Physical Activity and DNA Methylation in Humans
Abstract
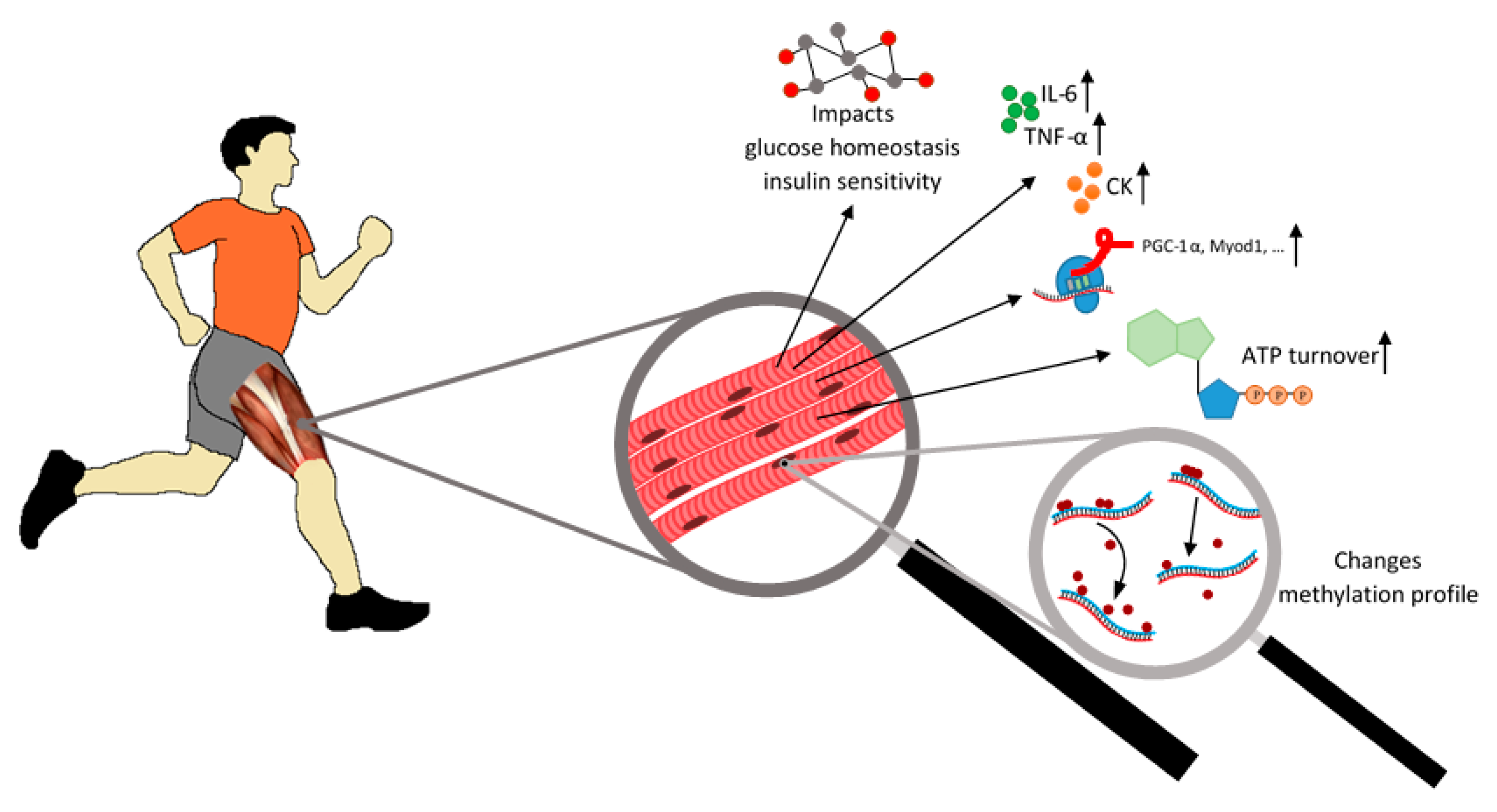
:1. Introduction
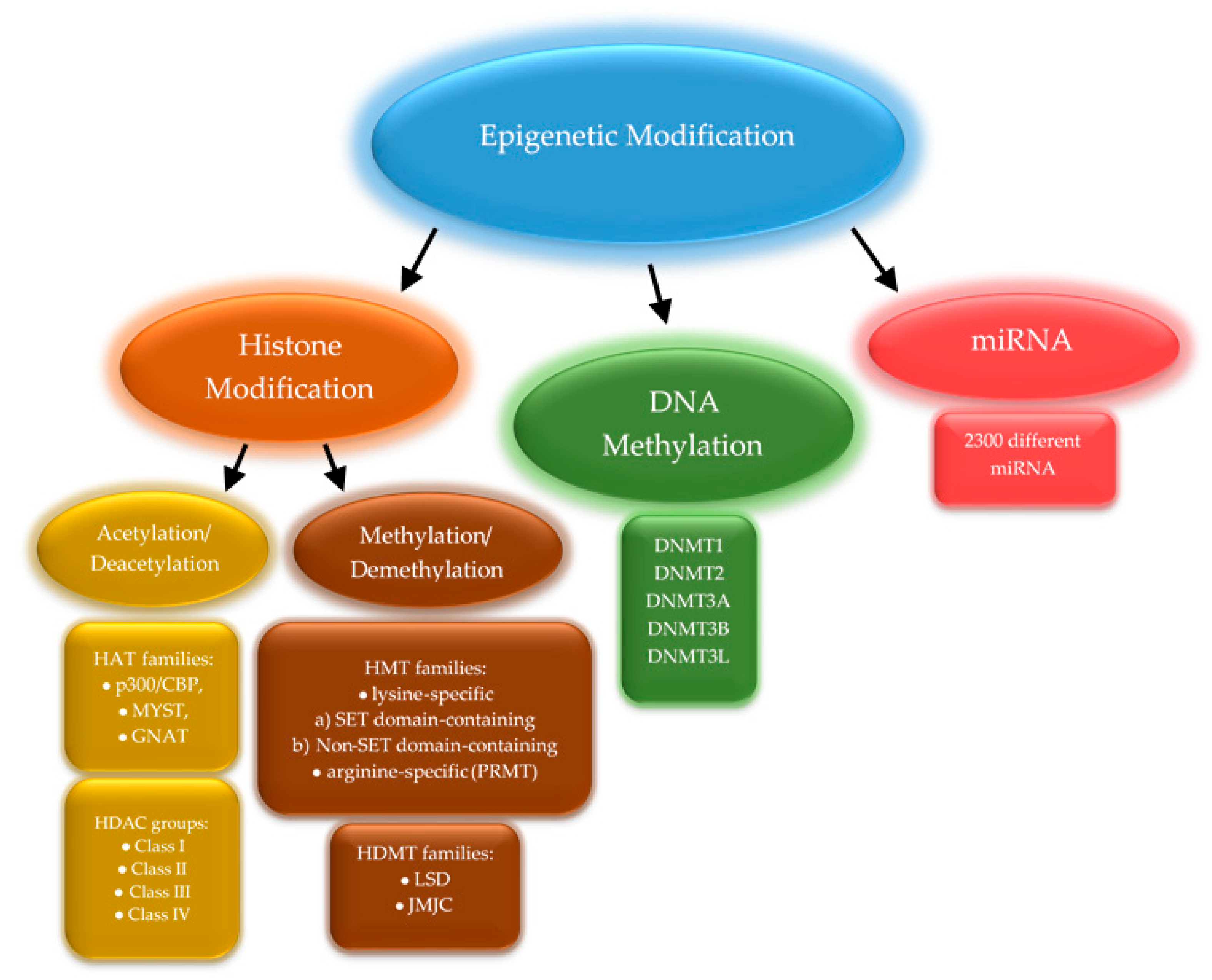
2. Global Methylation
3. Gene Methylation
3.1. In Muscle
3.2. In Fat Tissue
3.3. In Blood Cells
3.4. In Diseased Tissues
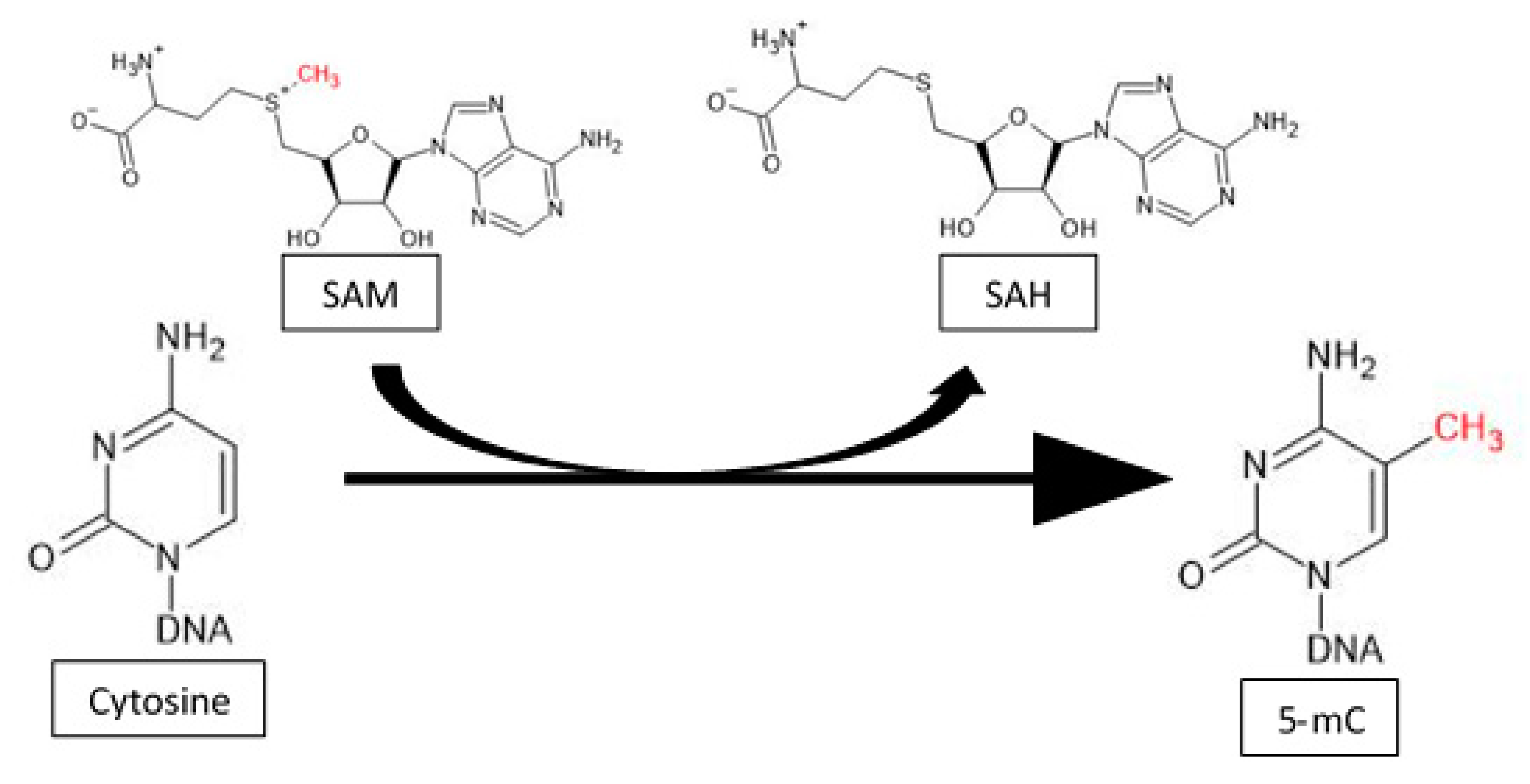
4. Expression of Methyltransferases
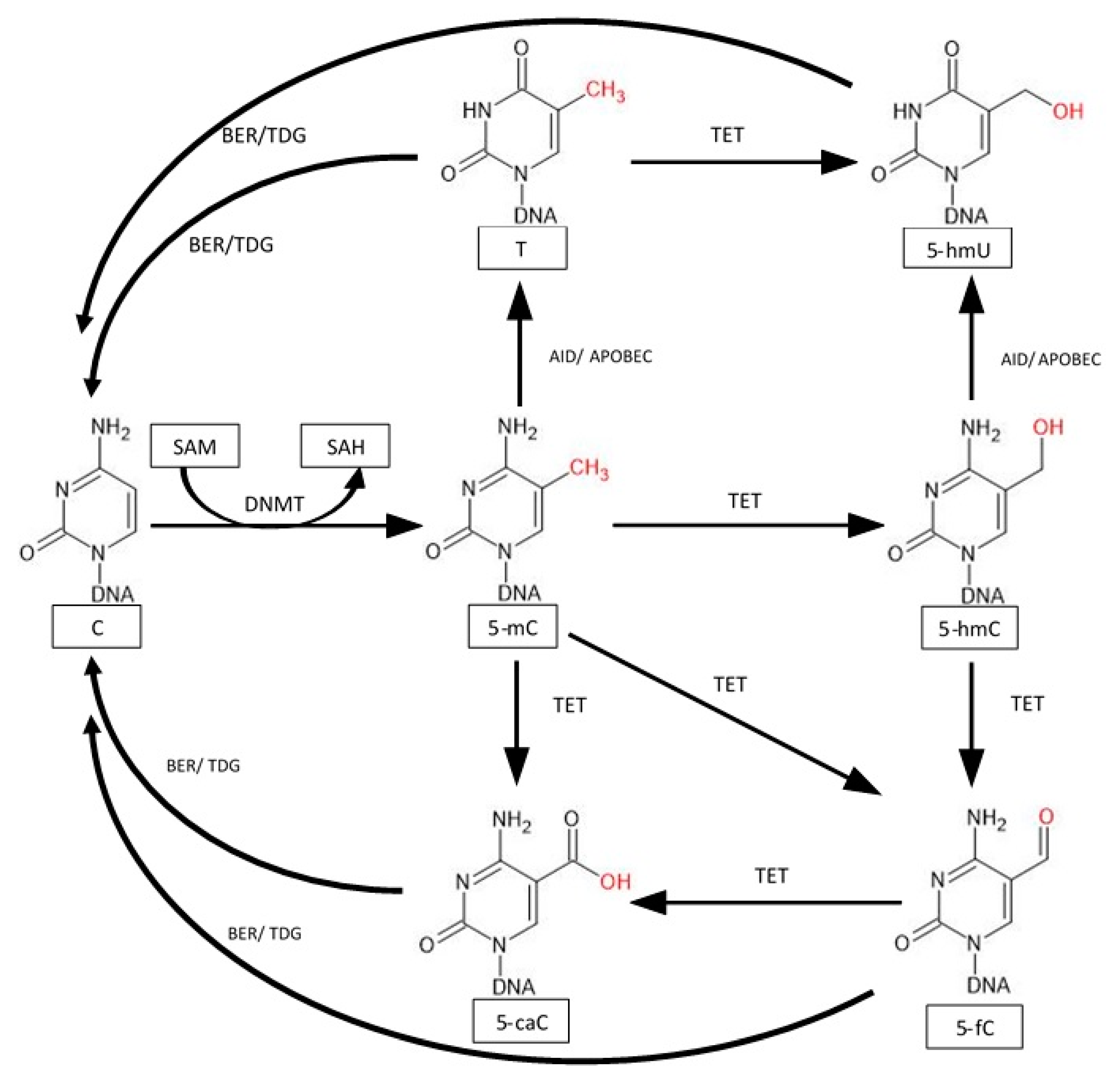
5. Active Demethylation
6. Summary
Funding
Conflicts of Interest
References
- Egan, B.; Zierath, J.R. Exercise Metabolism and the Molecular Regulation of Skeletal Muscle Adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Z. Exercise, PGC-1α and Metabolic Adaptation in Skeletal Muscle. Appl. Physiol. Nutr. Metab. 2009, 34, 424–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mokdad, A.H.; Marks, J.S.; Stroup, D.F.; Gerberding, J.L. Actual Causes of Death in the United States, 2000. J. Am. Med. Assoc. 2004, 291, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, K.; Rohde, T.; Zacho, M.; Asp, S.; Pedersen, B.K. Evidence That Interleukin-6 Is Produced in Human Skeletal Muscle during Prolonged Running. J. Physiol. 1998, 508, 949. [Google Scholar] [CrossRef]
- Coffey, V.G.; Hawley, J.A. The Molecular Bases of Training Adaptation. Sports Med. 2007, 37, 737–763. [Google Scholar] [CrossRef]
- Barrès, R.; Yan, J.; Egan, B.; Treebak, J.T.; Rasmussen, M.; Fritz, T.; Caidahl, K.; Krook, A.; O’Gorman, D.J.; Zierath, J.R. Acute Exercise Remodels Promoter Methylation in Human Skeletal Muscle. Cell Metab. 2012, 15, 405–411. [Google Scholar] [CrossRef] [Green Version]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P.; et al. An Estimate of the Total Number of True Human MiRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [Green Version]
- Lyko F The DNA Methyltransferase Family: A Versatile Toolkit for Epigenetic Regulation. Nat. Rev. Genet. 2018, 19, 81–92. [CrossRef]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and Biological Role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2017, 14, 1108. [Google Scholar] [CrossRef] [Green Version]
- Światowy, W.; Jagodzińśki, P.P. Molecules Derived from TRNA and SnoRNA: Entering the Degradome Pool. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 108, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Flitton, M.; Rielly, N.; Warman, R.; Warden, D.; Smith, A.D.; Macdonald, I.A.; Knight, H.M. Interaction of Nutrition and Genetics via DNMT3L-Mediated DNA Methylation Determines Cognitive Decline. Neurobiol. Aging 2019, 78, 64–73. [Google Scholar] [CrossRef]
- Beckerman, M. Cellular Signaling in Health and Disease; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef] [Green Version]
