lancet 2018, 391, 1186–1196. [Google Scholar] [CrossRef]
- Kato, H.; Perl, A. Blockade of Treg Cell Differentiation and Function by the Interleukin-21-Mechanistic Target of Rapamycin Axis Via Suppression of Autophagy in Patients With Systemic Lupus Erythematosus. Arthritis Rheumatol. 2018, 70, 427–438. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.L.; Lawrence, I.; Hoffman, M.; Elgindi, D.; Nadhan, K.; Potnis, M.; Jin, A.; Sershon, C.; Binnebose, R.; Lorenzini, A.; et al. Topical rapamycin reduces markers of senescence and aging in human skin: An exploratory, prospective, randomized trial. Geroscience 2019, 41, 861–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
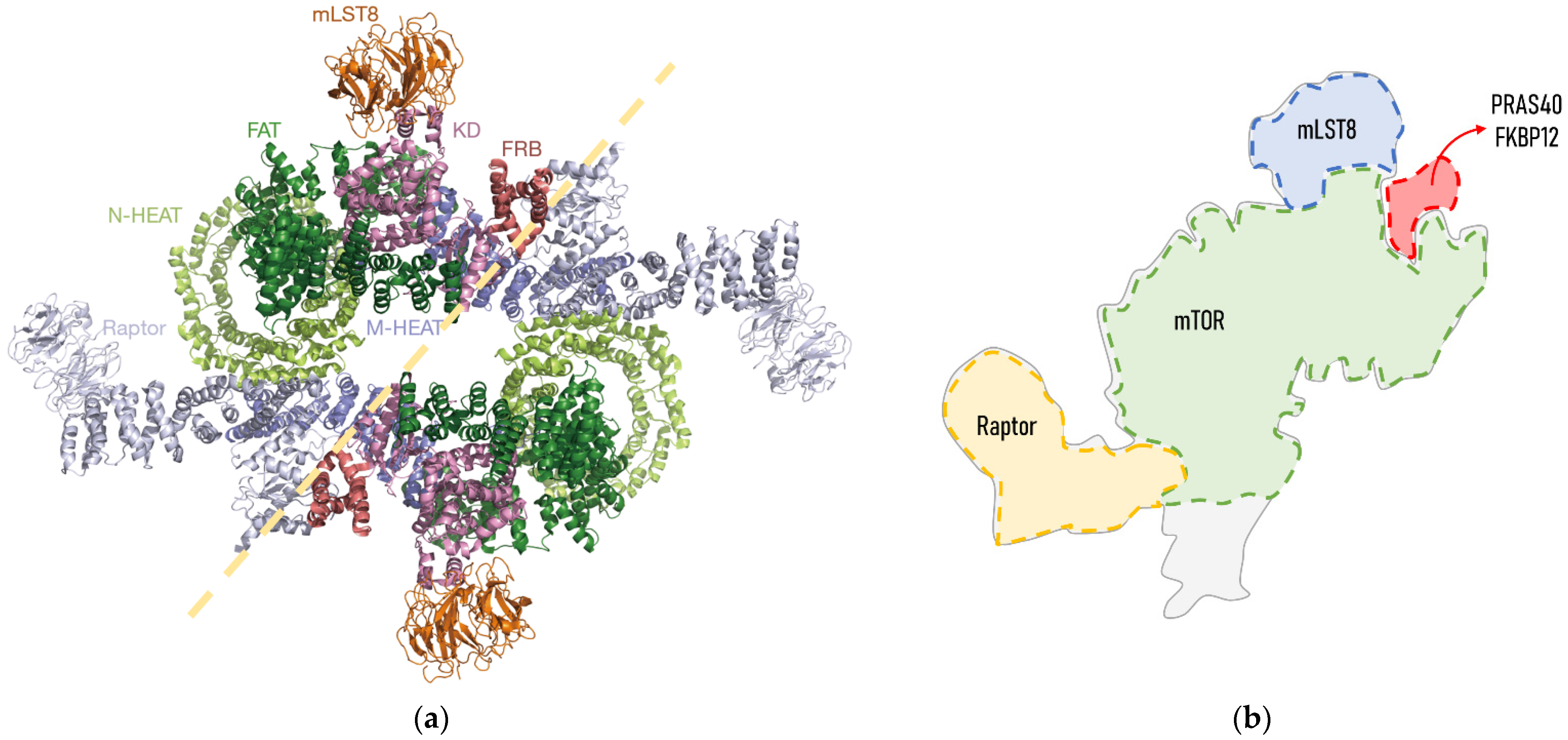
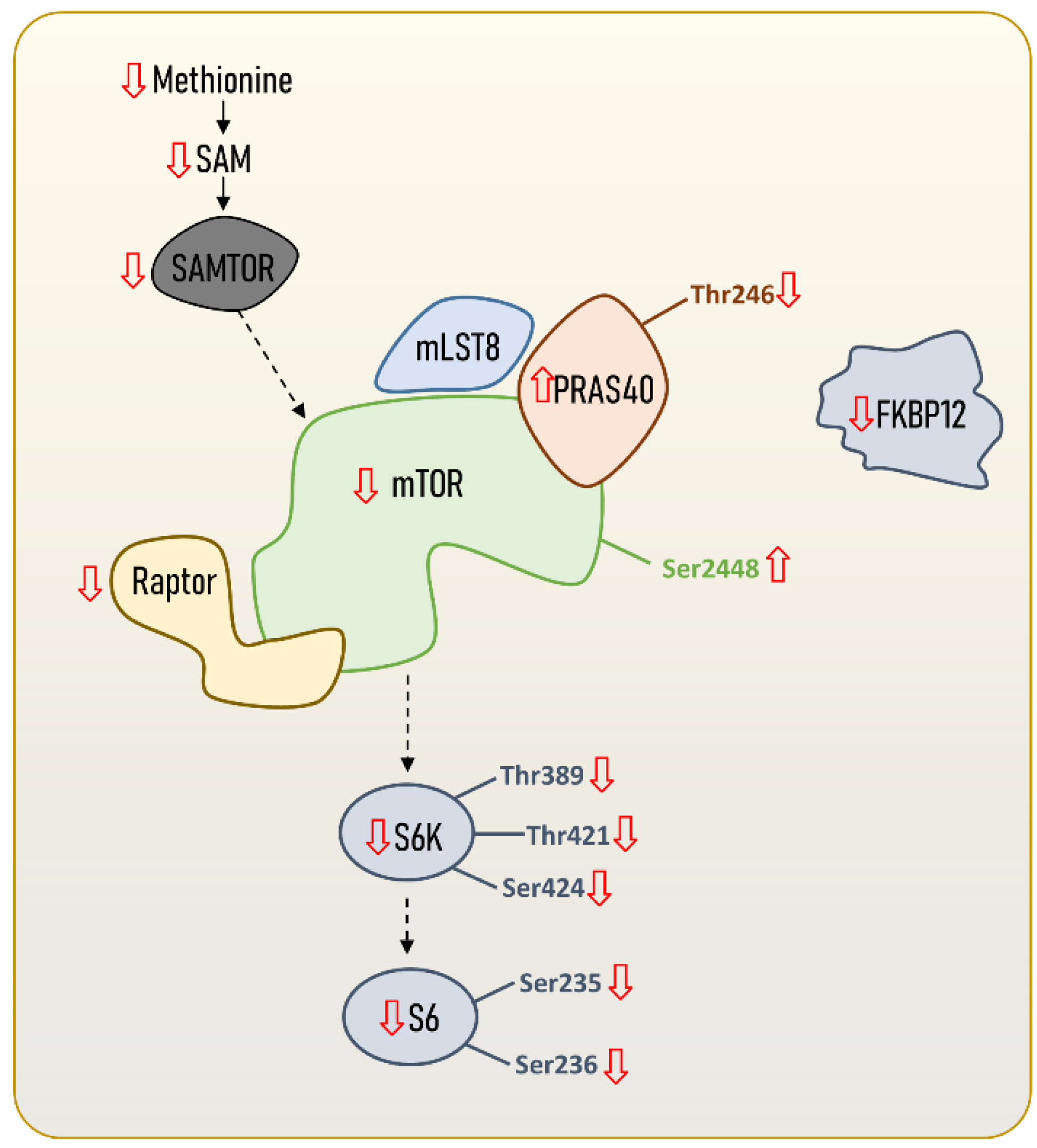
| Species (ML) | Sex | ML Range | mTORC1 | Tissue | Phenotype | Ref. |
|---|---|---|---|---|---|---|
| Ames dwarf mice (5) vs. Wild type mice (3.5) | F | 3.5 to 5 | Reduced # | Liver, skeletal muscle | Longevous | [22] |
| Naked mole rat (37) vs. Wild type mice (3.5) | F | 3.5 to 30 | Reduced # | Liver | Longevous | [23] |
| 8 mammals | M | 3.5 to 46 | Reduced * | Heart | Longevous | [14] |
| 26 mammals | n.d. | 2.1 to 37 | Reduced * | Brain, heart, liver, kidney, lung and limb | Longevous | [25] |