- Illingworth, R.S.; Bird, A.P. CpG Islands—‘A Rough Guide’. FEBS Lett. 2009, 583, 1713–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gromova, E.S.; Khoroshaev, A.V. Prokaryotic DNA Methyltransferases: The Structure and the Mechanism of Interaction with DNA. Mol. Biol. 2003, 37, 300–314. [Google Scholar] [CrossRef]
- Bird A DNA Methylation Patterns and Epigenetic Memory. Genes Dev. 2002, 16, 6–21. [CrossRef] [Green Version]
- Sharples, A.P.; Seaborne, R.A. Chapter Ten—Exercise and DNA Methylation in Skeletal Muscle. In Sports, Exercise, and Nutritional Genomics; Barh, D., Ahmetov, I.I., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 211–229. ISBN 978-0-12-816193-7. [Google Scholar]
- Hunter, D.J.; James, L.; Hussey, B.; Wadley, A.J.; Lindley, M.R.; Mastana, S.S. Impact of Aerobic Exercise and Fatty Acid Supplementation on Global and Gene-Specific DNA Methylation. Epigenetics 2019, 14, 294. [Google Scholar] [CrossRef] [Green Version]
- Robson-Ansley, P.J.; Saini, A.; Toms, C.; Ansley, L.; Walshe, I.H.; Nimmo, M.A.; Curtin, J.A. Dynamic Changes in Dna Methylation Status in Peripheral Blood Mononuclear Cells Following an Acute Bout of Exercise: Potential Impact of Exercise-Induced Elevations in Interleukin-6 Concentration. J. Biol. Regul. Homeost. Agents 2014, 28, 407–417. [Google Scholar]
- Zhang, F.F.; Cardarelli, R.; Carroll, J.; Zhang, S.; Fulda, K.G.; Gonzalez, K.; Vishwanatha, J.K.; Morabia, A.; Santella, R.M. Physical Activity and Global Genomic DNA Methylation in a Cancer-Free Population. Epigenetics 2011, 6, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bollati, V.; Schwartz, J.; Wright, R.; Litonjua, A.; Tarantini, L.; Suh, H.; Sparrow, D.; Vokonas, P.; Baccarelli, A. Decline in Genomic DNA Methylation through Aging in a Cohort of Elderly Subjects. Mech. Ageing Dev. 2009, 130, 234–239. [Google Scholar] [CrossRef] [Green Version]
- Hughes, L.A.E.; Simons, C.C.J.M.; van den Brandt, P.A.; Goldbohm, R.A.; de Goeij, A.F.; de Bruïne, A.P.; van Engeland, M.; Weijenberg, M.P. Body Size, Physical Activity and Risk of Colorectal Cancer with or without the CpG Island Methylator Phenotype (CIMP). PLoS ONE 2011, 6, e18571. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Zhu, J.; Liu, X.D.; Luo, M.Y.; Xu, N.J. Roles of Physical Exercise in Neurodegeneration: Reversal of Epigenetic Clock. Transl. Neurodegener. 2021, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sailani, M.R.; Halling, J.F.; Møller, H.D.; Lee, H.; Plomgaard, P.; Pilegaard, H.; Snyder, M.P.; Regenberg, B. Lifelong Physical Activity Is Associated with Promoter Hypomethylation of Genes Involved in Metabolism, Myogenesis, Contractile Properties and Oxidative Stress Resistance in Aged Human Skeletal Muscle. Sci. Rep. 2019, 9, 3272. [Google Scholar] [CrossRef] [Green Version]