| Human | M/F | 89 to 102 | Reduced * | Whole blood | Longevous | [26] |
| Species | Sex | Genotype | mTORC1 | Tissue | Phenotype | Longevity | Ref. |
|---|---|---|---|---|---|---|---|
| Worm | - | daf-15−/− | Reduced # | Whole | n.d. | +28% | [31] |
| Worm | - | let-363−/− | Reduced * | Whole | n.d. | +250% | [32] |
| Fly | M | dTOR | Reduced # | Whole | n.d. | +22–24% | [27] |
| Fly | M | dS6K | Reduced # | Whole | n.d. | +22–24% | [27] |
| Fly | M | Tsc1/Tsc2 | Reduced # | Whole | n.d. | +12–14% | [27] |
| Mouse | F | hTsc1 | Reduced # | Heart, liver, kidney, skeletal muscle | Improved health | +12.3% | [28] |
| Mouse | F | mtor+/−mlst8+/− | Reduced * | Liver | n.d. | +14.4 | [33] |
| Mouse | F | mTORΔ/Δ | Reduced * | Heart, liver, kidney, brain | Delayed ageing | +19% † | [34] |
| Mouse | M | mTORΔ/Δ | Reduced * | Heart, liver, kidney, brain | Delayed ageing | +22% † | [34] |
| Mouse | F | s6k1−/− | Reduced # | Liver, thymus, fibroblasts | Improved health | +19% | [29,30] |
| Mouse | n.d. | fkbp12−/− | Increased * | Hippocampus | Altered behaviour and neuronal function | n.d. | [35] |
| Species | Sex | mTORC1 | Tissue | Phenotype | Ref. |
|---|---|---|---|---|---|
| Mouse | F | Increased # | Liver | Aged | [39] |