- Turner, D.C.; Gorski, P.P.; Maasar, M.F.; Seaborne, R.A.; Baumert, P.; Brown, A.D.; Kitchen, M.O.; Erskine, R.M.; Dos-Remedios, I.; Voisin, S.; et al. DNA Methylation across the Genome in Aged Human Skeletal Muscle Tissue and Muscle-Derived Cells: The Role of HOX Genes and Physical Activity. Sci. Rep. 2020, 10, 15360. [Google Scholar] [CrossRef]
- Coyle, Y.M.; Xie, X.J.; Lewis, C.M.; Bu, D.; Milchgrub, S.; Euhus, D.M. Role of Physical Activity in Modulating Breast Cancer Risk as Defined by APC and RASSF1A Promoter Hypermethylation in Nonmalignant Breast Tissue. Cancer Epidemiol. Biomark. Prev. A Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2007, 16, 192–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, M.V.; Toffoli, L.V.; Arruda, D.W.; Soldera, L.M.; Pelosi, G.G.; Neves-Souza, R.D.; Freitas, E.R.; Castro, D.T.; Marquez, A.S. Age-Related Changes in the Global DNA Methylation Profile of Leukocytes Are Linked to Nutrition but Are Not Associated with the MTHFR C677T Genotype or to Functional Capacities. PLoS ONE 2012, 7, e52570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.F.; Cardarelli, R.; Carroll, J.; Fulda, K.G.; Kaur, M.; Gonzalez, K.; Vishwanatha, J.K.; Santella, R.M.; Morabia, A. Significant Differences in Global Genomic DNA Methylation by Gender and Race/Ethnicity in Peripheral Blood. Epigenetics 2011, 6, 623–629. [Google Scholar] [CrossRef] [Green Version]
- Luttropp, K.; Nordfors, L.; Ekström, T.J.; Lind, L. Physical Activity Is Associated with Decreased Global DNA Methylation in Swedish Older Individuals. Scand. J. Clin. Lab. Investig. 2013, 73, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Dimauro, I.; Scalabrin, M.; Fantini, C.; Grazioli, E.; Beltran Valls, M.R.; Mercatelli, N.; Parisi, A.; Sabatini, S.; Di Luigi, L.; Caporossi, D. Resistance Training and Redox Homeostasis: Correlation with Age-Associated Genomic Changes. Redox Biol. 2016, 10, 34–44. [Google Scholar] [CrossRef] [Green Version]
- White, A.J.; Sandler, D.P.; Bolick, S.C.; Xu, Z.; Taylor, J.A.; DeRoo, L.A. Recreational and Household Physical Activity at Different Time Points and DNA Global Methylation. Eur. J. Cancer 2013, 49, 2199–2206. [Google Scholar] [CrossRef] [Green Version]
- Machado, O.A.S.; Diniz, V.L.S.; Passos, M.E.P.; de Oliveira, H.H.; Santos-Oliveira, L.C.; Alecrim, A.L.; Bertola Lobato, T.; Manoel, R.; Correa, I.; Silva, E.B.; et al. Physical Exercise Increases Global and Gene-Specific (Interleukin-17 and Interferon-γ) DNA Methylation in Lymphocytes from Aged Women. Exp. Physiol. 2021, 106, 1878–1885. [Google Scholar] [CrossRef]
- Denham, J.; Marques, F.Z.; Bruns, E.L.; O’Brien, B.J.; Charchar, F.J. Epigenetic Changes in Leukocytes after 8 Weeks of Resistance Exercise Training. Eur. J. Appl. Physiol. 2016, 116, 1245–1253. [Google Scholar] [CrossRef]