| Mouse | M | Increased # | Liver | Aged | [39] |
| Mouse | M | Increased # | Liver | Aged | [36] |
| Mouse | M | Increased * | Hematopoietic stem cells | Aged | [37] |
| Rat | M | Increased * | Skeletal muscle | Aged | [38] |
| Human | F/M | Increased * | Frontal cortex | Aged | [41] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mota-Martorell, N.; Jové, M.; Pamplona, R. mTOR Complex 1 Content and Regulation Is Adapted to Animal Longevity. Int. J. Mol. Sci. 2022, 23, 8747. https://doi.org/10.3390/ijms23158747
Mota-Martorell N, Jové M, Pamplona R. mTOR Complex 1 Content and Regulation Is Adapted to Animal Longevity. International Journal of Molecular Sciences. 2022; 23(15):8747. https://doi.org/10.3390/ijms23158747
Chicago/Turabian StyleMota-Martorell, Natalia, Mariona Jové, and Reinald Pamplona. 2022. "mTOR Complex 1 Content and Regulation Is Adapted to Animal Longevity" International Journal of Molecular Sciences 23, no. 15: 8747. https://doi.org/10.3390/ijms23158747
APA StyleMota-Martorell, N., Jové, M., & Pamplona, R. (2022). mTOR Complex 1 Content and Regulation Is Adapted to Animal Longevity. International Journal of Molecular Sciences, 23(15), 8747. https://doi.org/10.3390/ijms23158747