- Lindholm, M.E.; Marabita, F.; Gomez-Cabrero, D.; Rundqvist, H.; Ekström, T.J.; Tegnér, J.; Sundberg, C.J. An Integrative Analysis Reveals Coordinated Reprogramming of the Epigenome and the Transcriptome in Human Skeletal Muscle after Training. Epigenetics 2014, 9, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Nitert, M.D.; Dayeh, T.; Volkov, P.; Elgzyri, T.; Hall, E.; Nilsson, E.; Yang, B.T.; Lang, S.; Parikh, H.; Wessman, Y.; et al. Impact of an Exercise Intervention on DNA Methylation in Skeletal Muscle from First-Degree Relatives of Patients with Type 2 Diabetes. Diabetes 2012, 61, 3322–3332. [Google Scholar] [CrossRef] [Green Version]
- Rönn, T.; Volkov, P.; Davegårdh, C.; Dayeh, T.; Hall, E.; Olsson, A.H.; Nilsson, E.; Tornberg, A.; Dekker Nitert, M.; Eriksson, K.-F.; et al. A Six Months Exercise Intervention Influences the Genome-Wide DNA Methylation Pattern in Human Adipose Tissue. PLoS Genet. 2013, 9, e1003572. [Google Scholar] [CrossRef]
- Kashimoto, R.K.; Toffoli, L.V.; Manfredo, M.H.F.; Volpini, V.L.; Martins-Pinge, M.C.; Pelosi, G.G.; Gomes, M.V. Physical Exercise Affects the Epigenetic Programming of Rat Brain and Modulates the Adaptive Response Evoked by Repeated Restraint Stress. Behav. Brain Res. 2016, 296, 286–289. [Google Scholar] [CrossRef]
- Ntanasis-Stathopoulos, I.; Tzanninis, I.-G.; Philippou, A.; Koutsilieris, M. Epigenetic Regulation on Gene Expression Induced by Physical Exercise. J. Musculoskelet. Neuronal. Interact. 2013, 13, 133–146. [Google Scholar]
- Grazioli, E.; Dimauro, I.; Mercatelli, N.; Wang, G.; Pitsiladis, Y.; Luigi, L.D.; Caporossi, D. Physical Activity in the Prevention of Human Diseases: Role of Epigenetic Modifications. BMC Genom. 2017, 18, 802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyne, D.J.; O’Sullivan, D.E.; Olij, B.F.; King, W.D.; Friedenreich, C.M.; Brenner, D.R. Physical Activity, Global DNA Methylation, and Breast Cancer Risk: A Systematic Literature Review and Meta-Analysis. Cancer Epidemiol. Biomark. Prev. 2018, 27, 1320–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajpeyi, S.; Covington, J.D.; Taylor, E.M.; Stewart, L.K.; Galgani, J.E.; Henagan, T.M. Skeletal Muscle PGC1α -1 Nucleosome Position and -260 Nt DNA Methylation Determine Exercise Response and Prevent Ectopic Lipid Accumulation in Men. Endocrinology 2017, 158, 2190–2199. [Google Scholar] [CrossRef]
- Petrie, M.A.; Sharma, A.; Taylor, E.B.; Suneja, M.; Shields, R.K. Impact of Short- and Long-Term Electrically Induced Muscle Exercise on Gene Signaling Pathways, Gene Expression, and PGC1a Methylation in Men with Spinal Cord Injury. Physiol. Genom. 2020, 52, 71–80. [Google Scholar] [CrossRef]
- Lochmann, T.L.; Thomas, R.R.; Bennett, J.P.; Taylor, S.M. Epigenetic Modifications of the PGC-1α Promoter during Exercise Induced Expression in Mice. PLoS ONE 2015, 10, e0129647. [Google Scholar] [CrossRef]
- Seaborne, R.A.; Strauss, J.; Cocks, M.; Shepherd, S.; O’Brien, T.D.; van Someren, K.A.; Bell, P.G.; Murgatroyd, C.; Morton, J.P.; Stewart, C.E.; et al. Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. Sci. Rep. 2018, 8, 1898. [Google Scholar] [CrossRef]
- Alibegovic, A.C.; Sonne, M.P.; Højbjerre, L.; Bork-Jensen, J.; Jacobsen, S.; Nilsson, E.; Faerch, K.; Hiscock, N.; Mortensen, B.; Friedrichsen, M.; et al. Insulin Resistance Induced by Physical Inactivity Is Associated with Multiple Transcriptional Changes in Skeletal Muscle in Young Men. Am. J. Physiology. Endocrinol. Metab. 2010, 299, E752–E763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, M.I. Genomics, Type 2 Diabetes, and Obesity. N. Engl. J. Med. 2010, 363, 2339–2350. [Google Scholar] [CrossRef] [Green Version]
- Kazakova, E.V.; Chen, M.; Jamaspishvili, E.; Lin, Z.; Yu, J.; Sun, L.; Qiao, H. Association between RBMS1 Gene Rs7593730 and BCAR1 Gene Rs7202877 and Type 2 Diabetes Mellitus in the Chinese Han Population. Acta Biochim. Pol. 2018, 65, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.S.; Salehzadeh, F.; Fritz, T.; Zierath, J.R.; Krook, A.; Osler, M.E. Mitochondrial Regulators of Fatty Acid Metabolism Reflect Metabolic Dysfunction in Type 2 Diabetes Mellitus. Metab. Clin. Exp. 2012, 61, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Rowlands, D.S.; Page, R.A.; Sukala, W.R.; Giri, M.; Ghimbovschi, S.D.; Hayat, I.; Cheema, B.S.; Lys, I.; Leikis, M.; Sheard, P.W.; et al. Multi-Omic Integrated Networks Connect DNA Methylation and MiRNA with Skeletal Muscle Plasticity to Chronic Exercise in Type 2 Diabetic Obesity. Physiol. Genom. 2014, 46, 747–765. [Google Scholar] [CrossRef]
- Kanzleiter, T.; Jähnert, M.; Schulze, G.; Selbig, J.; Hallahan, N.; Schwenk, R.W.; Schürmann, A. Exercise Training Alters DNA Methylation Patterns in Genes Related to Muscle Growth and Differentiation in Mice. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E912–E920. [Google Scholar] [CrossRef] [Green Version]
- Blüher, S.; Käpplinger, J.; Herget, S.; Reichardt, S.; Böttcher, Y.; Grimm, A.; Kratzsch, J.; Petroff, D. Cardiometabolic Risk Markers, Adipocyte Fatty Acid Binding Protein (AFABP) and the Impact of High-Intensity Interval Training (HIIT) in Obese Adolescents. Metab. Clin. Exp. 2017, 68, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, S.; Sagara, J. Regulatory Molecules Involved in Inflammasome Formation with Special Reference to a Key Mediator Protein, ASC. Semin. Immunopathol. 2007, 29, 231–238. [Google Scholar] [CrossRef]
- Radom-Aizik, S.; Zaldivar, F., Jr.; Leu, S.-Y.; Adams, G.R.; Oliver, S.; Cooper, D.M. Effects of Exercise on MicroRNA Expression in Young Males Peripheral Blood Mononuclear Cells. Clin. Transl. Sci. 2012, 5, 32. [Google Scholar] [CrossRef] [Green Version]
- Radom-Aizik, S.; Zaldivar, F.; Oliver, S.; Galassetti, P.; Cooper, D.M. Evidence for MicroRNA Involvement in Exercise-Associated Neutrophil Gene Expression Changes. J. Appl. Physiol. 2010, 109, 252–261. [Google Scholar] [CrossRef]
- Nishida, Y.; Hara, M.; Higaki, Y.; Taguchi, N.; Nakamura, K.; Nanri, H.; Horita, M.; Shimanoe, C.; Yasukata, J.; Miyoshi, N.; et al. Habitual Light-Intensity Physical Activity and ASC Methylation in a Middle-Aged Population. Int. J. Sports Med. 2019, 40, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Takeoka, M.; Mori, M.; Hashimoto, S.; Sakurai, A.; Nose, H.; Higuchi, K.; Itano, N.; Shiohara, M.; Oh, T.; et al. Exercise Effects on Methylation of ASC Gene. Int. J. Sports Med. 2010, 31, 671–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke-Harris, R.; Wilkin, T.J.; Hosking, J.; Pinkney, J.; Jeffery, A.N.; Metcalf, B.S.; Godfrey, K.M.; Voss, L.D.; Lillycrop, K.A.; Burdge, G.C. PGC1α Promoter Methylation in Blood at 5ߝ7 Years Predicts Adiposity from 9 to 14 Years (EarlyBird 50). Diabetes 2014, 63, 2528–2537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morabia, A.; Zhang, F.F.; Kappil, M.A.; Flory, J.; Mirer, F.E.; Santella, R.M.; Wolff, M.; Markowitz, S.B. Biologic and Epigenetic Impact of Commuting to Work by Car or Using Public Transportation: A Case-Control Study. Prev. Med. 2012, 54, 229–233. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.F.; Santella, R.M.; Wolff, M.; Kappil, M.A.; Markowitz, S.B.; Morabia, A. White Blood Cell Global Methylation and IL-6 Promoter Methylation in Association with Diet and Lifestyle Risk Factors in a Cancer-Free Population. Epigenetics 2012, 7, 606–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuki, S.; Nishida, K.; Hashimoto, S.; Morikawa, M.; Takasugi, S.; Nagata, M.; Taniguchi, S.; Rokutan, K.; Nose, H. Effects of Milk Product Intake on Thigh Muscle Strength and NFKB Gene Methylation during Home-Based Interval Walking Training in Older Women: A Randomized, Controlled Pilot Study. PLoS ONE 2017, 12, e0176757. [Google Scholar] [CrossRef]
- Streese, L.; Khan, A.W.; Deiseroth, A.; Hussain, S.; Suades, R.; Tiaden, A.; Kyburz, D.; Cosentino, F.; Hanssen, H. High-Intensity Interval Training Modulates Retinal Microvascular Phenotype and DNA Methylation of P66Shc Gene: A Randomized Controlled Trial (EXAMIN AGE). Eur. Heart J. 2020, 41, 1514–1519. [Google Scholar] [CrossRef] [Green Version]
- Gillman, A.S.; Helmuth, T.; Koljack, C.E.; Hutchison, K.E.; Kohrt, W.M.; Bryan, A.D. The Effects of Exercise Duration and Intensity on Breast Cancer-Related DNA Methylation: A Randomized Controlled Trial. Cancers 2021, 13, 4128. [Google Scholar] [CrossRef]
- Zeng, H.; Irwin, M.L.; Lu, L.; Risch, H.; Mayne, S.; Mu, L.; Deng, Q.; Scarampi, L.; Mitidieri, M.; Katsaros, D.; et al. Physical Activity and Breast Cancer Survival: An Epigenetic Link through Reduced Methylation of a Tumor Suppressor Gene L3MBTL1. Breast Cancer Res. Treat. 2012, 133, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, Y.; Nagasaki, H.; Akiyama, Y.; Hashimoto, Y.; Takizawa, T.; Kojima, K.; Kawano, T.; Sugihara, K.; Imai, K.; Nakachi, K. DNA Methylation Status Is Inversely Correlated with Green Tea Intake and Physical Activity in Gastric Cancer Patients. Int. J. Cancer 2009, 124, 2677–2682. [Google Scholar] [CrossRef] [PubMed]
- Pirola, C.J.; Gianotti, T.F.; Burgueño, A.L.; Rey-Funes, M.; Loidl, C.F.; Mallardi, P.; Martino, J.S.; Castaño, G.O.; Sookoian, S. Epigenetic Modification of Liver Mitochondrial DNA Is Associated with Histological Severity of Nonalcoholic Fatty Liver Disease. Gut 2013, 62, 1356–1363. [Google Scholar] [CrossRef]
- Gomez-Pinilla, F.; Zhuang, Y.; Feng, J.; Ying, Z.; Fan, G. Exercise Impacts Brain-Derived Neurotrophic Factor Plasticity by Engaging Mechanisms of Epigenetic Regulation. Eur. J. Neurosci. 2011, 33, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Laye, M.J.; Pedersen, B.K. Acute Exercise and Ca2+ Stimulation Regulate Enzymes Involved in DNA Methylation in Human Skeletal Muscle. Med. Sci. Sports Exerc. 2010, 42, 23. [Google Scholar] [CrossRef]
- Horsburgh, S.; Todryk, S.; Toms, C.; Moran, C.N.; Ansley, L. Exercise-Conditioned Plasma Attenuates Nuclear Concentrations of DNA Methyltransferase 3B in Human Peripheral Blood Mononuclear Cells. Physiol. Rep. 2015, 3, e12621. [Google Scholar] [CrossRef]
- Sølvsten, C.A.E.; de Paoli, F.; Christensen, J.H.; Nielsen, A.L. Voluntary Physical Exercise Induces Expression and Epigenetic Remodeling of VegfA in the Rat Hippocampus. Mol. Neurobiol. 2018, 55, 567–582. [Google Scholar] [CrossRef]
- Damal Villivalam, S.; Ebert, S.M.; Lim, H.W.; Kim, J.; You, D.; Jung, B.C.; Palacios, H.H.; Tcheau, T.; Adams, C.M.; Kang, S. A Necessary Role of DNMT3A in Endurance Exercise by Suppressing ALDH1L1-Mediated Oxidative Stress. EMBO J. 2021, 40, e106491. [Google Scholar] [CrossRef]
- Fuso, A.; Raia, T.; Orticello, M.; Lucarelli, M. The Complex Interplay between DNA Methylation and MiRNAs in Gene Expression Regulation. Biochimie 2020, 173, 12–16. [Google Scholar] [CrossRef]
- Morales, S.; Monzo, M.; Navarro, A. Epigenetic Regulation Mechanisms of MicroRNA Expression. Biomol. Concepts 2017, 8, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Takai, D. Disruption of the Expression and Function of MicroRNAs in Lung Cancer as a Result of Epigenetic Changes. Front. Genet. 2013, 4, 275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, H.; Maruyama, R.; Yamamoto, E.; Kai, M. DNA Methylation and MicroRNA Dysregulation in Cancer. Mol. Oncol. 2012, 6, 567–578. [Google Scholar] [CrossRef] [Green Version]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 Family Reverts Aberrant Methylation in Lung Cancer by Targeting DNA Methyltransferases 3A and 3B. Proc. Nat. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef] [Green Version]
- Glaich, O.; Parikh, S.; Bell, R.E.; Mekahel, K.; Donyo, M.; Leader, Y.; Shayevitch, R.; Sheinboim, D.; Yannai, S.; Hollander, D.; et al. DNA Methylation Directs MicroRNA Biogenesis in Mammalian Cells. Nat. Commun. 2019, 10, 5657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, B.; Lin, M.H.; Chen, C.C.; Peng, K.L.; Wu, M.S.; Tseng, M.C.; Chen, Y.J.; Shen, C.J. DNA Demethylation by DNMT3A and DNMT3B in Vitro and of Methylated Episomal DNA in Transiently Transfected Cells. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 1048–1061. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, X.; Clark, E.; Mulcahey, M.; Huang, S.; Shi, Y.G. TET1 Is a DNA-Binding Protein That Modulates DNA Methylation and Gene Transcription via Hydroxylation of 5-Methylcytosine. Cell Res. 2010, 20, 1390–1393. [Google Scholar] [CrossRef] [Green Version]
- Rawłuszko-Wieczorek, A.A.; Siera, A.; Jagodziński, P.P. TET Proteins in Cancer: Current “State of the Art”. Crit. Rev. Oncol. Hematol. 2015, 96, 425–436. [Google Scholar] [CrossRef]
- Jessop, P.; Toledo-Rodriguez, M. Hippocampal TET1 and TET2 Expression and DNA Hydroxymethylation Are Affected by Physical Exercise in Aged Mice. Front. Cell Dev. Biol. 2018, 6, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chen, Y.; Xu, Z.; Wu, Y.; Zhang, Y.; Shi, L. Chronic Exercise Mediates Epigenetic Suppression of L-Type Ca2+ Channel and BKCa Channel in Mesenteric Arteries of Hypertensive Rats. J. Hypertens. 2020, 38, 1763–1776. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Światowy, W.J.; Drzewiecka, H.; Kliber, M.; Sąsiadek, M.; Karpiński, P.; Pławski, A.; Jagodziński, P.P. Physical Activity and DNA Methylation in Humans. Int. J. Mol. Sci. 2021, 22, 12989. https://doi.org/10.3390/ijms222312989
Światowy WJ, Drzewiecka H, Kliber M, Sąsiadek M, Karpiński P, Pławski A, Jagodziński PP. Physical Activity and DNA Methylation in Humans. International Journal of Molecular Sciences. 2021; 22(23):12989. https://doi.org/10.3390/ijms222312989
Chicago/Turabian StyleŚwiatowy, Witold Józef, Hanna Drzewiecka, Michalina Kliber, Maria Sąsiadek, Paweł Karpiński, Andrzej Pławski, and Paweł Piotr Jagodziński. 2021. "Physical Activity and DNA Methylation in Humans" International Journal of Molecular Sciences 22, no. 23: 12989. https://doi.org/10.3390/ijms222312989
APA StyleŚwiatowy, W. J., Drzewiecka, H., Kliber, M., Sąsiadek, M., Karpiński, P., Pławski, A., & Jagodziński, P. P. (2021). Physical Activity and DNA Methylation in Humans. International Journal of Molecular Sciences, 22(23), 12989. https://doi.org/10.3390/ijms222312989






